Abstract
Bottlenecks in protein expression and secretion often limit the development of industrial processes. By manipulating chaperone and foldase levels, improvements in yeast secretion were found for a number of proteins. Recently, sustained endoplasmic reticulum stress, occurring due to recombinant protein production, was reported to cause oxidative stress in yeast. Saccharomyces cerevisiae cells are able to trigger an adaptive response to oxidative-stress conditions, resulting in the upregulation of both primary and secondary antioxidant defenses. SOD1 encodes for a superoxide dismutase that catalyzes the dismutation of superoxide anions (O2−) into oxygen and hydrogen peroxide. It is a Cu2+/Zn2+ metalloenzyme and represents an important antioxidant defense in nearly all aerobic and aerotolerant organisms. We found that overexpression of the Kluyveromyces lactis SOD1 (KlSOD1) gene was able to increase the production of two different heterologous proteins, human serum albumin (HSA) and glucoamylase from Arxula adeninivorans. In addition, KlSOD1 overexpression led to a significant decrease in the amount of reactive oxygen species (ROS) that originated during protein production. The yield of HSA also increased when K. lactis cells were grown in the presence of the antioxidant agent ascorbic acid and decreased when cells were challenged with menadione, a ROS generator compound. Moreover, we observed that, in high-osmolarity medium, cells overexpressing KlSOD1 showed higher growth rates than control cells. Our results thus further support the notion that the production of some heterologous proteins may be improved by manipulating genes involved in general stress responses.
The development of industrial processes based on the successful secretion of recombinant proteins at high levels may be hampered at a number of different steps, such as folding, glycosylation, intracellular translocation, and release from the cells. Data from literature pointed out that several genes, when overexpressed, stimulate the release of heterologous proteins outside the cells; improvements in yeast secretion were found for a number of proteins by manipulating chaperone and foldase levels. Robinson et al. (27) demonstrated that in Saccharomyces cerevisiae, the overexpression of a single integrated copy of the protein disulfide isomerase gene led to 4- and 10-fold increases in the secretion yields of acid phosphatase and human platelet-derived growth factor B, respectively. In addition, the secretion capability of recombinant yeast strains producing a single-chain antibody fragment increased twofold when the BiP (immunoglobulin heavy-chain binding protein) or protein disulfide isomerase genes were overexpressed separately and eightfold when both chaperones were co-overexpressed (32). Similarly, Harmsen et al. (12) reported a 20-fold increase in the amount of extracellular prochymosin when the BiP gene was overexpressed in S. cerevisiae.
It has been reported that sustained endoplasmic reticulum (ER) stress causes oxidative stress both in yeast and mammalian cells (11, 14). This seems to be a “side effect” of cellular responses against ER stress; as oxidative protein folding is intensified by the loading of resident or heterologous proteins to the ER, this also results in the production of reactive oxygen species (ROS) (14). The heterologous production of cutinase in S. cerevisiae cells, as an example, resulted in protein aggregation in the ER and oxidative stress, as well as the triggering of the unfolded protein response (UPR) and ER-associated degradation pathways (28).
Kluyveromyces lactis was one of the first alternative yeasts for which an efficient transformation system was developed (7). After that, it was proficiently exploited as a host for heterologous protein expression with more than 40 produced proteins of different origins: bacteria, fungi, plants, and mammals (39). The success of K. lactis as an expression system is attributable to several distinctive characteristics. First, it has GRAS (generally regarded as safe) status, which allows food and pharmaceutical applications of this microorganism and of its derivatives. Second, the completely sequenced genome and the availability of both the episomal and integrative expression vectors make the genetic manipulation of this host straightforward. K. lactis can grow on cheap lactose-based media, such as residual whey in dairy industries (21), and does not require expensive explosion-proof plants like those necessary for methylotrophic yeasts. Finally, K. lactis represents a valuable alternative host for heterologous gene expression due to its outstanding secretory capabilities displayed in several studies.
It is well established that the heterologous overexpression of proteins is connected with different stress reactions. A prominent player is the ER where the protein quality control system is often rate limiting. The heterologous expression of secreted proteins can saturate the cell capacity to properly fold protein, triggering the unfolded protein response and resulting in the reduction of protein expression. Moreover, sustained ER stress causes oxidative stress both in yeast and mammalian cells with the production of ROS (14). Some success has been achieved in alleviating such bottlenecks via co-overexpressing ER chaperones; however, additional improvements are still highly demanded (25).
The first line of defense against ROS includes the superoxide dismutase (SOD) enzymes. Cytosolic SODs catalyze the dismutation of superoxide anions (O2·−) into oxygen and hydrogen peroxide. They are copper-zinc metalloenzymes and represent one of the major lines of defense against oxidative stress for nearly all aerobic and aerotolerant organisms, which evolved mechanisms to prevent or repair oxidative damage caused by oxygen exposure (9). In eukaryotes, Cu2+/Zn2+-binding metallochaperones, such as Ccs1, facilitate Cu2+/Zn2+ insertion into target metalloenzymes. Ccs1 activates Sod1 by distributing Cu2+ into the newly synthesized apoprotein and catalyzing the formation of an essential disulfide bond (4, 5). The purpose of this study was to isolate the K. lactis SOD1 (KlSOD1) and K. lactis CCS1 (KlCCS1) genes and to evaluate the effect of their increased dosage on the secretion of heterologous proteins. In S. cerevisiae, SOD1 codes for cytosolic Cu2+/Zn2+ SOD (EC 1.15.1.1), while CCS1 codes for the corresponding copper chaperone. The working hypothesis of this study was that increasing intracellular SOD activity would help the cell to alleviate the stress associated with heterologous protein secretion and would ameliorate the yields of secreted proteins. The effect on the secretion of two different recombinant proteins, glucoamylase (GAA) from the yeast Arxula adeninivorans and human serum albumin (HSA), was investigated. The modulation of some phenotypic behaviors was also assessed: K. lactis cells effectively improved their resistance to oxidative and osmotic stresses as a consequence of the increased cytosolic SOD activity.
MATERIALS AND METHODS
Strains and growth conditions.
The Kluyveromyces lactis strain used in this work was MW278-20C (MATα, ade2, leu2, uraA). Yeast cells were grown in YPD complex medium (10 g liter−1 yeast extract, 10 g liter−1 peptone, 20 g liter−1 glucose) and minimal medium YKK (6.7 g liter−1 yeast nitrogen base without amino acids, 8.5 g liter−1 KH2PO4 and 3.4 g liter−1 K2HPO4, 20 g liter−1 glucose). A total of 0.1 mM CuCl2 and 0.1 mM ZnCl2 were also added to the culture media in order to improve metal cofactor availability, without any effect on strain growth. For the maintenance of recombinant plasmids, supplements of 0.1 or 0.3 g liter−1 antibiotic G418 were added in complex and minimal media, respectively. Cultivation in the absence of oxygen was performed under magnetic stirring conditions in an anaerobic jar in YKK medium at 30°C for 72 h.
Osmotic stress sensitivity was determined by growing cells at 30°C, in a shake flask, in YKK medium supplemented with 1.8 M KCl or 1.8 M sorbitol.
When specified in the text, ascorbic acid was directly added to the growth medium at 30 or 60 μg/ml. Escherichia coli DH5α (ϕ80lacZΔM15, recA1, endA1, gyrA96, thi-1, hsdR17, relA1) was used for general cloning purposes according to standard procedures (30).
PCR amplification.
The entire KlSOD1 and KlCCS1 coding regions were amplified from K. lactis genomic DNA using the primer pairs KlSOD1-for (GCAAGCTTATGGTTAATGCAGTTGCA) and KlSOD1-rev (GCAAGCTTTTAAGCGTTAGAGATACC) and KlCCS1-for (GCAAGCTTATGTCTGGTACAGATGAG) and KlCCS1-rev (GCAAGCTTTCAGAATCTGATGTTGTG), respectively. The fragments encoding the whole KlSod1 and KlCcs1 proteins of K. lactis were amplified by using primers designed on the basis of the nucleotide sequences deposited in EMBL under the accession numbers CAG99284 and CAG98985, respectively. All primers contained the recognition site for the restriction endonuclease HindIII (italicized in the sequences above). DNA amplification was performed using a thermocycler (Robocycler Gradient 96, Bio-Rad, La Jolla, CA) programmed with the following temperature profile: 94°C for 5 min (1 cycle); 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min (30 cycles); and 72°C for 7 min (1 cycle).
Plasmid and DNA manipulation.
The PCR fragments encoding the KlSod1 and KlCcs1 proteins were cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions, giving pCR-KlSOD1 and pCR-KlCCS1, and the gene correctness was confirmed by DNA sequencing (MWG Biotech, Martinsried, Germany). The genes KlSOD1 and KlCCS1 were excised by HindIII digestion from pCR-KlSOD1 and pCR-KlCCS1, respectively, and were ligated into the vector p426SD11 (37), linearized with HindIII. The pGM-GAM and pYG132 plasmids were utilized for the expression of the reporter proteins. The K. lactis-E. coli shuttle vector pGM-GAM contains the GAA gene from the yeast Arxula adeninivorans under the control of the S. cerevisiae GAPDH (glyceraldehyde-3-phosphate dehydrogenase) promoter and of the S. cerevisiae PHO5 terminator (23). The plasmid pYG132 contains an expression cassette for the secretion of the recombinant HSA (rHSA), driven by the native signal sequence, under the control of the ethanol-inducible K. lactis ADH4 promoter and of the K. lactis PGK terminator (29). K. lactis strains were transformed by electroporation with a Bio-Rad gene-pulser apparatus, as described by Wesolowsky-Louvel et al. (40).
SOD activity.
SOD activity was assayed by measuring the enzymatic inhibition of cytochrome c reduction, as described in reference 22. One unit of SOD was defined as the amount of enzyme that inhibited the reduction of cytochrome c by 50% in a coupled system, using xanthine and xanthine oxidase to generate superoxide anion at pH 7.8 and 25°C. To determine the Mn-SOD activity, Cu2+/Zn2+ SOD was inactivated by adding 30 mM KCN to the samples. Cu2+/Zn2+ SOD activity was calculated as the difference between the total and Mn2+-SOD activities.
Western blot analysis of Cu2+/Zn2+ SOD and recombinant HSA.
Proteins from total extracts (20 μg, each lane) or from cell-free culture broths (10 μl of a 1:10 dilution) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using the buffer system of Laemmli (15) and 12% acrylamide gels. After they were electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad), target proteins were detected with specific rabbit polyclonal antibodies: antibodies raised against rat Cu2+/Zn2+ SOD (Stressgene, Victoria, Canada) at a dilution of 1:1,000 for KlSOD1 detection and anti-HSA primary antibodies used at a dilution of 1:10,000 (Sigma) for rHSA identification. The secondary antibodies used for both analyses were monoclonal anti-rabbit immunoglobulin G conjugated with peroxidase (Promega). Immunologically active proteins were visualized with an enhanced chemiluminescence detection system (GE Healthcare) according to the manufacturer's instructions. The densitometric analysis was done with an image analyzer (Phoretix 1D; Non Linear Dynamics Ltd.).
GAA activity.
GAA activity was determined as the starch-hydrolyzing activity of the cell-free culture broths, according to Morlino et al. (23), with minor modifications: GAA activity was estimated at 37°C, and samples were not cooled on ice. One unit of GAA activity was defined as the quantity of enzyme needed to decrease the absorbance at 580 nm by one absorbance unit per minute.
Northern blot analysis.
Total RNA was prepared by extraction with hot acidic phenol (1). Northern blot analysis was performed as described previously (30). The A. adeninivorans GAA probe corresponded to the 2.1-kb HindIII region derived from the pGM-GAM plasmid. The KlKAR2 and KlHSP60 probes were PCR amplified from the K. lactis DNA genome. To amplify the 1,000-bp fragment of KlKAR2 and the 1,700-bp PCR product of KlHSP60, the following primer pairs were used: 5′-AAAAAGTTCAGTGGGATGGC-3′ and 5′-CATGGCTTTGTCATTCTTGG-3′ and 5′-GTACCGTAAAGCCAGGCAAT-3′ and 5′-GCATACCTGGCATACCACCTG-3′, respectively.
All probes were labeled with [α-32P]dATP by using the Ready Prime DNA labeling system (GE Healthcare) according to the manufacturer's instructions.
Oxidative and heat-shock stress treatments.
Yeast cells were grown for 48 h on YKK supplemented with 0.1 mM Cu2+ and 0.1 mM Zn2+, harvested from cultures, and washed twice with bidistilled water. The cells were then resuspended in 100 mM potassium phosphate buffer (pH 7.4) and treated or not with a cytotoxic level of H2O2 (50 mM) or high temperature (48°C). Cell survival was monitored by taking samples 15 min after the treatment; the samples were diluted in 100 mM phosphate buffer (pH 7.4) and then plated onto YPD agar to obtain viable counts. The experiment was repeated three times, and plate counts were made in triplicate.
ROS production.
Flow cytometric analysis was used to assess the production of free intracellular radicals. K. lactis cells, treated or not for 1 h either with 50 mM H2O2 or 5 mM menadione, were incubated with dihydrorhodamine 123 (DHR) for 2 h and analyzed by using a FACSCalibur system (BD Biosciences, San Jose, CA) at a low flow rate with excitation and emission settings at 488 and 525 to 550 nm (filter FL1), respectively.
rHSA production in the presence of oxidative stress.
K. lactis cells harboring pYG132 for recombinant HSA production, grown for 48 h in YKK medium, were treated or not with 5 mM menadione for 1 h, washed twice with bidistilled water, and then resuspended in YKK medium supplemented with 2% ethanol to induce rHSA expression. Samples were taken at 3 and 6 h after the treatment, and proteins from 10 μl of cell-free medium were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis in order to detect HSA secretion levels by Western blot analysis.
RESULTS
Isolation, sequence analysis, and overexpression of the K. lactis SOD1 gene.
We cloned the KlSOD1 gene and the deduced amino acid sequence was compared with the Swiss-Prot protein database. The analysis showed strong sequence similarity with Sod1p from other yeasts; among them, Sod1p from S. cerevisiae resulted in 75% identity and 81% similarity. The closeness with human Sod1p was also fairly high (57% identity and 68% similarity). The KlSod1p contained the highly conserved histidine residues (H-47, H-49, H-64, H-72, H-81, and H-121) involved in the interaction with the metallic cofactors, which are essential for activity and folding in all of the Sod1 enzymes (13). In addition, the residues involved in critical interactions were present in the invariant positions: these include the aspartic residue D-84 involved in Cu2+ binding, the arginine residue R-144 that participated in leading the substrate to the active site, and the two cysteine residues C-58 and C-147 involved in the formation of a disulfide bond. In the C-terminal region, relevant for the Sod1 activation mechanism (5), KlSod1p presented an alanine in position 143 and a histidine in position 145, instead of the conserved residues of serine S-143 and leucine L-145 of mammalian Sod1.
The K. lactis strain overexpressing KlSOD1 was obtained by transforming the MW278-20C strain with the vector pKlSOD1. The amounts of KlSod1p produced by the wild-type and recombinant strains were determined by Western blot analysis. After the optimization of culture parameters (data not shown), the strains were grown at 30°C for 48 h in YKK medium supplemented with 0.1 mM Cu2+ and 0.1 mM Zn2+, the conditions that better maximized the SOD specific activity of the cells without affecting the growth performances. The presence in K. lactis of the expression vector pKlSOD1 led to an increased production of immunologically active Sod1p compared with the wild-type strain (Fig. 1A) and also resulted in an effective enhancement of intracellular SOD activity (Fig. 1B).
FIG. 1.
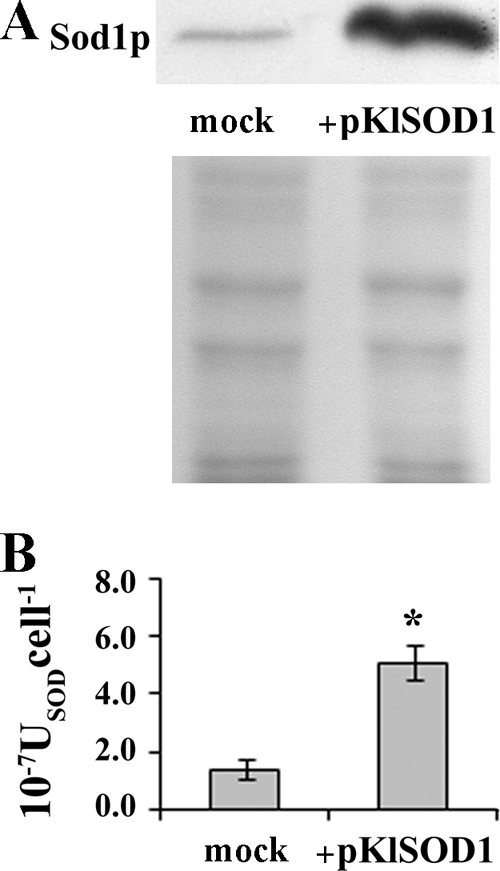
(A) Western blot analysis of KlSod1p content in K. lactis MW278-20C (mock) and recombinant strain transformed with the pKlSOD1 plasmid (+pKlSOD1). Each well was loaded with 20 μg of total extract proteins; a Coomassie blue-stained electrophoresis gel of the same samples is shown at the bottom of the panel. (B) Comparison of cytoplasmic SOD activities (USODcell−1) from the above-mentioned strains, measured by spectrophotometric assay. *, sample is significantly different (P ≤ 0.01) compared to the mock.
Effect of the increased cytoplasmic SOD activity on the secretion of rHSA.
K. lactis MW278-20C cells carrying either the empty or the pKlSOD1 plasmid were transformed with the vector pYG132 containing the cDNA coding for the rHSA. A Western blot analysis with anti-HSA antibody of cell-free cultures was performed after 48 h of growth in YKK supplemented with 0.1 mM Cu2+ and 0.1 mM Zn2+ (Fig. 2A). Densitometric analysis of the Western blot films revealed a fourfold increase in the production of HSA from cells overexpressing KlSod1p, in comparison with the cells carrying the empty vector and grown under the same conditions. In the same strains, cytoplasmic SOD activities were compared, confirming that the cells carrying the plasmid pKlSOD1 contained an increased level of SOD activity (Fig. 2B). The specific growth rate was not affected by the presence of the recombinant vectors (mock, 0.30 h−1; strains with pGM-GAM, 0.26 h−1; strains with pGM-GAM and pKlSOD1, 0.28 h−1).
FIG. 2.
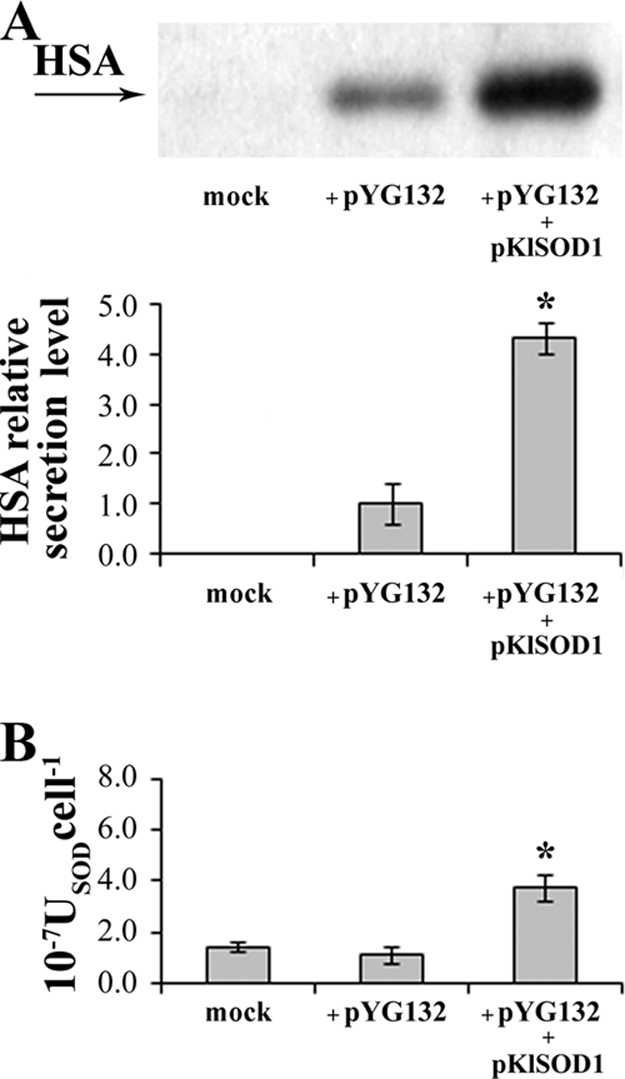
HSA production in the control strain K. lactis MW278-20C carrying the empty vector p426SD11 (mock) and in the strains harboring pYG132 transformed with either p426SD11 (+pYG132) or pKlSOD1 (+pYG132 +pKlSOD1). (A) Western blot analysis of HSA secretion level in cell-free supernatants from 48-h cultures grown on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2. Quantification of the chemiluminescent signal on the blot is shown at the bottom of the panel. The signal from the control strain was set at 1. (B) SOD activities (USODcell−1) of total cell extracts from the same cultures. *, sample is significantly different (P ≤ 0.01) compared to the strain harboring pYG132.
Effect of the increased cytoplasmic SOD activity on the secretion of GAA.
In order to determine the effect of KlSOD1 overexpression on heterologous protein secretion, A. adeninivorans GAA was also used as a reporter protein. K. lactis MW278-20C carrying p426SD11 or pKlSOD1 was transformed with the plasmid pGM-GAM. The GAA activity in the cell-free supernatants of batch cultures was analyzed (Fig. 3A). SOD overexpression doubled GAA activity and led to a higher level of cytoplasmic SOD activity (Fig. 3C). The biomass yields of the strains, expressed as grams of cell dry weight per gram of substrate, were comparable (0.19 and 0.20, respectively). A northern analysis was also performed to evaluate the amounts of GAA mRNA transcripted in the strains carrying the plasmid pGM-GAM together with either p426SD11 or pKlSOD1 (Fig. 3B). This experiment indicates that the secretion improvement of GAA in the KlSOD1-overexpressing strain was not associated with an enhanced transcription of the GAA gene, which was instead slightly reduced compared to the control (Fig. 3B; compare lanes 3 and 2).
FIG. 3.
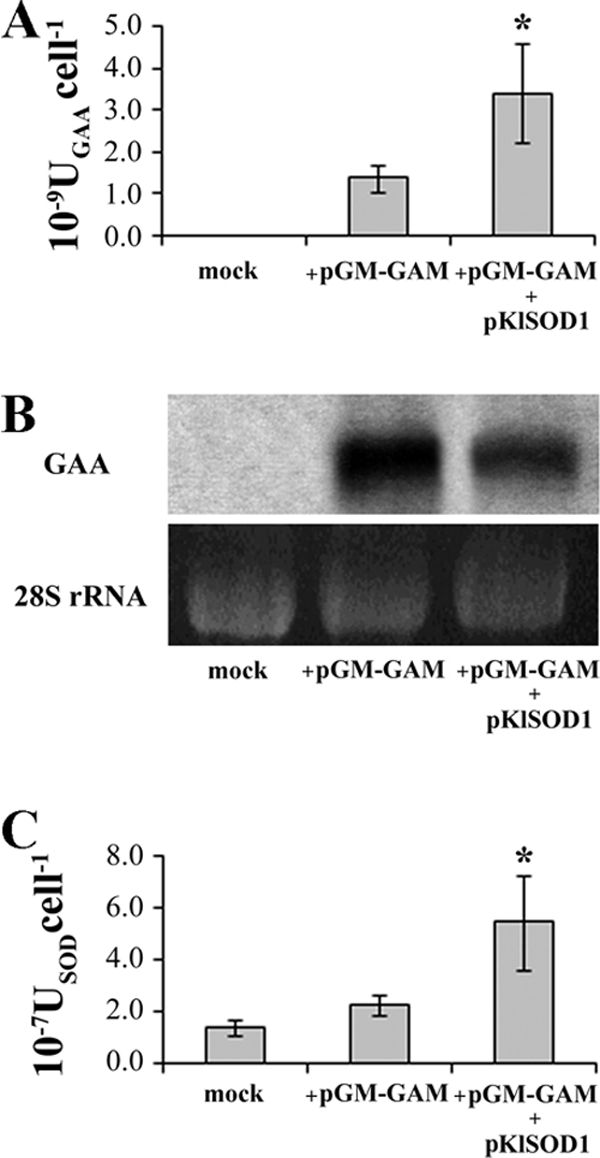
GAA production in the control strain K. lactis MW278-20C carrying the empty vector p426SD11 (mock) and in the strains harboring pGM-GAM transformed with either p426SD11 (+pGM-GAM) or pKlSOD1 (+pGM-GAM +pKlSOD1). (A) Extracellular GAA activities detected in the cell-free supernatant from 48-h cultures grown on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2. (B) Northern blot analysis of GAA mRNA from the total RNA extract from the biomass of the above-mentioned cultures. (C) SOD activities (USODcell−1) of total cell extracts from the same cultures. *, sample is significantly different (P ≤ 0.05) compared to the strain harboring pGM-GAM.
To further support the notion that the heterologous protein production in K. lactis can be ameliorated by overexpressing KlSOD1, we tried to improve the secretory capabilities of the vga3 K. lactis strain, altered in the outer-chain extension of N-glycoproteins and already reported to be a hypersecretory mutant (36). The vga3 cells carrying the pGM-GAA plasmid were transformed with either p426SD11 or pKlSOD1 and grown for 48 h in YKK medium supplemented with 0.1 mM Cu2+ and 0.1 mM Zn2+. The resulting GAA activity detected in the culture medium of cells carrying the pKlSOD1 plasmid was threefold higher than that of the cells transformed with the empty vector and sevenfold higher with respect to wild-type cells expressing GAA (data not shown).
Effects of the increased dosage of KlSOD1 in K. lactis.
To evaluate whether the overexpression of KlSOD1 affected resistance to hyperosmosis, batch cultures containing high concentrations of organic or ionic osmolytes were carried out. In the presence of 1.8 M sorbitol or 1.8 M potassium chloride, K. lactis overexpressing KlSOD1 and GAA grew better than the cells overexpressing only GAA (Fig. 4A).
FIG. 4.
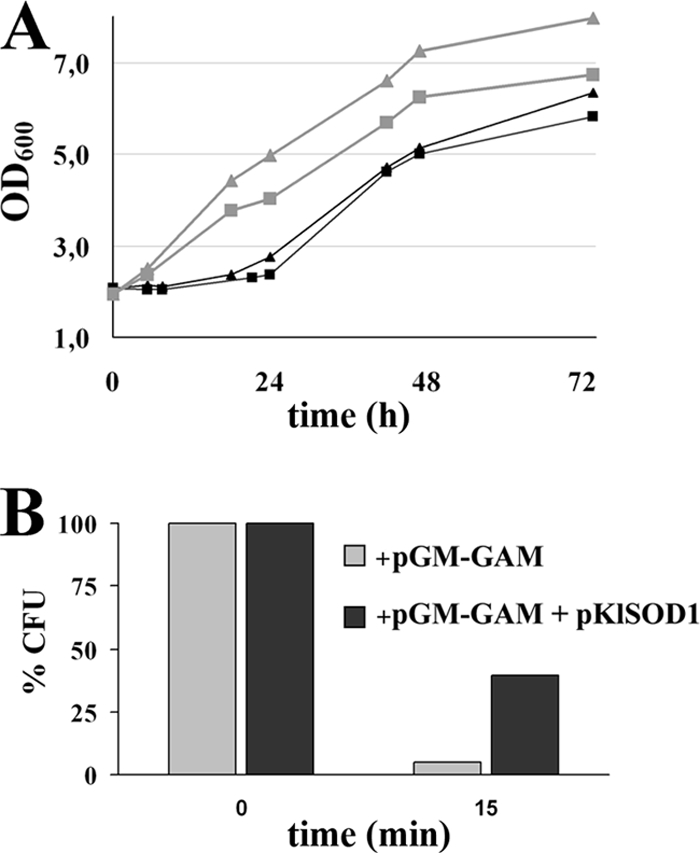
(A) Growth curves under hyperosmotic stress conditions (black symbols, 1.8 M sorbitol; gray symbols, 1.8 M KCl) of the K. lactis MW278-20C strains harboring pGM-GAM and either the empty vector p426SD11 (squares) or the pKlSOD1 plasmid (triangles) on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2. (B) Cell viability after H2O2 exposure: the above-mentioned strains grown on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2 for 48 h were challenged with 50 mM H2O2 for 15 min. The viability was evaluated by plating the samples on YPD medium and was expressed as the CFU percentage of the corresponding untreated cultures. The values are the means of three independent experiments and show an SD of <10%.
Phenotypes related to oxidative stress also occur when cells are driven to express and secrete high levels of heterologous proteins. The resistance to hydrogen peroxide of the GAA-overexpressing cells was analyzed comparing the behaviors of strains carrying either p426SD11 or pKlSOD1. The survival of the cells after a challenge with 50 mM hydrogen peroxide for 15 or 30 min was determined as a percentage of the CFU (Fig. 4B). An increase in survival rate was observed in cells expressing recombinant GAA and carrying the plasmid pKlSOD1 (40%) with respect to the control (5%).
Oxidative stress in K. lactis cells originated by heterologous protein as a limiting step.
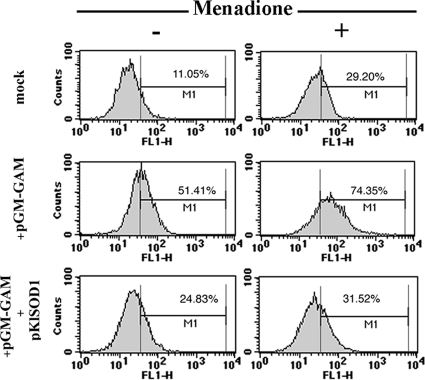
The accumulation of ROS in the wild-type strain and in the cells expressing GAA was analyzed by flow cytometry after incubation of the cells with the fluorescent dye DHR. This compound accumulates inside the cells, and it is oxidized to the corresponding fluorescent chromophore by ROS. Cells expressing GAA showed an increased fluorescence of the population compared with the marginal fluorescence presented by the wild-type cells (Fig. 5). The DHR measurement was reduced by increasing the SOD activity in the recombinant strain, in agreement with the enhanced resistance to the H2O2 challenge observed in these cells (Fig. 4B). A suppression of ROS accumulation by KlSOD1 was also observed when cells were challenged with menadione, an intracellular superoxide-generator compound. Similar results were obtained when the cells were treated with H2O2 (data not shown).
FIG. 5.
Indicated strains grown on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2 for 48 h were challenged with 5 mM menadione for 1 h; ROS were analyzed by DHR staining with flow cytometry. M1, marker; FL1-H, fluorescence intensity.
To determine whether ROS play a role in the limitation of heterologous protein production, we analyzed the level of HSA secreted by cells grown in the presence or absence of the antioxidant ascorbic acid (Fig. 6A). K. lactis cells expressing HSA produced up to 3.5-fold more heterologous protein than the control cells. However, the effects of the external antioxidant agent and the increase in the internal defense mechanism relying on Sod1p were not additive. In fact, the yield of the recombinant protein was not further increased when the strain producing the HSA and overexpressing KlSOD1 was grown in the presence of ascorbic acid (not shown).
FIG. 6.
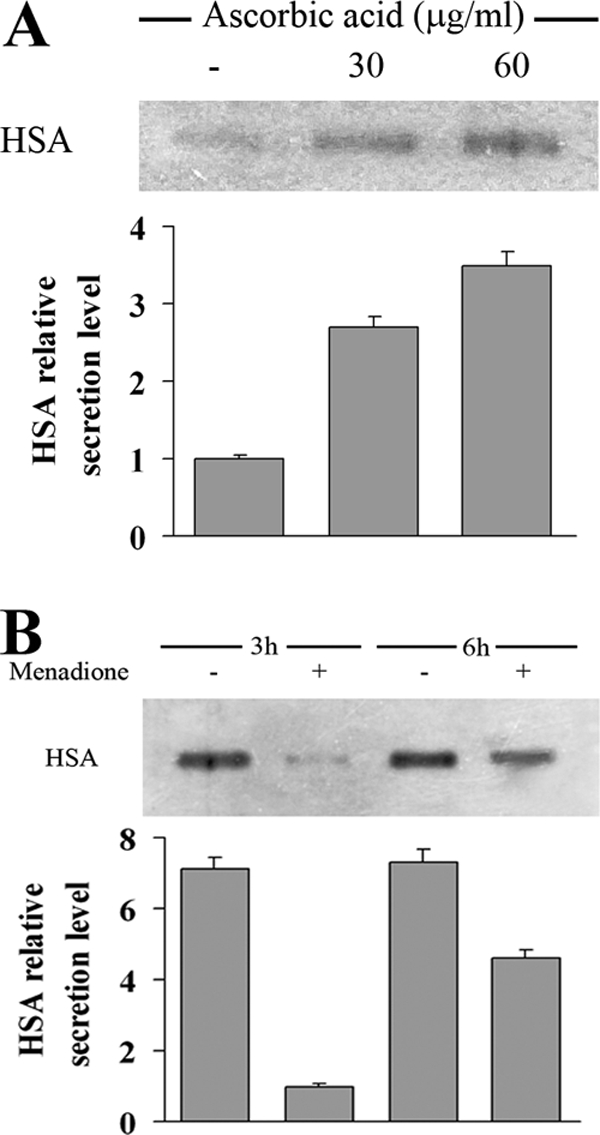
(A) HSA production of K. lactis harboring pYG132 vector treated or not (−) for 48 h with 30 and 60 μg/ml of ascorbic acid. Gel lanes were loaded with 5 μl of a 1:10 dilution of cell-free medium. Quantification of the chemiluminescent signal on the blot is shown at the bottom of the panel. The signal from the untreated cells was set at 1. (B) HSA secretion level of the same strain was monitored 3 and 6 h after the challenge with 5 mM menadione for 1 h. A total of 15 μl of cell-free medium was loaded in the gel. Quantification of the chemiluminescent signal on the blot is shown at the bottom of the panel. The signal from the challenged cells 3 h after treatment was set at 1.
To support the notion that ROS production is a limiting factor in the yield of recombinant proteins, K. lactis cells expressing HSA were challenged with menadione: a reduction of sevenfold in the secretion of HSA was found in the cell-free supernatant of the treated culture (Fig. 6B). A partial recovery of the HSA secretion level was observed 6 h after the treatment, possibly indicating the complete activation of antioxidant cell defenses.
We propose that the enhancement of recombinant protein production by the increased dosage of KlSOD1 was based on the detoxification of ROS. GAA production was measured in the culture supernatants of K. lactis carrying pGM-GAA or both pGM-GAA and pKlSOD1 by growing the cells under anaerobic conditions. The results, normalized by cell number, revealed that cells overexpressing KlSOD1 produced the same GAA activity (P = 0.08) as the control cells (2.34 × 10−9 and 2.08 × 10−9 UGAA cell−1) when the growth conditions were not favorable to ROS generation.
KlCCS1 overexpression in K. lactis cells.
Sod1 relies on a copper chaperone (CCS) to acquire its essential copper cofactor, and in S. cerevisiae cells, the increased dosage of CCS1 alone is able to enhance SOD activity (30). CCS1-related sequences were then searched in the K. lactis genome database to investigate if the overexpression of this chaperone could enhance heterologous protein production by itself.
The KlCCS1 gene was cloned and the deduced amino acid sequence showed a relevant closeness with Ccs1p from other yeasts, such as that from S. cerevisiae (55% identity and 75% similarity) and from Candida glabrata. The KlCcs1p presented, as did the homologous ScCcs1p, three domains: domain I, the Atx1p-like N-terminal region with the Met-X-Cys-X2-Cys motif essential for the in vivo activation of apo-Sod1p under strict copper limitation conditions (30); domain II, the central SOD1-like region that it is known to participate in Ccs1-Sod1 protein-protein interaction with the larger contacts area (16); and domain III, the most highly conserved region among CCS molecules that presents the Cys-X-Cys motif critical for copper ion binding and its direct transfer to Sod1p (31).
To investigate whether the overexpression of KlCCS1 would be able to increase GAA production, the multicopy plasmid pKlCCS1 was constructed. The K. lactis MW278-20C strains carrying or not carrying the pGM-GAM plasmid were transformed with either p426SD11 or pKlCCS1, and the culture medium was assayed for GAA activity. The presence of the expression vector pKlCCS1 did not increase GAA production (Fig. 7A). The SOD activity was also analyzed in the total extracts prepared from the cells treated as described in Fig. 7A. Consistently, the presence of pKlCCS1 did not enhance the intracellular SOD activity (Fig. 7B).
FIG. 7.
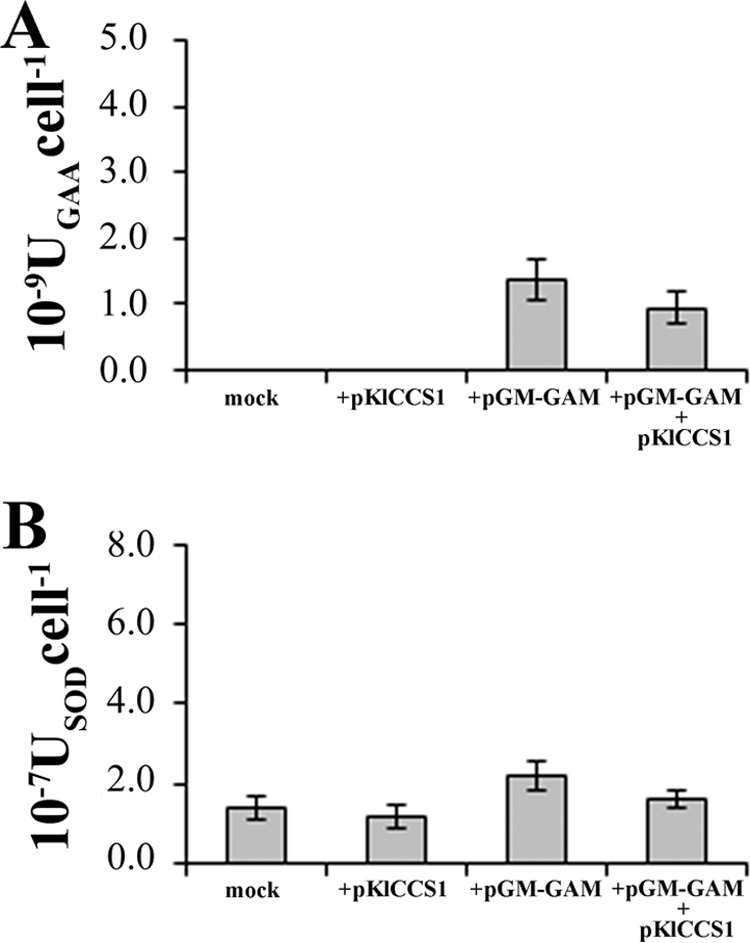
(A) GAA production in batch cultures of K. lactis MW278-20C carrying either the empty vector p426SD11 (mock) or the pKlCCS1 plasmid (+pKlCCS1) and from both strains additionally transformed with pGM-GAM (+pGM-GAM and +pGM-GAM +pKlCCS1). (B) Cytoplasmic SOD activities (USODcell−1) detected in the cell extracts related to the above-mentioned cultures.
UPR activation in K. lactis cells producing recombinant proteins.
When a protein overload enters the secretory pathway, the cell chaperone activities may be insufficient, triggering ER stress. K. lactis cells producing a heterologous protein were therefore analyzed for the occurrence of ER stress. To this aim, we measured the cells' growth capability in the presence of tunicamycin, a molecule known to induce the UPR. Indeed, the K. lactis MW278-20C cells transformed with the pGM-GAM plasmid showed an increased sensitivity to this compound compared to the control (data not shown). However, the increased dosage of KlSOD1 in those cells was not able to relieve such sensitivity. To further confirm the activation of the UPR in cells overexpressing GAA, a northern analysis of KlKAR2 was performed. This chaperone, which mediates protein folding in the ER, upregulated in cells transformed with the pGM-GAM plasmid compared to the control cells that do not produce the heterologous protein (Fig. 8A). Furthermore, the increased SOD activity was not able to significantly decrease the level of KlKAR2 transcription, in agreement with the growth behavior of the strains in the presence of tunicamycin.
FIG. 8.
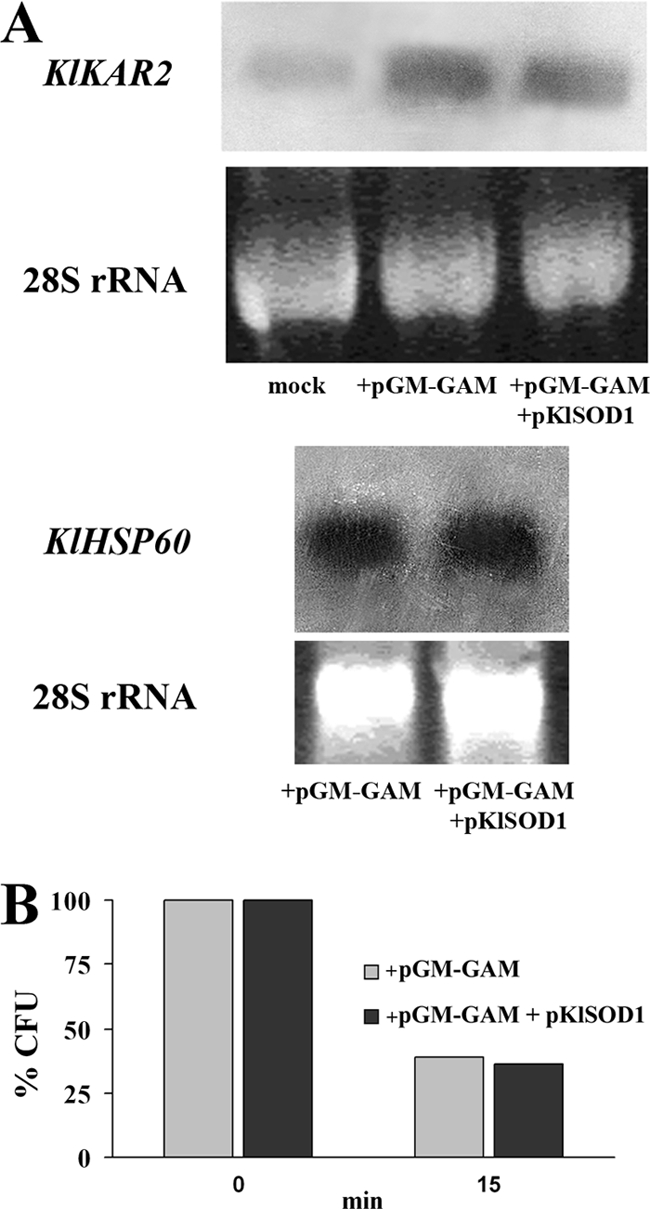
(A) Northern blot analysis of KlKAR2 and KlHSP60 mRNA in the total RNA extracted from K. lactis MW278-20C strains harboring pGM-GAM (mock) and either the empty vector p426SD11 (+pGM-GAM) or the pKlSOD1 plasmid (+pGM-GAM +pKlSOD1), grown on YKK medium supplemented with 0.1 mM CuCl2 and 0.1 mM ZnCl2 for 48 h. (B) Heat stress resistance of the above-mentioned strains. The viability of the cells, challenged at 48°C for 15 min, was evaluated by plating samples on YPD medium and was expressed as the CFU percentage of the corresponding untreated cultures. The values are the means of three independent experiments and show an SD of <10%.
Recently, it has been reported that the activation of heat shock signaling cooperates with the UPR to mitigate ER stress (18). We thus investigated whether the increased dosage of KlSOD1 could activate such a stress response pathway. To this aim, we used, as a reporter of heat-shock signaling pathway activation, the expression of the mitochondrial chaperone KlHSP60, which we previously demonstrated to be one of its targets in K. lactis (35). As shown in Fig. 8A, under these conditions, the level of KlHSP60 mRNA expression was similar between KlSOD1-overexpressing cells and the strain expressing only GAA. To confirm the lack of activation of the heat stress response in the same cells, a heat shock tolerance study was performed. Also, in this case, no protection against a 48°C challenge was observed in the cells with an increased dosage of SOD activity (Fig. 8B).
DISCUSSION
Kluyveromyces lactis has been studied for decades, and its safe use in many food industry applications, as well as its use as a host for recombinant protein production, has been reported (3).
Protein expression and secretion in yeast is a complex, multistep process. Improvements in protein production by K. lactis have been explored at many major points of protein synthesis, folding, and secretion. Several genes, when overexpressed, enhance the release of heterologous proteins outside the cell. These helper genes code for proteins involved in the glycosylation pathway (37, 38), in the oxidative folding machinery (19), in the UPR (2), in the ER-Golgi membrane trafficking and exocytosis (34).
Recently, proteomic studies were applied to the identification of unobvious bottlenecks in Kluyveromyces lactis protein secretion, revealing that the overexpression of heterologous proteins induces the upregulation of stress response proteins, such as Hsp26p and Sod2p (39). These data are in agreement with our findings: in the present study, DHR staining of K. lactis transformed with the pGM-GAM plasmid indicated that an increased amount of ROS was effectively present in cells expressing high levels of recombinant GAA and suggested that the cells were driven to increase the antioxidant defense mechanisms. Moreover, the decreased secretion of GAA was obtained when the cells were previously challenged with menadione, whereas the addition of the antioxidant ascorbic acid to the growth medium was able to increase GAA production. These results confirmed that ROS played an important role in the limitation of heterologous protein production.
Cellular responses against ER stress led to oxidative stress in mammalian cells. Also in yeast, the expression of the transcription factor GCN4, the S. cerevisiae homolog of mammalian ATF4, is activated during ER stress, upregulates glutathione biosynthesis, and thereby protects the cells against the accumulation of ROS coming from ER-associated degradation and the oxidative protein folding machinery in the ER (11). The cultivation of methylotrophic yeasts like P. pastoris on methanol leads to significant oxidative stress, which may be relieved by the addition of antioxidants like ascorbic acid (41). Similarly, the expression of antioxidant enzymes like S. cerevisiae Sod1 in P. pastoris was reported to relieve oxidative stress (17).
In the present study, the dosage of KlSOD1 was increased and resulted in higher cytoplasmic SOD activity that relieved the ongoing oxidative stress taking place in K. lactis cells producing recombinant proteins. Furthermore, a higher level of SOD improved the secretion of two different heterologous proteins, HSA and GAA; this was effective only when the cells were grown under aerobic conditions. On the other hand, the increased SOD activity was not able to alleviate the ER stress present in those cells.
Hyperosmotic stress is often regarded as a typical problem connected with high-cell-density fermentations, as the medium formulation requires a high solute concentration in such systems. The batch medium may have an osmolarity around 1,000 mosmol/liter, which induces osmotic stress in yeasts (20). The K. lactis cells overexpressing KlSOD1 showed an enhanced production of heterologous proteins and were more resistant to additional osmotic and oxidative stresses, two gainful industrial features.
Recently, Gasser et al. (10) identified, by transcriptomic analysis, new S. cerevisiae helper factors improving heterologous protein secretion as efficiently as, or even more efficiently than, known candidates. In particular, the overproduction of Cup5p, the subunit c of the yeast vacuolar (H+)-ATPase (V0 domain), increased the secretion of a human antibody Fab fragment as much as Pdi1. Cup5p is involved in the homeostasis of iron and copper (8, 33); the latter is also relevant for SOD activity.
Consistent with the fact that there is little intracellular free copper available in the cytoplasm of the yeast cell (26), specialized metallochaperones have been identified, including Ccs1p (6), that bind copper after the ions enter the cell. This chaperone subsequently transfers its copper cargo to Sod1p (24). The S. cerevisiae enzyme can receive copper only through Ccs1p, and this absolute dependence on Ccs1p is correlated with the presence of two residues of proline near the C terminus (P-143 and P-145) (5). K. lactis Sod1p does not present any conservation of the indicated residues, and those positions are embedded in an otherwise exceptionally conserved region of the compared enzymes.
The lack of effect of the overexpression of the copper chaperone KlCcs1 on SOD activity shows that this component is not limiting under standard conditions. These data and the significant changes observed in the KlSod1p primary structure described above may suggest that in Kluyveromyces yeasts, different mechanism(s) for cation handling are present.
This study supports the notion that it is important to relieve, at least in part, the oxidative stress occurring when cells produce recombinant proteins to enhance their production yield.
Acknowledgments
We thank F. Castelli for helpful technical assistance and the anonymous reviewers who helped us in improving the paper.
This work was partially supported by the PRIN 2006 research program “Yeast as Sources of Biodiversity for the Production of Molecules of Agro-Alimentary and Pharmaceutical Interest,” by Ateneo research funding La Sapienza 2008, and by Istituto Pasteur-Fondazione Cenci Bolognetti.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Wiley Press, Hoboken, NJ.
- 2.Bao, W., and H. Fukuhara. 2001. Secretion of human proteins from yeast: stimulation by duplication of polyubiquitin and protein disulfide isomerase genes in Kluyveromyces lactis. Gene 272:103-110. [DOI] [PubMed] [Google Scholar]
- 3.Bonekamp, F. J., and J. Oosterom. 2004. On the safety of Kluyveromyces lactis: a review. Appl. Microbiol. Biotechnol. 41:1-3. [Google Scholar]
- 4.Brown, N. M., A. S. Torres, P. E. Doan, and T. V. O'Halloran. 2004. Oxygen and the copper chaperone CCS regulate posttranslational activation of Cu, Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 101:5518-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, M. C., J. B. Girouard, J. L. Ulloa, J. R. Subramaniam, P. C. Wong, J. S. Valentie, and V. C. Culotta. 2004. Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc. Natl. Acad. Sci. USA 101:5964-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culotta, V. C., L. W. Klomp, J. Strain, R. L. Casareno, B. Krems, and J. D. Gitlin. 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 272:23469-23472. [DOI] [PubMed] [Google Scholar]
- 7.Das, S., and C. P. Hollenberg. 1982. A high-frequency transformation system for the yeast Kluyveromyces lactis. Curr. Genet. 6:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Eide, D. J., J. T. Bridgham, Z. Zhao, and J. R. Mattoon. 1993. The vacuolar H+-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 241:447-456. [DOI] [PubMed] [Google Scholar]
- 9.Fridovich, I. 1978. The biology of oxygen radicals. Science 201:875-880. [DOI] [PubMed] [Google Scholar]
- 10.Gasser, B., M. Sauer, M. Maurer, G. Stadlmayr, and D. Mattanovich. 2007. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl. Environ. Microbiol. 73:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 12.Harmsen, M. M., M. I. Bruyne, and H. A. Raué. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 13.Hart, P. J., M. M. Balbirnie, N. L. Ogihara, A. M. Nersissian, M. S. Weiss, J. S. Valentine, and D. Eisenberg. 1999. A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry 38:2167-2178. [DOI] [PubMed] [Google Scholar]
- 14.Haynes, C. M., E. A. Titus, and A. A. Cooper. 2004. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell 15:767-776. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, A., A. S. Torres, T. V. O'Halloran, and A. C. Rosenzweig. 2001. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8:751-755. [DOI] [PubMed] [Google Scholar]
- 17.Li, J. R., and P. Yu. 2007. Expression of Cu,Zn-superoxide dismutase gene from Saccharomyces cerevisiae in Pichia pastoris and its resistance to oxidative stress. Appl. Microbiol. Biotechnol. 136:127-139. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y., and A. Chang. 2008. Heat shock response relieves ER stress. EMBO J. 27:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodi, T., B. Neglia, and C. Donnini. 2005. Secretion of human serum albumin by Kluyveromyces lactis overexpressing KlPDI1 and KlERO1. Appl. Environ. Microbiol. 71:4359-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattanovich, D., B. Gasser, H. Hohenblum, and M. Sauer. 2004. Stress in recombinant protein producing yeasts. J. Biotechnol. 113:121-135. [DOI] [PubMed] [Google Scholar]
- 21.Maullu, C., G. Lampis, A. Desogus, A. Ingianni, G. M. Rossolini, and R. Pompei. 1999. High-level production of heterologous protein by engineered yeasts grown in cottage cheese whey. Appl. Environ. Microbiol. 65:2745-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 23.Morlino, G. B., L. Tizzani, M. M. Bianchi, and L. Frontali. 1999. Inducible amplification of gene copy number and heterologous protein production in the yeast Kluyveromyces lactis. Appl. Environ. Microbiol. 65:4808-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Halloran, T. V., and V. C. Culotta. 2000. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275:25057-25060. [DOI] [PubMed] [Google Scholar]
- 25.Raden, D., S. Hildebrandt, P. Xu, E. Bell, F. J. Doyle III, and A. S. Robinson. 2005. Analysis of cellular response to protein overexpression. Syst. Biol. (Stevenage) 152:285-289. [DOI] [PubMed] [Google Scholar]
- 26.Rae, T. D., P. J. Schmidt, R. A. Pufahl, V. C. Culotta, and T. V. O'Halloran. 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284:805-808. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, A. S., V. Hines, and D. Wittrup. 1994. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology 12:381-384. [DOI] [PubMed] [Google Scholar]
- 28.Sagt, C. M., W. H. Müller, L. van der Heide, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 2002. Impaired cutinase secretion in Saccharomyces cerevisiae induces irregular endoplasmic reticulum (ER) membrane proliferation, oxidative stress, and ER-associated degradation. Appl. Environ. Microbiol. 68:2155-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saliola, M., C. Mazzoni, N. Solimando, A. Crisà, C. Falcone, G. Jung, and R. Fleer. 1999. Use of the KlADH4 promoter for ethanol-dependent production of recombinant human serum albumin in Kluyveromyces lactis. Appl. Environ. Microbiol. 65:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 31.Schmidt, P. J., C. Kunst, and V. C. Culotta. 2000. Copper activation of superoxide dismutase 1 (SOD1) in vivo. Role for protein-protein interactions with the copper chaperone for SOD1. J. Biol. Chem. 275:33771-33776. [DOI] [PubMed] [Google Scholar]
- 32.Shusta, E. V., R. T. Raines, A. Plückthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 33.Szczypka, M. S., Z. Zhu, P. Silar, and D. J. Thiele. 1997. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13:1423-1435. [DOI] [PubMed] [Google Scholar]
- 34.Toikkanen, J. H., L. Sundqvist, and S. Keränen. 2004. Kluyveromyces lactis SSO1 and SEB1 genes are functional in Saccharomyces cerevisiae and enhance production of secreted proteins when overexpressed. Yeast 21:1045-1055. [DOI] [PubMed] [Google Scholar]
- 35.Uccelletti, D., F. Farina, P. Pinton, P. Goffrini, P. Mancini, C. Talora, R. Rizzuto, and C. Palleschi. 2005. The Golgi Ca2+-ATPase KlPmr1p function is required for oxidative stress response by controlling the expression of the heat shock element HSP60 in Kluyveromyces lactis. Mol. Biol. Cell 16:4636-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uccelletti, D., F. Farina, S. Rufini, P. Magnelli, C. Abeijon, and C. Palleschi. 2006. The Kluyveromyces lactis α1,6-mannosyltransferase KlOch1p is required for cell-wall organization and proper functioning of the secretory pathway. FEMS Yeast Res. 6:449-457. [DOI] [PubMed] [Google Scholar]
- 37.Uccelletti, D., D. Staneva, S. Rufini, P. Venkov, and C. Palleschi. 2005. Enhanced secretion of heterologous proteins in Kluyveromyces lactis by over-expression of the GDP-mannose pyrophosphorylase, KlPsa1p. FEMS Yeast Res. 5:735-746. [DOI] [PubMed] [Google Scholar]
- 38.Uccelletti, D., S. Anticoli, and C. Palleschi. 2007. The apyrase KlYnd1p of Kluyveromyces lactis affects glycosylation, secretion, and cell wall properties. FEMS Yeast Res. 7:731-739. [DOI] [PubMed] [Google Scholar]
- 39.Van Ooyen, A. J. J., P. Dekker, M. Huang, M. M. Olsthoorn, D. I. Jacobs, P. A. Colussi, and C. Taron. 2006. Heterologous protein production in the yeast Kluyveromyces lactis. FEMS Yeast Res. 6:381-392. [DOI] [PubMed] [Google Scholar]
- 40.Wesolowsky-Louvel, M., K. D. Breunig, and H. Fukuhara. 1996. Kluyveromyces lactis, p. 139-201. In K. Wolf (ed.), Nonconventional yeasts in biotechnology. Springer-Verlag, Berlin, Germany.
- 41.Xiao, A., X. Zhou, L. Zhou, and Y. Zhang. 2006. Improvement of cell viability and hirudin production by ascorbic acid in Pichia pastoris fermentation. Appl. Microbiol. Biotechnol. 72:837-844. [DOI] [PubMed] [Google Scholar]