Abstract
Colonization by the fungal pathogen Candida albicans varies significantly, depending upon the pH and availability of oxygen. Because of our interest in extracellular molecules as potential quorum-sensing molecules, we examined the physiological conditions which regulate the production of the aromatic alcohols, i.e., phenethyl alcohol, tyrosol, and tryptophol. The production of these fusel oils has been well studied for Saccharomyces cerevisiae. Our data show that aromatic alcohol yields for C. albicans are determined by growth conditions. These conditions include the availability of aromatic amino acids, the pH, oxygen levels, and the presence of ammonium salts. For example, for wild-type C. albicans, tyrosol production varied 16-fold merely with the inclusion of tyrosine or ammonium salts in the growth medium. Aromatic alcohol production also depends on the transcription regulator Aro80p. Our results are consistent with aromatic alcohol production—aromatic transaminases (gene products for ARO8 and ARO9), aromatic decarboxylase (ARO10), and alcohol dehydrogenase (ADH)—via the fusel oil pathway. The expression of ARO8, ARO9, and ARO10 is also pH dependent. ARO8 and ARO9 were alkaline upregulated, while ARO10 was alkaline downregulated. The alkaline-dependent change in expression of ARO8 was Rim101 independent, while the expression of ARO9 was Rim101 dependent.
The dimorphic fungal pathogen Candida albicans grows and colonizes different niches of human hosts (41), which differ significantly both physically and chemically. The pH of the oral cavity varies according to diet, the metabolism of other microflora, and salivary flow. The stomach pH is less than 3, while the duodenum pH is 5 and the large intestine pH is 7.7 (4). The pH of the blood is around 7.4. C. albicans can thrive in all of these pH conditions. Similarly, C. albicans can adapt to aerobic, anaerobic, or hypoxic microenvironments, as is evident from its ability to exist in the anaerobic gastrointestinal tract (14, 15), and still cause infections ranging from superficial skin infections to deep-seated generalized infections where multiple internal tissues and host cells are invaded and colonized. The interiors of biofilms may also be partially anaerobic environments (31). The ability to adapt in all these different conditions is one of the most important attributes of severe fungal pathogens. Our long-standing interest is in extracellular molecules produced by C. albicans (31). Does their production differ under these different growth conditions, and do they have a role in cellular adaptations from one condition to another?
It has been well established that Saccharomyces cerevisiae secretes fusel oils, fusel being from the old German word Fousel, meaning bad spirit (40). The components of fusel oils and the mechanism of formation of these higher alcohols from amino acids have been well characterized for S. cerevisiae (18, 34, 40). Fusel oil formation from amino acids proceeds through the Ehrlich pathway, which was first proposed 100 years ago (16, 30). This pathway consists of three enzymatic steps: transamination to form an α-keto acid, which would then be decarboxylated to an aldehyde and reduced to the fusel alcohol (Fig. 1). This pathway has been confirmed by more recent work (34, 39), including elegant studies using 13C-labeled amino acids and 13C nuclear magnetic resonance spectroscopy (13, 18). Ehrlich also showed that the addition of ammonium salts and asparagine inhibited the formation of fusel oils (16, 40) and that yeasts produced tyrosol (p-hydroxyphenyl ethanol) if tyrosine was added to the fermenting mixture and tryptophol if tryptophan was added (16, 40). S. cerevisiae can use tryptophan, phenylalanine, or tyrosine as the only source of cellular nitrogen (8), with the main products of their catabolism being tryptophol, phenethyl alcohol, or tyrosol, respectively (22, 27, 34, 36).
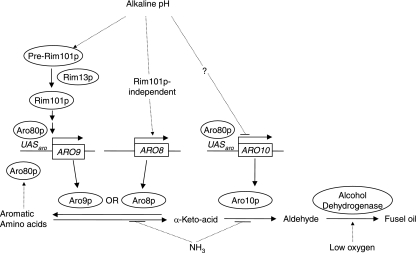
FIG. 1.
Regulation of the production of aromatic alcohols from aromatic amino acids. C. albicans can use the aromatic amino acids tryptophan, phenylalanine, and tyrosine as cellular nitrogen sources. This results in the production of tryptophol, phenylethanol and tyrosol, respectively, which are known collectively as fusel oils. Fusel oil production depends on environmental factors, including the availability of aromatic amino acids, the presence of ammonia, the oxygen level, and an alkaline pH (indicated by dotted lines). Aromatic amino acids stimulate Aro80p, a transcription activator required for full expression of ARO8 and ARO9 (encoding aromatic transaminases) and ARO10 (aromatic decarboxylase). Genes are in boxes; enzymes/proteins are in ellipses. The scheme is based on our findings as well as on pathways reported previously for both S. cerevisiae (6, 13, 16, 21) and C. albicans (10, 11, 33). UAS, upstream activation sequence.
The present paper examines aromatic alcohol production in C. albicans and finds that the production characteristics fit those expected of fusel oils. That is, aromatic alcohol production uses homologous genes and enzymes as in S. cerevisiae, is dependent on the availability of amino acid precursors, and is regulated by pH, oxygen availability, and nitrogen repression by ammonium. For instance, tyrosol production per gram (dry weight) of cells varied over 16-fold for wild-type cells. Studies with null mutants revealed that transaminases and decarboxylase are under the dual control of the Aro80p and pH pathways. The ARO8 and ARO9 gene products (transaminases) were alkaline upregulated, while the ARO10 gene product (decarboxylase) was alkaline downregulated. ARO8 was Rim 101p independent, while ARO9 was Rim101p dependent.
MATERIALS AND METHODS
Strains and growth media.
The C. albicans strains SC5314, CAI4 (ura3Δ::imm434/ura3Δ::imm434), and BWP17 (ura3Δ::imm434/ura3Δ::imm434, arg4::hisG/arg4::hisG, his1::hisG/his1::hisG) (42) were obtained from Alexander Johnson, University of California at San Francisco. The aro80 (orf19.3012) and rim13 (orf19.3995) insertion mutants were obtained from Aaron Mitchell's collection (32); they were derived from strain BWP17. CAR2 (rim101::hisG/rim101::hisG-URA3-hisG ura3::imm434/ura3::imm434) (33) was obtained from Fritz A. Muhlschlegel, Canterbury, United Kingdom.
For routine growth of strains, yeast extract-peptone-dextrose medium (10 g of yeast extract per liter, 20 g of peptone per liter, 20 g of glucose per liter) was used. To quantify aromatic alcohols in culture supernatants, GPP, GPA, or GPP+A medium (23) was used. GPP medium contains the following (per 900 ml of distilled water): 4.0 g of KH2PO4, 3.2 g of Na2HPO4, 1.2 g of l-proline, and 0.7 g of MgSO4·7H2O. After the medium was autoclaved, 100 ml of 20% (wt/vol) glucose, 1 ml of a vitamin mix, and 0.25 ml of a mineral mix were added. The vitamin mix contains the following (per 100 ml of 20% ethanol): 2 mg of biotin, 20 mg of thiamine-HCl, and 20 mg of pyridoxine-HCl. The mineral mix contains the following (per 100 ml of 0.1 N HCl): 0.5 g of CuSO4·5H2O, 0.5 g of ZnSO4·H2O, 0.8 g of MnCl2·4H2O, and 0.5 g of FeSO4. The vitamin mix and the mineral mix were filter sterilized through 0.2-μm-pore-size Whatman (Maidstone, United Kingdom) cellulose nitrate filters and stored at 4°C. For GPA medium, ammonium sulfate (10 mM) replaced proline, and for GPP+A, ammonium sulfate was added to GPP. Anaerobic growth employed the Hungate technique for growing stringent anaerobes as adapted for C. albicans by Dumitru et al. (14). Thus, our regular GPP medium (50 ml) was supplemented with 200 μl of 1 mM oleic acid in 100% methanol, 200 μl of 4 mM nicotinic acid, and 1 ml of 500 mM NH4Cl (14). The cells were harvested after 5 days at 30°C.
Quantification of excreted aromatic alcohols.
C. albicans strains were cultured for 24 to 28 h in 50 ml of defined medium (GPP, pH 6.8) with shaking at 250 rpm. Cells were grown at 30 or 37°C . When necessary, pH values were adjusted using 1 N HCl or 1 N NaOH. After growth, the fungal cultures were harvested by centrifugation at 6,000 rpm for 10 min. The supernatants were filtered through 0.2-μm Millipore filters, extracted with ethyl acetate, filtered, and concentrated by rotary evaporation to 50 μl. One microliter of sample was then injected into a HP6890 gas chromatograph-mass spectrometer (GC-MS) with an HP 19091B-005 50-m capillary column. The flow rate was 1.0 ml/min. GC used an inlet temperature of 280°C and a temperature program of 80°C for 2 min followed by increases of 60°C/min until 160°C was reached and held for 2 min and then 10°C/min until 300°C was reached and held for 5 min, with a total run time of 24.33 min. MS used a 5-min solvent delay. Ethyl acetate extraction is suitable for the three aromatic alcohols as well as farnesol, whereas hexane or 1:4 ethylacetate/hexane extraction is suitable for farnesol (20) but not for the aromatic alcohols. The phenethyl alcohol was purchased from Aldrich Chemical Co., Milwaukee, WI; tyrosol from Avocado Research Chemicals, Ltd., Heysham, United Kingdom; and tryptophol from TCI-EP, Tokyo, Japan.
Northern analysis.
The C. albicans total RNA used for mRNA accumulation was extracted by the hot phenol extraction method using yeast cells harvested at mid-log phase (24). Equal amounts of RNA (15 μg) were resolved on 1.0% agarose-formaldehyde gels and transferred to GeneScreen Plus membranes (NEN Life Science Products, Boston, MA). A NorthernMax complete Northern blotting kit (Ambion, Austin, TX) was used for transfer and hybridization. The DNA templates for probe synthesis were prepared using PCR with C. albicans SC5314 genomic DNA and the following primers (5′ to 3′): ARO8, TATTCCAACACCGTCGTTCA and ACAAACTGGTCCAAGGCATC; ARO9, CAAAACTCCGCCTTCCAGTA and AGCCATCCATCAACACCTTT; ARO10, GTGCTTATGCTGCTGATGGA and TCTTTTTGGGTTCTGCTGCTG; RIM101, AGTCCATGTCCCATTGAAGC and ACACCGCCAAACTCTAATGC; and ACT1, AGTTATCGATAACGGTTCTG and AGATTTCCAGAATTTCACTC. The DNA templates were used to synthesize DNA probes labeled with 32P using an oligolabeling kit (Rad Prime DNA labeling system; Invitrogen Life Technologies, Carlsbad, CA), as described by Atkin et al. (3). Northern blots were phosphorimaged using a Storm phosphorimager (Amersham Pharmacia Biotech), and measurements of ARO8, ARO9, ARO10, and RIM101 mRNA were normalized with an ACT1 mRNA control.
RESULTS
Environmental control of aromatic alcohol production by C. albicans.
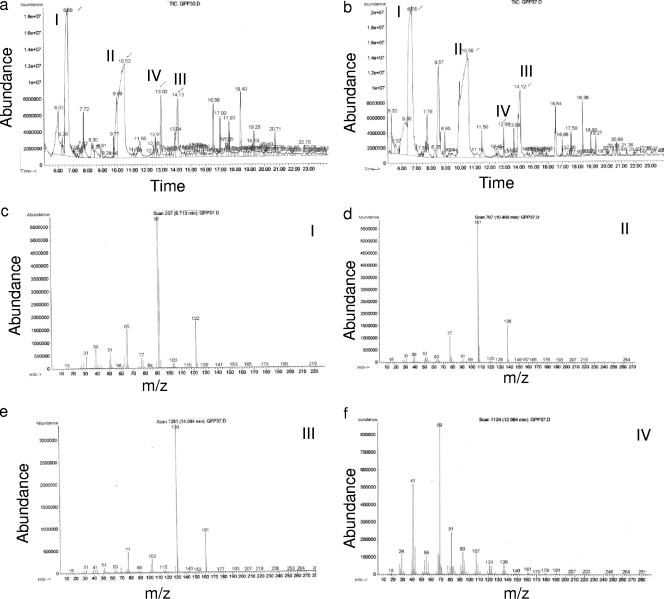
The aromatic alcohol yields for S. cerevisiae vary with different growth conditions (18). The rationale for the environmental variables tested here was to see if the control mechanisms operative in C. albicans paralleled those known for fusel oil production in S. cerevisiae (21). Temperature, anaerobiosis, precursor availability, ammonium ions, and pH were examined. In each case, supernatants from C. albicans SC5314 grown in GPP medium were analyzed for the three aromatic alcohols, phenethyl alcohol, tyrosol, and tryptophol (Fig. 2). Peaks were identified by comparing their retention times and MS spectra to those of the pure compounds (Fig. 2c to f).
FIG. 2.
GC-MS analysis of ethyl acetate extracts from cell-free supernatants of C. albicans. Cells were grown overnight at 30°C (a) or 37°C (b) prior to GC-MS analysis. The GC peaks labeled I, II, III, and IV were identified by MS as phenethyl alcohol (c), tyrosol (d), tryptophol (e), and farnesol (f), respectively.
Aromatic alcohol production was not affected by growth temperature.
With regard to temperature, there was little difference in levels of aromatic alcohol production between 30°C (Fig. 2a) and 37°C (Fig. 2b). The concentrations of phenethyl alcohol, tyrosol, and tryptophol were 830, 2,120, and 440 μg/g at 30°C and 1,030, 2,530, and 660 μg/g at 37°C, respectively (Fig. 3a). For comparison, farnesol was present at 17 and 20 μg/g at 30°C and 37°C, respectively (Fig. 2a and b). The identities of the other peaks in Fig. 2 remain unknown. Their mass values do not correspond with those expected for any of the other fusel oils.
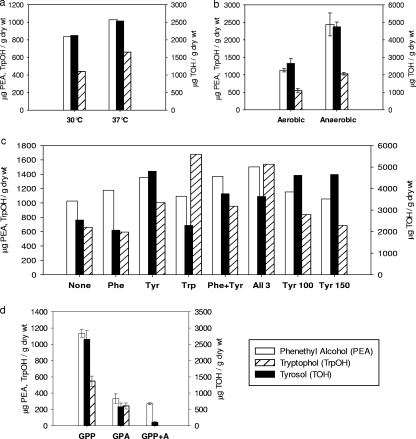
FIG. 3.
Effects of environmental conditions on production of aromatic alcohols. GC-MS analysis of cell-free supernatants of C. albicans: (a) 30°C and 37°C (from Fig. 2a and b, respectively); (b) grown at 30°C aerobically or anaerobically; (c) defined GPP medium (37°C) supplemented with the indicated amino acid(s), at 50 μg/ml unless otherwise indicated; (d) medium with proline (GPP) or ammonia (GPA) or both (GPP+A) as the nitrogen source. Throughout, phenethyl alcohol, tyrosol, and tryptophol are expressed as micrograms per gram (dry weight) of fungal cells. Data for panels b and d (all at 30°C and pH 7) are the averages from triplicate experiments, with bars representing standard errors, whereas panels a and c show the averages from two experiments which agreed within ±10%.
Increased aromatic alcohols are produced anaerobically.
Another of the environmental variables examined was anaerobiosis. Anaerobic growth conditions (14) should be relevant for C. albicans growing in animal gastrointestinal tracts (15) and biofilms (31). Alem et al. (1) recently reported that C. albicans biofilms produced 1.5-fold more tyrosol than did the corresponding planktonic cells. We found that cells of C. albicans grown anaerobically at 30°C produced roughly twice as much of each of the three aromatic alcohols as did aerobically grown cells (Fig. 3b).
Aromatic amino acid precursors elevate aromatic alcohol production.
We examined aromatic alcohol production by cells grown at 37°C in GPP medium supplemented with the aromatic amino acids phenylalanine, tyrosine, or tryptophan, which are precursors for aromatic alcohol production (Fig. 3c). In each case, the expected alcohol increased in abundance. At 37°C, tyrosol production was increased 2-fold in the presence of tyrosine, and tryptophol production was increased 2.5-fold in the presence of tryptophan (Fig. 3c). No significant further changes in tyrosol levels were observed as tyrosine was increased from 50 to 150 μg/ml (Fig. 3c). Throughout, the cell morphologies remained unchanged by the amino acid additions; the cells were 90 to 95% hyphal.
Ammonia suppresses aromatic alcohol production.
Another environmental variable expected to influence fusel oils is the ammonia effect (6, 21), whereby aromatic alcohol production is inhibited by ammonia. C. albicans SC5314 produced five to seven times less aromatic alcohols when grown at 30°C in GPA medium than when grown in GPP medium (Fig. 3d). The two media differ only in whether ammonium sulfate or l-proline (each at 10 mM) is the nitrogen source. Aromatic alcohol production was also five- to sevenfold lower when the cells were grown in medium with both l-proline and ammonium sulfate (Fig. 3d), showing that the ammonia effect is operative even in the presence of proline. In the upstream regions for C. albicans ARO8 to -10, we found several GAT(A/T)(A/G) sequences, putative binding sites for the GATA transcription factors Gln3 and Gat1 (9, 25). These conserved regulators mediate nitrogen catabolite repression by activating genes whose products are required for nitrogen catabolism.
Decreased production of aromatic alcohols by an aro80 mutant.
By analogy with aromatic alcohol production in S. cerevisiae (21), aromatic alcohol production in C. albicans is expected to depend on Aro80p. S. cerevisiae Aro80p is a member of the Zn2Cys6 transcription activator family which increases synthesis of Aro9p (aromatic transaminase) and Aro10p (aromatic decarboxylase). Aro80p is activated by the three aromatic amino acids (21). The C. albicans Aro80p is 32% identical with its S. cerevisiae homolog. Importantly, however, the conservation is much higher (67% identical) at the N terminus that contains the zinc binuclear DNA binding domain (Fig. 4a). Thus, both S. cerevisiae and C. albicans Aro80p are bimetal thiolate cluster proteins/transcription regulators (28, 38), and they probably recognize the same or similar sequences. Aro80p is a transcription activator of ARO8, -9, and -10 in S. cerevisiae (21) and, by analogy, it may be in C. albicans also. We also chose the ARO8, -9, and -10 genes for study because ARO9 was consistently upregulated when S. cerevisiae was grown in a glucose-limited chemostat with phenylalanine as the sole nitrogen source (5) while ARO10 was the only decarboxylase gene whose transcript profile correlated strongly with α-ketoacid decarboxylase activity in chemostat culture (5, 18, 39).
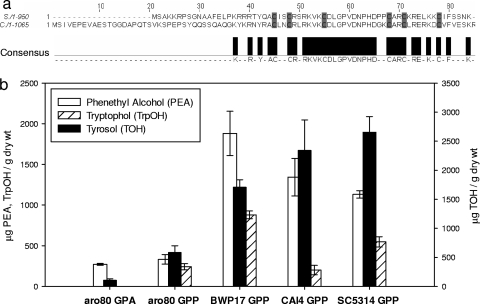
FIG. 4.
Effects of Aro80p on production of aromatic alcohols. (a) Comparison of the N-terminal portions of Aro80p from C. albicans and S. cerevisiae, showing the bimetal thiolate cluster expected in Zn2Cys6-type transcription activators. The six cysteine residues are shown in grey, and the other conserved amino acids in the DNA binding domain are shown in black. (b) GC-MS analyses of C. albicans (30°C) supernatants from SC5314, CAI4, BWP17, and aro80 grown in proline (GPP)- or ammonia (GPA)-containing medium. Tryptophol was below the detection limit for aro80 GPA medium. Bars represent standard errors.
When grown in GPP medium, C. albicans aro80 produced ca. 3.5 times less phenethyl alcohol, 4.5 times less tyrosol, and 2.5 times less tryptophol than did the wild-type SC5314 and ca. five times less phenethyl alcohol, 2.5 times less tyrosol, and 3.5 times less tryptophol than did the parent BWP17 (Fig. 4b). In the presence of ammonia (GPA instead of GPP medium), the aro80 production levels for phenethyl alcohol, tyrosol, and tryptophol were reduced a further 1.2-, 4-, and 20-fold, respectively (Fig. 3b). The fact that the ammonia effect is still observed for aro80 suggests that the ammonia effect on the ARO8 and ARO9 (transaminases) and AR010 (decarboxylase) genes and/or proteins (Fig. 1) is independent of aro80. These findings are consistent with the conclusion that aromatic amino acid metabolism in C. albicans, like that in S. cerevisiae (21), is both stimulated by transcription activation by Aro80p and subject to nitrogen catabolite repression by ammonia.
Alkaline pH elevates aromatic alcohol yield.
The final environmental variable we explored was pH. C. albicans grows over a pH range of ca. 1.5 to 10, and culture pH is strongly influenced by the nitrogen source. For both C. albicans and Ceratocystis ulmi (23), the pH remains constant at ca. 6.5 throughout growth in both proline- and arginine-containing media. Indeed, GPP and GPR media were designed to study fungal dimorphism without concurrent changes in pH (23). In contrast, with ammonium salts as the nitrogen source, the pH drops to 2 to 3 with (NH4)2SO4 or NH4Cl but remains at pH 6.5 with ammonium tartrate (23). Many bacteria respond to pH extremes by synthesizing amino acid decarboxylases at a low external pH and amino acid deaminases at a high external pH (17). C. albicans may have similar mechanisms. In unbuffered medium 199 (a glutamine-containing medium purchased from Sigma, St. Louis, MO), cultures that started at pH values ranging from 4 to 10 returned to pH 7 within 6 h (M. C. Lorenz, personal communication). Accordingly, production of the aromatic alcohols by wild-type cells was examined for highly buffered cultures grown in GPP medium at pH 3, 7, or 8, in which pH 3 constitutes acid stress and pH 7 and 8 constitute more-alkaline conditions (11). Production of the three aromatic alcohols by SC5314 was two- to threefold higher for cells grown at pH 7 (Fig. 5a) or pH 8 (data not shown) than by cells grown under acid stress (Fig. 5a). Northern blots for these cells showed that ARO8 and ARO9 were alkaline upregulated while ARO10 was alkaline downregulated (Fig. 5b, lanes 1 and 2).
FIG. 5.
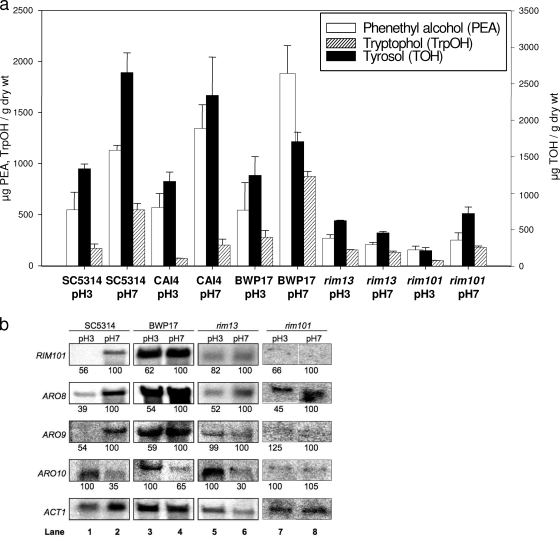
Effects of pH on production of aromatic alcohols. (a) GC-MS analysis of C. albicans (30°C). SC5314, BWP17, CAI4, rim13, and rim101 grown in GPP medium at pH 3 or pH 7. Values are the averages from triplicate experiments. Bars represent standard errors. (b) Northern blot analysis of RNA prepared from mid-log phase cells grown at 30°C. Blots were probed for the transcripts indicated along the left side. The numbers below represent the signal quantified with a phosphorimager, normalized for the ACT1 loading control (average results from three replicate experiments). Lane numbers are at the bottom. Note that lanes 7 and 8 are from the same blot and exposed for the same time.
Rim101p is required for maximal aromatic alcohol production.
Alkaline pH responses can be either Rim101p dependent or Rim101p independent (11). For the Rim101-dependent alkaline response, Rim101p needs to be proteolytically cleaved to its active form by Rim13p (Fig. 1), a calpain protease (10). Rim101p then activates transcription of a variety of genes, including PHR1 and PRA1, as well as its own gene, RIM101. Thus, in the absence of RIM13, Rim101p would remain inactive and genes such as PHR1 and PRA1 would not be expressed at alkaline pH (11).
The rim13 and rim101 mutant cells grew as yeasts in GPP medium at 37°C at pH values from 3 to 8, thus confirming that the RIM101 pathway is required for alkaline-induced filamentation (4). However, the absence of filamentation was not due to growth defects. The rim13 and rim101 dry weights after 24 h were very close to those for wild-type cells, and in the GlcNAc-induced filamentation assay (20), histidine-supplemented rim13 and uridine-supplemented rim101 produced germ tubes just like wild-type cells (data not shown). Thus, RIM101 is not essential for filamentation, and other pathways are available for hypha formation.
With wild-type SC5314, the aromatic alcohol yields in vitro were elevated at pH 7 compared to pH 3 (Fig. 5a); thus, we wanted to see if this pH effect was Rim101p dependent. We tested aromatic alcohol production of rim13 and rim101 at pH 3 and pH 7 (Fig. 5a) and then compared these values with those of their parent strains, BWP17 and CAI4, respectively, as well as with wild-type SC5314 (Fig. 5a). The observation that two independent mutants in the RIM101 pathway, i.e., rim13 and rim101, both curtail fusel alcohol synthesis implicates this pathway in its synthesis.
For the wild-type SC5314 and the two parental strains, BWP17 and CAI4, the aromatic alcohol yields in vitro were pH dependent, being at least 2.0-fold higher at pH 7 than at pH 3. In contrast, pH regulation was lost for rim13 (Fig. 5a). Two independent rim13 mutants were tested, and they behaved similarly. Although the rim13 and rim101 mutants produced less aromatic alcohol, the pH regulation was not completely lost for rim101 (Fig. 5a). Farnesol production by rim13 and rim101, which was similar to that of wild-type SC5314 and parent strains BWP17 and CAI4 at both pH values (data not shown), was used as a control.
We further tested C. albicans with regard to whether the genes for aromatic alcohol production were pH regulated, and if so, whether the pH effects observed were Rim101p dependent or Rim101p independent. The latter distinction was made using rim13 and rim101 mutants; rim13 is the mutant in which Rim101p cannot be activated. For wild-type SC5314, ARO8 and ARO9 were alkaline upregulated while ARO10 was alkaline downregulated (Fig. 5b, lanes 1 and 2). This pattern was also seen for BWP17 (Fig. 5b, lanes 3 and 4). ARO9 was regulated in a RIM101p-dependent manner, because its alkaline upregulation was lost in both rim13 (lanes 5 and 6) and rim101 (lanes 7 and 8). In contrast, ARO8 was regulated in a Rim101p-independent manner, because it was still alkaline regulated in both rim13 and rim101 (Fig. 5b, lanes 5 to 8). The situation with ARO10 is more complicated in that its alkaline downregulation was lost in rim101 (lanes 7 and 8) but not in rim13 (lanes 5 and 6). These findings reinforce the microarray data of Bensen et al. (4), who found that ARO8 was alkaline upregulated and ARO10 was alkaline downregulated. Alkaline induction of RIM101 can also be seen in Fig. 5b (lanes 1 to 2).
DISCUSSION
Fusel alcohols are the natural products of amino acid catabolism. Yeasts cannot use branched chain or aromatic amino acids as their sole carbon source (8). However, they can be used as nitrogen sources under otherwise nitrogen-limiting conditions, with the consequent production of fusel alcohols as potentially toxic or regulatory by-products (2, 19). Our studies of aromatic alcohol production showed that C. albicans produced three aromatic alcohols, phenethyl alcohol, tyrosol, and tryptophol, and they are consistent with the use of the same pathway as in S. cerevisiae, i.e., transamination (ARO8, ARO9), decarboxylation (ARO10), and then reduction by alcohol dehydrogenase (ADH) (18, 34). This pathway is summarized in Fig. 1. We found that C. albicans produced the three expected aromatic alcohols in roughly constant proportions under all conditions studied. Previously, Lingappa et al. (26) reported the production of phenethyl alcohol and tryptophol, whereas Chen et al. (7) detected tyrosol and Martins et al. (29) detected phenethyl alcohol and isoamyl alcohol. Isoamyl alcohol is the fusel alcohol derived from leucine (18).
Aromatic alcohol production was dependent on the transcription factor Aro80p. It was also repressed by ammonium ions and elevated under anaerobic conditions or whenever the appropriate amino acids, phenylalanine, tyrosine, or tryptophan, were provided in the growth medium. Aromatic alcohol production was determined by growth conditions. For example, for wild-type C. albicans, tyrosol production varied 16-fold merely with the inclusion of tyrosine (Fig. 3c) or ammonium ions (Fig. 3d) in the growth medium. Also, we found that cells of C. albicans grown anaerobically at 30°C produced roughly twice as much of each of the three aromatic alcohols as did aerobically grown cells (Fig. 3b). This increased production occurred despite the fact that our anaerobic growth medium is a modified GPP medium containing 10 mM ammonium salts (14). C. albicans upregulates three alcohol dehydrogenase genes (ADH1, ADH2, and ADH5) during hypoxic growth (35), a finding which is consistent with the fact that larger amounts of aromatic alcohols are secreted under anaerobic conditions (Fig. 3b). Aromatic alcohol production would be energetically favorable under anaerobic conditions. The aromatic aldehydes would be electron acceptors and substrates for one or more of the alcohol dehydrogenases (Fig. 1). Higher aromatic alcohol production under anaerobic conditions would also explain the observation of Alem et al. (1) that, on a per weight basis, biofilm cells secreted 50% more tyrosol than did planktonic cells. This 50% increase would be expected if 30 to 40% of the biofilm cells were growing in anaerobic conditions.
Because aromatic alcohols are formed from aromatic amino acids by a pathway which includes decarboxylation, we also considered whether their production was part of a pH response by C. albicans. All microbes have an optimal pH for growth, and many use pH-regulated genes to bring the external pH close to this optimal range (37). These studies were pioneered by Ernest Gale and Helen Epps (17), who showed that in an amino acid- or protein-rich environment, many bacteria made amino acid decarboxylases at low external pHs and amino acid deaminases at high external pHs, in each case acting to neutralize the pH of the growth medium. C. albicans is also capable of neutralizing unbuffered growth media. However, compensating for pH extremes is clearly not the dominant reason for aromatic alcohol production by C. albicans. Aromatic alcohol production was actually reduced during growth at low pH (Fig. 5a), and the transaminase and decarboxylase genes were regulated in an opposite manner by pH (Fig. 5b). ARO8 and ARO9 were alkaline upregulated, whereas ARO10 was alkaline downregulated. This pH regulation in opposite directions is consistent with aromatic alcohol production being maximal at pH 7 (Fig. 5a).
Finally, aromatic alcohol production was insensitive to pH in the rim13 mutant (Fig. 5a), and the pH-dependent upregulation of ARO9 was lost in both rim13 and rim101 (Fig. 5b). Thus, we suggest that ARO9 should be added to the list of Rim101p-regulated genes. Dual regulation of ARO9 by Aro80p and Rim101p suggests that Aro9p is a critical step for the regulation of fusel oils, a reasonable possibility because the decarboxylation step that follows is effectively irreversible (12, 18, 34). For ARO10, regulation was lost in rim101 but not in rim13 (Fig. 5b). This juxtaposition could mean that ARO10 expression is dependent on Rim101p but not on the activation/processing of that protein by Rim13p.
Acknowledgments
We thank Raluca Dumitru for preparing the supernatants from anaerobically grown cells.
This work was supported by the University of Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, the John C. and Nettie V. David Memorial Trust Fund, and the Farnesol and Candida albicans Research Fund of the University of Nebraska Foundation.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Alem, M. A. S., M. D. Y. Oteef, T. H. Flowers, and L. J. Douglas. 2006. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot. Cell 5:1770-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe, M. P., J. W. Slaven, S. K. DeLong, S. Ibrahimo, and A. B. Sachs. 2001. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 20:6464-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin, A. L., L. R. Schenkman, M. Eastman, J. N. Dahlseid, M. J. Lelivelt, and M. R. Culbertson. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272:22163-22172. [DOI] [PubMed] [Google Scholar]
- 4.Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in C. albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335-1351. [DOI] [PubMed] [Google Scholar]
- 5.Boer, V. M., S. L. Tai, Z. Vuralhan, Y. Arifin, M. C. Walsh, M. D. Piper, J. H. de Winde, J. T. Pronk, and J. M. Daran. 2007. Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose-limited chemostat cultures. FEMS Yeast Res. 7:604-620. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., and G. R. Fink. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20:1150-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., M. Fujita, Q. Feng, J. Clardy, and G. R. Fink. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. 101:5048-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, T. G. 1981. Nitrogen metabolism in S. cerevisiae, p. 39-99. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast S. cerevisiae. Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY.
- 9.Dabas, N., and J. Morschhäuser. 2007. Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot. Cell 6:875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, D. 2003. Adaptation to environmental pH in C. albicans and its relation to pathogenesis. Curr. Genet. 44:1-7. [DOI] [PubMed] [Google Scholar]
- 11.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in C. albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrick, S., and P. J. Large. 1993. Activities of the enzymes of the Ehrlich pathway and formation of branched-chain alcohols in Saccharomyces cerevisiae and Candida utilis grown in continuous culture on valine or ammonium as sole nitrogen source. J. Gen. Microbiol. 139:2783-2792. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson, J. R., L. E. J. Salgado, and M. J. E. Hewlins. 2003. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278:8028-8034. [DOI] [PubMed] [Google Scholar]
- 14.Dumitru, R., J. M. Hornby, and K. W. Nickerson. 2004. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 48:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumitru, R., D. H. M. L. P. Navarathna, C. P. Semighini, C. G. Elowsky, R. V. Dumitru, D. Dignard, M. Whiteway, A. L. Atkin, and K. W. Nickerson. 2007. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot. Cell 6:465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich, F. 1907. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Ber. Dtsch. Chem. Ges. 40:1027-1047. [Google Scholar]
- 17.Gale, E. F., and H. M. R. Epps. 1942. The effect of the pH of the medium during growth on the enzymatic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem. J. 36:600-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazelwood, L. A., J.-M. Daran, A. J. A. van Maris, J. T. Pronk, and J. R. Dickinson. 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407-450. [DOI] [PubMed] [Google Scholar]
- 20.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iraqui I., S. Vissers, B. André, and A. Urrestarazu. 1999. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3360-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kradolfer, P., P. Niederberger, and R. Hütter. 1982. Tryptophan degradation in S. cerevisiae: characterization of two aromatic aminotransferases. Arch. Microbiol. 133:242-248. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni, R. K., and K. W. Nickerson. 1981. Nutritional control of dimorphism in Ceratocystis ulmi. Exp. Mycol. 5:148-154. [Google Scholar]
- 24.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in S. cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limjindaporn, T., R. A. Khalaf, and W. A. Fonzi. 2003. Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50:993-1004. [DOI] [PubMed] [Google Scholar]
- 26.Lingappa, B. T., M. Prasad, Y. Lingappa, D. F. Hunt, and K. Biemann. 1969. Phenethyl alcohol and tryptophol: autoantibiotics produced by the fungus Candida albicans. Science 163:192-194. [DOI] [PubMed] [Google Scholar]
- 27.Lingens, F., W. Goebel, and H. Uesseler. 1967. Regulation of the biosynthesis of aromatic amino acids in S. cerevisiae. 2. Repression, induction and activation. Eur. J. Biochem. 1:363-374. (In German.) [DOI] [PubMed] [Google Scholar]
- 28.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 29.Martins, M., M. Henriques, J. Azeredo, S. M. Rocha, M. A. Coimbra, and R. Oliveira. 2007. Morphogenesis control in Candida albicans and Candida dubliniensis through signaling molecules produced by planktonic and biofilm cells. Eukaryot. Cell 6:2429-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neubauer, O., and K. Fromherz. 1911. Über den Abbau der Aminosaüren bei der Hefegärung. Hoppe-Seyler's Z. Physiol. Chem. 70:326-350. [Google Scholar]
- 31.Nickerson, K. W., A. L. Atkin, and J. M. Hornby. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72:3805-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobile, C. J., V. M. Bruno, M. L. Richard, D. A. Davis, and A. P. Mitchell. 2003. Genetic control of chlamydospore formation in C. albicans. Microbiology 149:3629-3637. [DOI] [PubMed] [Google Scholar]
- 33.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sentheshanuganathan, S. 1960. The mechanism of the formation of higher alcohols from amino acids by S. cerevisiae. Biochem. J. 74:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 361:399-411. [DOI] [PubMed] [Google Scholar]
- 36.Shin, M., T. Shinguu, C. Sano, and K. Umezawa. 1991. Metabolic fates of l-tryptophan in Saccharomyces uvarum (Saccharomyces carlsbergensis). Chem. Pharm. Bull. (Tokyo) 39:1792-1795. [DOI] [PubMed] [Google Scholar]
- 37.Slonczewski, J. L. 1992. pH-regulated genes in enteric bacteria. ASM News 58:140-144. [Google Scholar]
- 38.Vashee, S., H. Xu, S. A. Johnston, and T. Kodadek. 1993. How do “Zn2 Cys6” proteins distinguish between similar upstream activation sites? J. Biol. Chem. 268:24699-24706. [PubMed] [Google Scholar]
- 39.Vuralhan, Z., M. A. H. Luttik, S. L. Tai, V. M. Boer, M. A. Morais, D. Schipper, M. J. H. Almering, P. Kötter, J. R. Dickinson, J.-M. Daran, and J. T. Pronk. 2005. Physiological chracterization of the ARO10-dependent, broad-substrate specificity 2-oxo acid decarboxylase activity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:3276-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb, A. D., and J. L. Ingraham. 1963. Fusel oil. Adv. Appl. Microbiol. 5:317-353. [Google Scholar]
- 41.Whiteway, M., and C. Bachewich. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]