Abstract
Thermococcus kodakarensis possesses two chaperonins, CpkA and CpkB, and their expression is induced by the downshift and upshift, respectively, of the cell cultivation temperature. The expression levels of the chaperonins were examined by using specific antibodies at various cell growth temperatures in the logarithmic and stationary phases. At 60°C, CpkA was highly expressed in both the logarithmic and stationary phases; however, CpkB was not expressed in either phase. At 85°C, CpkA and CpkB were expressed in both phases; however, the CpkA level was decreased in the stationary phase. At 93°C, CpkA was expressed only in the logarithmic phase and not in the stationary phase. In contrast, CpkB was highly expressed in both phases. The results of reverse transcription-PCR experiments showed the same growth phase- and temperature-dependent profiles as observed in immunoblot analyses, indicating that the expression of cpkA and cpkB is regulated at the mRNA level. The cpkA or cpkB gene disruptant was then constructed, and its growth profile was monitored. The cpkA disruptant showed poor cell growth at 60°C but no significant defects at 85°C and 93°C. On the other hand, cpkB disruption led to growth defects at 93°C but no significant defects at 60°C and 85°C. These data indicate that CpkA and CpkB are necessary for cell growth at lower and higher temperatures, respectively. The logarithmic-phase-dependent expression of CpkA at 93°C suggested that CpkA participates in initial cell growth in addition to lower-temperature adaptation. Promoter mapping and quantitative analyses using the Phr (Pyrococcus heat-shock regulator) gene disruptant revealed that temperature-dependent expression was achieved in a Phr-independent manner.
Living microorganisms face various kinds of stress and protect themselves by inducing several stress-responsive proteins. Under temperature stress, which is a major form of stress in nature, cells produce a group of proteins called heat shock proteins (HSPs) (13). The predominant HSPs are classified by their molecular masses as HSP104, HSP90, HSP70, HSP60, and small HSP (3, 13). The best-studied chaperone is HSP60. HSP60s, called chaperonins, assist in refolding denatured proteins and folding newly synthesized proteins. Chaperonins are classified into two groups, I and II (14). Group I chaperonins consist of two protein components, GroEL and GroES, with molecular masses of approximately 60 and 10 kDa, respectively, and are found in bacteria, mitochondria, and chloroplasts. Group II chaperonins, which have a structure similar to that of the GroEL/GroES complex, are found in archaea and the cytosol of eukaryotes and can facilitate protein folding in the absence of cochaperonin GroES. Cytosolic group II chaperonins are closely related to cell growth and markedly enhanced at the early S phase of the cell cycle in mouse and human cultured cells (29). The cytosolic group II chaperonin that is upregulated around the early S phase is associated with tubulin in vivo. In archaea, it is unclear whether or not chaperonin expression is under cell cycle control; however, some homologues involved in cell cycle regulation have been identified in archaea (15). A cell cycle-dependent variation in Orc1/Cdc6 levels in Sulfolobus species has been demonstrated, reminiscent of the variation in cyclin levels during the eukaryotic cell cycle (19). The three Orc1/Orc6 replication initiation proteins of Sulfolobus acidocaldarius have been shown to vary in abundance over the cell cycle in a cyclin-like fashion, providing a clear example of cell cycle-specific events among archaea (19). In addition, a CDC48/VCP homologue has also been identified in Thermococcus kodakarensis, and growth phase-specific fragmentation was observed only in the stationary phase (11). These findings led us to examine whether the hyperthermophilic archaeal chaperonin is under cell cycle or growth phase regulation.
Thermococcus kodakarensis is a sulfur-reducing hyperthermophilic archaeon isolated from Kodakara Island, Japan (1, 17). It has two kinds of group II chaperonin genes, cpkA and cpkB, in its genome, and CpkA and CpkB were upregulated by growth temperature down- and upshifts, respectively (9, 28). Based on the phylogenetic similarities between members of the Archaea and Eukarya, CpkA and CpkB are also expected to have a cell cycle-dependent mechanism in their expression. However, the details are not available because a synchronized cultivation has not been established for an anaerobic hyperthermophile such as T. kodakarensis. Cell cycle regulation is correlated with growth phase regulation because the gap period is prolonged in the stationary phase due to an extended generation time. Hence, we have investigated in more detail whether or not two chaperonins, CpkA and CpkB, are expressed under growth phase-dependent regulation.
In the present study, the growth phase dependencies of the expression of these chaperonins were examined at various cell cultivation temperatures, 60°C, 85°C, and 93°C, by quantitative reverse transcription-PCR (RT-PCR) and immunoblot analyses. In addition, to obtain precise information on the temperature-dependent expression, the promoter region was analyzed by determining the transcriptional start points. In archaea, only limited information is available regarding the temperature-responsive factor. The heat shock regulation of members of the Archaea is predicted to be different from that of prokaryotes and eukaryotes because no sigma-like factor, such as sigma factor σH (σ32) in Escherichia coli (31), and no homologues of heat shock factor (HSF) in Drosophila and mammals have been identified in the genomes (18). Phr (Pyrococcus heat shock regulator) is the only known factor that has been identified as a heat shock regulator in Pyrococcus furiosus (25). Phr is involved in the heat-dependent expression of several heat shock genes by repressing transcription at low temperatures in P. furiosus. It has been suggested that the expression of the thermosome in P. furiosus is controlled by Phr (25). Recently, Phr has been shown to be involved in the heat-dependent induction of prefoldins in T. kodakarensis (4). In this report, the transcriptional levels of cpkA and cpkB were analyzed in a phr disruptant of T. kodakarensis, and the Phr dependency on chaperonin induction was also examined.
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
Thermococcus kodakarensis KOD1 (wild type) and its derivatives were cultivated anaerobically in a nutrient-rich medium (MA-YT or ASW-YT) with an addition of 5.0 g/liter of elemental sulfur or pyruvate or a synthetic medium (ASW-AA) containing amino acids and elemental sulfur (22). The construction procedure for the phr gene (TK2291) disruptant (T. kodakarensis KHR1) was described elsewhere (5). To obtain cells for RT-PCR and immunoblot analyses, growth curves at 60°C, 85°C, and 93°C in MA-YT medium with pyruvate were carefully monitored and cells were harvested at middle logarithmic and stationary phases. The optical densities at 660 nm of T. kodakarensis cells at 60°C, 85°C, and 93°C were 0.07, 0.2, and 0.2 in logarithmic phase and 0.3, 0.7, and 0.7 in stationary phase, respectively. E. coli TG-1 was used for general DNA manipulation and sequencing. E. coli cells were routinely cultivated at 37°C in Luria-Bertani (LB) medium. Ampicillin (50 μg/ml) was added to the medium when needed.
DNA manipulation and sequencing.
DNA manipulations were carried out by standard techniques as described previously by Sambrook and Russell (20). Restriction enzymes and other modifying enzymes were purchased from Takara or Toyobo. Plasmid DNA was isolated with a Wizard Plus miniprep DNA purification system (Promega). GFX PCR DNA and a gel band purification kit (GE Healthcare) were used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed with a BigDye Terminator cycle-sequencing ready reaction kit, version 3.1, and a model 3130 capillary DNA sequencer (Applied Biosystems).
RT-PCR.
Total RNAs extracted from cells of T. kodakarensis KOD1 and the Δphr mutant (5) that were cultivated at 60°C, 85°C, and 93°C were purified by using an RNeasy midi kit (Qiagen). Each RNA (0.25 μg) in 10 μl of the reaction mixture was reverse transcribed at 55°C for 30 min with Transcriptor reverse transcriptase (Roche) using the primer for each (cpkA-IR, 5′-CAT GCC CAT GTC CAT TCC GC-3′, and cpkB-IR, 5′-TGA GGA TCA TTA TGG CAG CC-3′). The cDNA obtained was then amplified by PCR using the primer pair for each (for cpkA, cpkA-IF, 5′-CGT CAA AGT CAT CAG CGA GC-3′, and cpkA-IR; for cpkB, cpkB-IF, 5′-GAC AAG ATC AAG GAG GTC GG-3′, and cpkB-IR). The reactions were carried out by using KOD Plus polymerase (Toyobo) for 20 cycles according to the reported procedure. The RT-PCR products were analyzed by 2% agarose gel electrophoresis. The amplified products for cpkA and cpkB were 256 bp and 712 bp, respectively. As a control to ensure that the signal intensities reflected the initial levels of each transcript directly, RT-PCRs were performed with the same total RNAs and the 16S rRNA gene-specific primers 16SrRNA-Fw, 5′-CCA TAG GCC TGA GGT ACT GG-3′, and 16SrRNA-Rv, 5′-CGC TCC CCT GGC CTT CGT CCC TCA CCG TCG-3′. The amplified product was 570 bp.
Immunoblot analyses.
The preparation of the specific antibodies anti-CpkA and anti-CpkB was described previously (9). Cell extracts were obtained from the T. kodakarensis cells cultivated at 60°C, 85°C, and 93°C and were subjected to electrophoresis in 0.1% sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gels and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked using 5% skim milk and incubated with the mouse anti-CpkA and anti-CpkB serum at dilutions of 1:5,000 and 1:10,000, respectively. The detection of immunocomplexes was performed by using alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Cappel) as the secondary antibody and a BCIP/NBT stock solution (Roche Diagnostics) according to the manufacturer's instructions.
Primer extension analysis.
The nonradioisotope primer extension analysis was carried out according to the reported procedure (8). Total RNA was isolated as described above. Cy5.5-labeled primer (cpkA-Cy5.5, 5′-TAA CAA CCG GCT GTC CAC TA-3′, or cpkB-Cy5.5, 5′-TAA CAA CTG GCT GGC CTG CG-3′) and total RNA (17 μg) were mixed and preincubated at 90°C for 5 min and then added to the reaction buffer. The reverse transcriptase reaction was carried out with Transcriptor reverse transcriptase (Roche) for 30 min at 50°C. The products were purified by ethanol precipitation and incubated for 3 min at room temperature in the presence of RNase. Then, the reverse transcripts were resuspended in loading buffer containing 95% formamide, 20 mM EDTA, and 0.05% bromophenol blue. After heat treatment at 90°C for 5 min, samples were loaded on 6% polyacrylamide gel containing 7 M urea. In order to prepare sequence ladders, the DNA fragments, including the upstream regions of cpkA and cpkB, respectively, were cloned into the plasmid pUC19. The fragments were amplified from the chromosomal DNA using primers 5′-ACT TCT CGA ATT CCC TGA CG-3′ and 5′-GTA TCC TGA GGA GTC CCT G-3′ for cpkA and 5′-CTT CCT TCC TTC AAA GAT TG-3′ and 5′-TGA GGA TCA TTA TGG CAG CC-3′ for cpkB under PCR conditions of 94°C for 2 min for the 1st cycle and 94°C for 15 s, 55°C for 30 s, and 68°C for 30 s for the 2nd to 25th cycles. The amplified products were cloned into pUC19, and the plasmids obtained were designated pUCcpkA and pUCcpkB, respectively. DNA sequence ladders using the same primers and pUCcpkA or pUCcpkB as a template were prepared according to the manufacturer's instructions provided with a Thermo Sequenase cycle sequencing kit (USB). The reverse transcripts were visualized by using an Odyssey infrared imaging system (LI-COR).
Construction of disruption vectors.
The vectors to disrupt cpkA and cpkB through single-crossover homologous recombination were constructed as follows. DNA fragments containing each target gene together with its flanking regions (ca. 800 bp each) were amplified by using T. kodakarensis genomic DNA as the template and primer sets (sense for cpkA disruption, cpkA-OF, 5′-TTC GAA TTC GAG TTC TCG CTT CCC TTA CG −3′; antisense for cpkA disruption, cpkA-OR, 5′-GAG GAA TTC CTC CCT TGA CCT GCT TAT CG-3′; sense for cpkB disruption, cpkB-OF, 5′-GCC GAA TTC CTC CCA GAA GTT CTT TCT GG-3′; antisense for cpkB disruption, cpkB-OR, 5′-AGC GAA TTC TTG AGG AGA ACG GCG TTA GG-3′ [underlined sequences indicate EcoRI site in the sense and antisense primers]). These DNA fragments were subcloned into pUD2 (21), and the constructs were named pUDcpkAfr and pUDcpkBfr. An inverse PCR was performed in order to amplify the entire pUDcpkAfr or pUDcpkBfr excluding the region of the disruption target (the primer sets for cpkA disruption were 5′-CAG AAA CGC TTC GTT AGT GCT CT-3′ and 5′-GAA CTT CTG CAT AGA GGC TGG CA-3′, and the primer sets for cpkB disruption were 5′-GAG GGT GGA TGA GTT TTA GTT AC-3′ and 5′-GAA TCT GCT CTT CTC TGG TCA GG-3′). After self-ligation, the plasmid containing the 5′- and 3′-flanking regions of the target gene was constructed. The DNA sequences of all vectors were confirmed by sequence determination.
Transformation of T. kodakarensis.
The theoretical background for specific gene disruption has been described previously (21). T. kodakarensis KU216 (pyrF disruptant, cpkA+ cpkB+) was cultivated in an ASW-YT liquid medium for 12 h, and cells were harvested (3,200 × g for 5 min) from 3 ml of culture. The cells were resuspended in 200 μl of 0.8× ASW and kept on ice for 30 min. Plasmid DNA (5 μg) was added to the cell suspension, and the mixtures were kept on ice for 1 h. The treated cells were cultured in 20 ml of ASW-AA liquid medium containing 5.0 g/liter of elemental sulfur in the absence of uracil for the period of 2 generations. They were then spread on an ASW-YT plate medium containing 7.5 mg/ml of 5-fluoroorotic acid in order to select transformants that had completed the second, pop-out recombination and incubated at 85°C for isolation of the cpkA disruptant or 60°C for isolation of the cpkB disruptant. Transformants with a host genotype (cpkA+ cpkB+) and those with the intended disruption (ΔcpkA or ΔcpkB; entire deletion of cpkA or cpkB gene) were distinguished by confirming the fragment length using PCR methods with specific primers that anneal outside of the target (for ΔcpkA confirmation, primers cpkA-OF and cpkA-OR, and for ΔcpkB confirmation, primers cpkB-OF and cpkB-OR). The 1,365-bp or 1,584-bp DNA was amplified by the above primer combinations from the cpkA disruptant or cpkB disruptant, respectively, indicating that both disruptants were satisfactorily constructed. (Primers cpkA-OF and cpkA-OR or primers cpkB-OF and cpkB-OR amplify 3,094 bp or 3,299 bp of DNA, respectively, from wild-type chromosomal DNA.) Furthermore, the construction of cpkA and cpkB disruptants was confirmed by no DNA amplification by primers cpkA-IF and cpkA-IR and primers cpkB-IF and cpkB-IR, respectively.
RESULTS
Quantitative analyses of protein expression.
The expression of CpkA and CpkB has been reported to be upregulated by down- and upshifts, respectively, in growth temperature (9, 28). It has not been clarified whether this temperature-dependent expression occurs in all growth phases. In the present study, the expression levels of CpkA and CpkB were examined by specific antibodies in both the logarithmic and stationary phases at the optimum growth temperature (85°C) and at lower (60°C) or higher (93°C) growth temperatures (Fig. 1). In the stationary phase, CpkA expression was detected only at 60°C and 85°C and not at 93°C. However, in the logarithmic phase, the levels of CpkA were not significantly changed at any of the temperatures examined. The temperature-dependent pattern of CpkB expression in the logarithmic phase was similarly observed in the stationary phases at all temperatures. The expression level increased as the cell growth temperature rose. It is noteworthy that the expression of CpkB is regulated depending on the cell growth temperature; however, that of CpkA is regulated depending on the growth phase in addition to the growth temperature. The growth phase-dependent manner of CpkA expression is a unique phenomenon that has not been observed in other chaperonins.
FIG. 1.
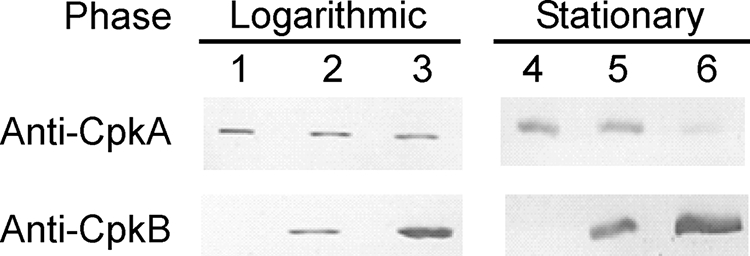
Protein levels of CpkA and CpkB in the logarithmic and stationary phases. Immunoblot analysis with anti-CpkA and anti-CpkB was performed using cytoplasmic extracts obtained from cells cultivated at 60°C (lanes 1 and 4), 85°C (lanes 2 and 5), and 93°C (lanes 3 and 6) as described in Materials and Methods. Cell extracts containing 1.0 μg of total protein were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto a polyvinylidene difluoride membrane, and detected with anti-CpkA and anti-CpkB.
Quantitative analyses of cpkA and cpkB transcripts.
It has been unclear whether CpkA and CpkB expression is regulated at the mRNA or the protein level. Most genes are regulated at the transcriptional level (mRNA level); however, some cold-inducible proteins are regulated at the posttranscriptional level (7). To investigate the amounts of mRNA of cpkA and cpkB, we performed quantitative analysis by RT-PCR using mRNAs isolated from T. kodakarensis grown in the logarithmic and stationary phases at 60°C, 85°C, and 93°C (Fig. 2). In cpkA during the logarithmic phase, no significant differences were observed in the patterns at all temperatures examined. However, at 85°C and 93°C, the signal intensity of the cpkA transcript decreased in the stationary phase. In the stationary phase in particular, the cpkA signal decreased as the cell cultivation temperature rose, and it almost disappeared at 93°C. As for cpkB in the logarithmic phase, its transcript was hardly detected at 60°C, but the level of the cpkB-specific transcript increased with upshifted growth temperatures, as shown in Fig. 2. A similar pattern was observed in the stationary phase. Growth phase-dependent expression was not observed in the transcriptional pattern of cpkB, but growth temperature-dependent expression was observed. We examined the cpkB-specific amount in the stationary phase by quantitative real-time PCR and observed that the transcriptional level at 93°C was 78-fold higher than that at 60°C (see Fig. S1 in the supplemental material). In the previous report, the expression levels were examined by using cells obtained from the stationary-phase condition (9). Focusing on both the logarithmic and stationary growth phases, the unique growth-dependent profiles of cpkA were observed, and regulation was shown to occur at the mRNA level.
FIG. 2.
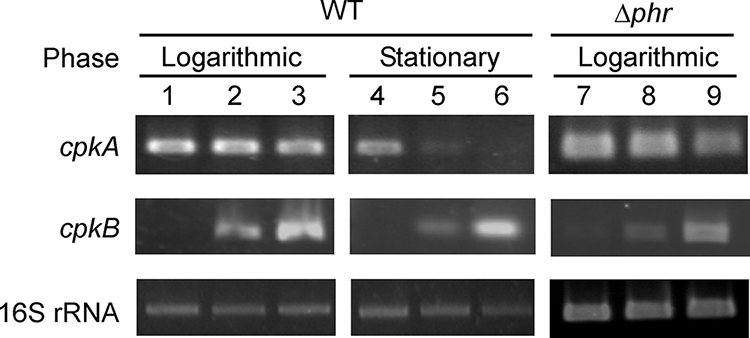
Comparison of mRNA levels of cpkA and cpkB in the logarithmic and stationary phases. RT-PCRs were performed with total RNAs isolated from the wild type (WT) and KHR1 cultivated at 60°C (lanes 1, 4, and 7), 85°C (lanes 2, 5, and 8), and 93°C (lanes 3, 6, and 9) as described in Materials and Methods. The levels of the cpkA and cpkB transcripts were evaluated as the signal intensities of fragments amplified with the respective gene-specific primers. As a control, to ensure that the intensities directly reflected the initial levels of each transcript, the levels of 16S rRNA were examined.
Promoter analyses of cpkA and cpkB.
As reported above, it has become clear that the expression of CpkA and CpkB is regulated at the transcriptional level, as for other known HSPs (31). We then compared the promoter regions of two chaperonin genes. The cpkA and cpkB genes show high sequence identity in their coding regions. However, the sequences of both genes vary in the region upstream from the initiation codon. Therefore, we expected that the difference between the upstream regions of cpkA and cpkB would contribute to their different manners of expression. To compare the promoter regions of cpkA and cpkB, their transcriptional start points were determined using RNA extracted from logarithmic-phase cells by the nonradioisotope-labeled primer extension method (8). As shown by the results in Fig. 3, the transcriptional start sites of cpkA and cpkB were identified at 32 and 67 nucleotides upstream of the initiation codon, respectively, indicating that cpkA and cpkB are transcribed from a single start point. The same start points were identified at all temperatures examined, and no additional points were identified. This indicates that multiple-promoter regulation was not involved in the temperature-dependent expression. The transcriptional machinery of archaea is generally similar to that of a eukaryote. The putative promoter region of cpkA and cpkB includes a transcription factor B (TFB)-binding element (CNANANC) and a TATA box (TTTAT/AA) (Fig. 4). In the heat shock response of a eukaryote, an HSF is known as a positive regulator of hsp genes that binds to the inverted repeat of heat shock element sequence NGAA (18). However, archaea do not have homologues of HSF- or heat shock element-like sequences. The regulation mechanisms of the heat shock responses of bacteria and eukarya are not applicable to archaea. The Phr-binding region is the only known archaeal heat-shock element that possesses three conserved regions, including the transcriptional start site (Fig. 4, conserved regions indicated by underlines). The Phr orthologue is also identified in T. kodakarensis as TK2291 (5). The effect of Phr on cpkB induction was examined by using a phr disruptant named KHR1.
FIG. 3.
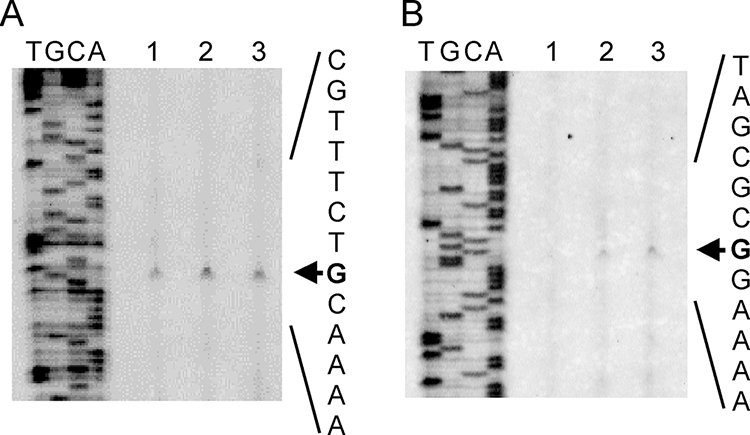
Results of primer extension analyses for cpkA (A) and cpkB (B). Primer extension reactions were performed with total RNAs isolated from T. kodakarensis bacteria cultivated at 60°C (lane 1), 85°C (lane 2), and 93°C (lane 3) as described in Materials and Methods. The extended products were analyzed with sequencing ladders generated using the same primer and template (lanes A, T, G, and C). The transcriptional start sites are indicated by arrows.
FIG. 4.
Sequence comparison of promoter regions of various heat shock elements. The transcription start sites that were determined are indicated by arrows. The positions of the TATA elements and TFB recognition element are indicated by boxes and dashed underlines, respectively. Phr recognition elements are underlined. Regions conserved between cpkB and hsp60s of Pyrococcus spp. are shaded. The transcriptional start sites are indicated by arrows. aaa+ ATPase, ATPase associated with diverse cellular activities; sHSP, small HSP; P. horikoshi, Pyrococcus horikoshii; P. abyssi, Pyrococcus abyssi.
Effect of Phr on chaperonin gene expression.
To investigate the effect of the phr gene disruption on the transcription of cpkA and cpkB, RT-PCR experiments were performed using a logarithmic-phase mRNA from KHR1. There was no significant difference between the mRNA amounts of cpkB in the wild type and KHR1 (Fig. 2). Phr is a heat-sensitive repressor and inhibits transcription by binding to DNA at lower temperatures (25). If the expression of cpkB were repressed by Phr binding, the level of cpkB would increase at 60°C in KHR1. The result obtained indicated that Phr is not involved in the heat-dependent expression of cpkB. The results of sequence analysis also indicate that Phr does not bind to the promoter region because the Phr-binding consensus was not identified (Fig. 4). The results of the experiments using the KHR strain also indicated that cpkA expression is not under Phr regulation.
Growth characteristics of cpkA or cpkB disruptants.
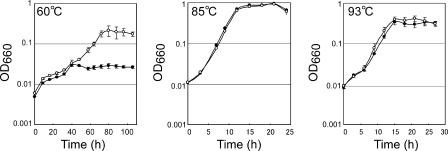
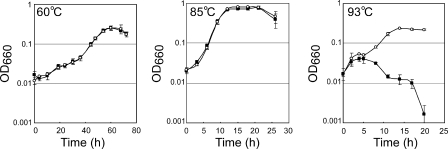
To examine the physiological roles of cpkA and cpkB in T. kodakarensis cells, we constructed cpkA or cpkB disruptants (harboring ΔcpkA or ΔcpkB, respectively) using the strategy described in Materials and Methods. The genotypes of the disruptants were first confirmed by PCR analysis with primer sets that anneal inside or outside of the disrupted region. The expected shortening in the length of the amplified DNA was confirmed as mentioned in Materials and Methods. An internal primer that anneals within the coding region was also used to confirm the deletion, indicating the absence of the target on the disruptant's chromosome. It was estimated that the gene disruptant obtained would show growth defects in response to the growth temperature. Therefore, the cpkA disruptant (ΔcpkA cpkB+) and cpkB disruptant (cpkA+ ΔcpkB) were precultivated at 93°C and 60°C, respectively, at which temperatures no growth defects were expected. These disruptants were then cultivated at 60°C, 85°C, and 93°C, and the growth profiles were compared with those of wild-type strain KU216 (cpkA+ cpkB+) (Fig. 5 and 6). As for the effect of cpkA disruption, no obvious difference was observed between the growth of strain KU216 and that of the cpkA disruptant at 85°C and 93°C (Fig. 5, center and right panels). It was expected that CpkA would be required for initial cell growth at all temperatures because it was expressed in the logarithmic growth phase at all temperatures. However, a clear growth defect was observed only at 60°C (Fig. 5, left panel), and no growth lag was observed at 85°C and 93°C for the cpkA disruptant. In the cpkA disruptant, CpkB could probably be substituted for the CpkA. In contrast, the cpkB disruptant showed a growth defect only at 93°C, and no defect was observed at 85°C or 60°C (Fig. 6). It was concluded that CpkB participates in cell survival in a higher-temperature environment. The results obtained indicate that CpkA and CpkB are important for cell growth at lower and higher growth temperatures, respectively, than the optimum one.
FIG. 5.
Growth characteristics of the cpkA disruptant and the parental strain. Representative growth curves of the cpkA disruptant (ΔcpkA cpkB+) and KU216 (cpkA+ cpkB+). Cells were cultivated at 60°C (left panel), 85°C (center panel), and 93°C (right panel), respectively. Error bars show standard deviations. ○, KU216; •, cpkA disruptant; OD660, optical density at 660 nm.
FIG. 6.
Growth characteristics of the cpkB disruptant and the parental strain. Representative growth curves of the cpkB disruptant (cpkA+ ΔcpkB) and KU216 (cpkA+ cpkB+). Cells were cultivated at 60°C (left panel), 85°C (center panel), and 93°C (right panel), respectively. Error bars show standard deviations. ○, KU216; •, cpkB disruptant; OD660, optical density at 660 nm.
DISCUSSION
The results of a previous immunoprecipitation study revealed that the subunit composition of the chaperonin in T. kodakarensis changes depending on the growth temperature (9). The results of the present study revealed that the subunit composition also changes depending on the growth phase because the expression level of CpkA is controlled by not only the growth temperature but also the growth phase. The increased ratio of CpkB to CpkA would enable the complex to be thermostable, because the biochemical analysis of the chaperonin of another Thermococcus sp., Thermococcus sp. strain KS-1, which possesses identical α and β chaperonin subunits and has amino acid sequences identical to those of CpkA and CpkB, respectively, has revealed that the homooligomer consisting of only β subunits was more thermostable than that consisting of only α subunits (30). A mildly hydrophobic Gly-Gly-Met (GGM) repeat sequence of GroEL exists in the C-terminal amino acid sequence of CpkA but not in that of CpkB (10, 30). It has been reported that the protrusion of the flexible and mildly hydrophobic C-terminal GGM repeat sequence and repulsion effect from the negatively charged wall inside the chaperonin molecule were required for the rapid folding of substrate proteins (23). Therefore, the change in the composition of the chaperonin may cause a change in the substrate preference. However, no growth defect was observed in cpkA and cpkB disruptants at the optimal temperature, at which both CpkA and CpkB were expressed in the wild type. These results suggested another possibility, namely, that CpkA and CpkB are alternatives for each other at 85°C and that specific substrates for CpkA or CpkB seem to be produced at 60°C or 93°C, respectively.
We speculated that the expression level of an archaeal chaperonin is related to the cell growth conditions. In the present study, it has been shown that the level of CpkA expression changed depending on the growth phases at 85°C and 93°C. The expression may be correlated with the cell cycle rather than the growth phase. A synchronized culture is required to examine cell cycle dependency; however, this has not been achieved for the hyperthermophilic anaerobic archaeon. Among archaea, cell cycle-specific gene expression has been analyzed in two microorganisms, the aerobic thermophilic archaeon S. acidocaldarius (16) and the halophilic archaeon Halobacterium salinarum (2). In H. salinarum, 87 genes out of 2,547 genes were found to be cell cycle regulated, and seven clusters were discovered in regulated genes (2). Among them, cluster five contained 34 genes with their highest transcriptional levels about 1.5 h after cell division. Several of them had a very low transcriptional level until the cell division was completed and were rapidly induced thereafter, similar to the M/G1-induced genes found in eukaryotes (2). In the cluster, the chaperonin homologues named thermosomes (OE3925R and OE4122R), members of the HSP60 family, were identified. H. salinarum OE3925R shows higher sequence similarity to CpkA than to CpkB, suggesting that CpkA is also under cell cycle regulation.
As for cpkB, growth phase-dependent expression was not observed at any of the temperatures examined. Instead, the typical heat dependency was observed. By transcription start site mapping, a single start site was identified at 67 nucleotides upstream of the initiation codon (Fig. 3B), indicating that multiple-promoter regulation was not involved in the temperature-dependent expression of cpkA and cpkB. In E. coli, rpoH, encoding the heat shock regulator σH (σ32), is transcribed in a complicated manner. rpoH transcription is regulated by multiple promoters and modulated by several factors, including two σ factors (σ70 [6] and σ24 [27]), the global regulator cyclic AMP receptor protein (12), and the DnaA protein (26). Such regulators are not involved in the archaeal system, because no homologues are found in the genomes. The transcriptional machinery of archaea is generally similar to that of a eukaryote. Both the TFB-binding element and a TATA box were identified in the promoter region (Fig. 4). The results of the RT-PCR experiment revealed that enhanced expression of cpkB at 93°C is not under Phr control (Fig. 1). When the promoter regions of cpkB and Pyrococcus HSP60 were compared, a typical conserved area was observed downstream of the TATA box, showing CAAANGAAC-N8-GGAA (Fig. 4, conserved regions indicated by shading). It has been reported that the nucleotide alignment of the 5′-flanking regions of archaeal chaperonin genes in Haloferax volcanii showed a high degree of sequence conservation between positions −37 and +1, especially in and immediately surrounding the TATA element of the putative core promoter (24). The results of mutation and deletion studies of the promoter region of H. volcanii chaperonin indicated that the regulatory sequences involved in heat-induced transcription lie within the promoter region (24). We speculate that the specific region containing CAAANGAAC-N8-GGAA participates in the heat-responsive regulation. The sequence of this element in cpkA is CTAANGAAG-N8-GCAA (different bases are indicated with underlining) (Fig. 4). The differences and the gap would be involved in heat inducibility. The unknown factor might bind this region to control the cpkB transcription.
The promoter region of cpkB shares sequence similarity with that of Pyrococcus HSP60, suggesting that they are all phylogenetically related and evolved from the same origin. However, the upstream region of cpkA shows less similarity with Pyrococcus elements. cpkA is considered to be a typical paralogue of cpkB acquired in the course of evolution to adapt to a lower-temperature environment. In fact, the cpkA disruptant shows a growth defect at lower growth temperatures. CpkA might participate in the maintenance of protein structures of stationary-phase cells at lower cell growth temperatures. However, the manner in which CpkA is involved in cell cycle regulation or growth phase regulation is still unclear. To investigate this issue in detail, a trial to develop a synchronized culture for T. kodakarensis is needed.
Supplementary Material
Acknowledgments
This study was supported by JSPS KAKENHI (20580090) and by a special grant from the Hyogo Science and Technology Association.
Footnotes
Published ahead of print on 3 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, A., C. Lange, and J. Soppa. 2007. Transcriptome changes and cAMP oscillations in an archaeal cell cycle. BMC Cell Biol. 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig, E. A., B. D. Gambill, and R. J. Nelson. 1993. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57:402-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danno, A., W. Fukuda, M. Yoshida, R. Aki, T. Tanaka, T. Kanai, T. Imanaka, and S. Fujiwara. 2008. Expression profiles and physiological roles of two types of prefoldins from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Mol. Biol. 382:298-311. [DOI] [PubMed] [Google Scholar]
- 5.Endoh, T., T. Kanai, and T. Imanaka. 2007. A highly productive system for cell-free protein synthesis using a lysate of the hyperthermophilic archaeon, Thermococcus kodakaraensis. Appl. Microbiol. Biotechnol. 74:1153-1161. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, J. W., and C. A. Gross. 1989. Identification of the σ E subunit of Escherichia coli RNA polymerase: a second alternate σ factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 7.Gualerzi, C. O., A. M. Giuliodori, and C. L. Pon. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527-539. [DOI] [PubMed] [Google Scholar]
- 8.Hiromoto, T., H. Matsue, M. Yoshida, T. Tanaka, H. Higashibata, K. Hosokawa, H. Yamaguchi, and S. Fujiwara. 2006. Characterization of MobR, the 3-hydroxybenzoate-responsive transcriptional regulator for the 3-hydroxybenzoate hydroxylase gene of Comamonas testosteroni KH122-3s. J. Mol. Biol. 364:863-877. [DOI] [PubMed] [Google Scholar]
- 9.Izumi, M., S. Fujiwara, M. Takagi, K. Fukui, and T. Imanaka. 2001. Two kinds of archaeal chaperonin with different temperature dependency from a hyperthermophile. Biochem. Biophys. Res. Commun. 280:581-587. [DOI] [PubMed] [Google Scholar]
- 10.Izumi, M., S. Fujiwara, M. Takagi, S. Kanaya, and T. Imanaka. 1999. Isolation and characterization of a second subunit of molecular chaperonin from Pyrococcus kodakaraensis KOD1: analysis of an ATPase-deficient mutant enzyme. Appl. Environ. Microbiol. 65:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon, S.-J., S. Fujiwara, M. Takagi, and T. Imanaka. 1999. Pk-cdcA, CDC48/VCP homologue from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1: transcriptional and enzymatic characterizations. Mol. Gen. Genet. 262:559-567. [DOI] [PubMed] [Google Scholar]
- 12.Kallipolitis, B. H., and P. Valentin-Hansen. 1998. Transcription of rpoH, encoding the Escherichia coli heat-shock regulator σ32, is negatively controlled by the cAMP-CRP/CytR nucleoprotein complex. Mol. Microbiol. 29:1091-1099. [DOI] [PubMed] [Google Scholar]
- 13.Lindquist, S., and E. A. Craig. 1988. The heat-shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 14.Lund, P. A., A. T. Large, and G. Kapatai. 2003. The chaperonins: perspectives from the Archaea. Biochem. Soc. Trans. 31:681-685. [DOI] [PubMed] [Google Scholar]
- 15.Lundgren, M., and R. Bernander. 2005. Archaeal cell cycle progress. Curr. Opin. Microbiol. 8:662-668. [DOI] [PubMed] [Google Scholar]
- 16.Lundgren, M., and R. Bernander. 2007. Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. USA 104:2939-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelham, H. R. 1982. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30:517-528. [DOI] [PubMed] [Google Scholar]
- 19.Robinson, N. P., I. Dionne, M. Lundgren, V. L. Marsh, R. Bernander, and S. D. Bell. 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116:25-38. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang, Y. C., H. C. Chang, A. Roeben, D. Wischnewski, N. Wischnewski, M. J. Kerner, F. U. Hartl, and M. Hayer-Hartl. 2006. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125:903-914. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, D. K., and C. J. Daniels. 1998. Heat shock inducibility of an archaeal TATA-like promoter is controlled by adjacent sequence elements. Mol. Microbiol. 27:541-551. [DOI] [PubMed] [Google Scholar]
- 25.Vierke, G., A. Engelmann, C. Hebbeln, and M. Thomm. 2003. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 278:18-26. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Q. P., and J. M. Kaguni. 1989. dnaA protein regulates transcriptions of the rpoH gene of Escherichia coli. J. Biol. Chem. 264:7338-7344. [PubMed] [Google Scholar]
- 27.Wang, Q. P., and J. M. Kaguni. 1989. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J. Bacteriol. 171:4248-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan, Z., S. Fujiwara, K. Kohda, M. Takagi, and T. Imanaka. 1997. In vitro stabilization and in vivo solubilization of foreign proteins by the beta subunit of a chaperonin from the hyperthermophilic archaeon Pyrococcus sp. strain KOD1. Appl. Environ. Microbiol. 63:785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota, S., H. Yanagi, T. Yura, and H. Kubota. 2001. Cytosolic chaperonin-containing t-complex polypeptide 1 changes the content of a particular subunit species concomitant with substrate binding and folding activities during the cell cycle. Eur. J. Biochem. 268:4664-4673. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, T., T. Kanzaki, R. Iizuka, T. Komada, T. Zako, R. Suzuki, T. Maruyama, and M. Yohda. 2006. Contribution of the C-terminal region to the thermostability of the archaeal group II chaperonin from Thermococcus sp. strain KS-1. Extremophiles 10:451-459. [DOI] [PubMed] [Google Scholar]
- 31.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:153-158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.