Abstract
We have observed that a soluble recombinant green fluorescent protein produced in Escherichia coli occurs in a wide conformational spectrum. This results in differently fluorescent protein fractions in which morphologically diverse soluble aggregates abound. Therefore, the functional quality of soluble versions of aggregation-prone recombinant proteins is defined statistically rather than by the prevalence of a canonical native structure.
The quality of recombinant proteins produced in bacteria and other host cells represents a major matter of concern in biological protein production and determines the potential use of target proteins for functional applications or structural analysis (2, 17). In contrast to what was formerly believed, the straightforward measurement of the soluble protein yield or the ratio between the soluble and total protein yields (usually given as a percentage of solubility) is not a useful indicator of quality. Properly folded and functional polypeptides often aggregate as inclusion bodies; therefore, an important fraction of functional protein species occurs in the insoluble cell fraction (7, 9, 18). Thus, the specific biological activity rather than the presence of the protein species in the soluble cell fraction reveals the conformational quality of the product and, therefore, its biotechnological potential. In this regard, an increasing number of structural analyses reveal the coexistence, in the embedded protein species, of a cross-β-sheet-based, amyloid-like organization (3) and also that of a native secondary structure (5). In inclusion bodies formed by enzymes, the associated enzymatic activity is sufficient for efficient in situ substrate processing. The proposal of biologically active inclusion bodies being usable as catalyzers (10) has resulted in the incorporation of diverse enzymes, in the form of inclusion bodies (including β-galactosidase, d-amino acid oxidase, maltodextrin phosphorylase, sialic acid aldolase, and polyphosphate kinase) (6, 11-15), into different types of enzymatic processes.
The molecular organization and quality of the soluble protein population, which is generally believed to adopt the native, functional conformation, have been studied much less. Therefore, in this conventional view, soluble proteins are expected to show a rather narrow conformational spectrum and to be highly functional. However, the recent finding that the solubility and conformational quality in recombinant bacteria are divergently controlled (8) seriously challenges such an assumption. Moreover, several independent observations clearly argue against a model picturing the soluble protein fraction as fully functional and structurally homogeneous. First, the specific activity of a recombinant β-galactosidase aggregated as inclusion bodies is, under defined production conditions, higher than that of its soluble counterpart (7). This strongly suggests that the activity of soluble protein species represents an average of the numbers of coexisting active and inactive protein forms. In the same context, reducing the growth temperature of recombinant Escherichia coli cells from 37°C to 16°C results in a significant increase of the specific emission of a recombinant green fluorescent protein (GFP) (19), indicating that at 37°C an important fraction of soluble species have not maturated to an optimal conformation for fluorescence emission. Finally, the finding of the so-called soluble aggregates, which are fibril-like structures in E. coli cells overproducing GFP variants (4), indicates that even soluble species can display an amyloidal organization, such as that found in inclusion bodies, and that GFP in these soluble aggregates may eventually fold into a form very different from the native conformation. Aggregation of soluble versions of other structurally diverse proteins, including β-galactosidase (1) and different maltose-binding fusion proteins (16), has also been reported. The functional properties of such soluble aggregates remain unexplored. Therefore, the meaning of “solubility” in both structural and functional terms is as yet essentially obscure.
To clarify the folding scenery of the soluble protein population in recombinant bacteria and the biological significance of solubility, we have examined (through fluorescence emission) the conformational quality in the soluble fraction of recombinant E. coli MC4100 cells producing a model GFP (mGFP) by conventional culture and induction of gene expression procedures (8). mGFP is a GFP fused to the VP1 capsid protein of foot-and-mouth disease virus that drives protein aggregation immediately upon production in E. coli. Consequently, most of the fusion protein is found in the form of inclusion bodies and only up to 45% of the total mGFP yield occurs in the soluble cell fraction (8).
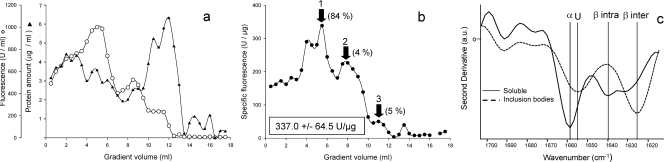
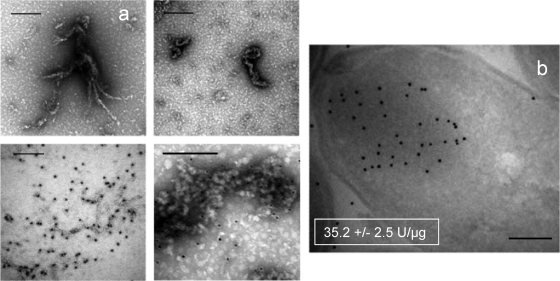
Cell pellets were obtained by centrifugation of bacterial cultures producing mGFP at 6,000 × g for 30 min at 4°C, frozen overnight at −80°C, and resuspended in lysis buffer (50 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA [pH 8]). Ice-jacketed samples were sonicated for 35 min at 50 W in cycles of 0.5 s. The soluble cell fraction was separated from cell debris and inclusion bodies by centrifugation at 15,000 × g for 15 min at 4°C and was submitted to density gradient ultracentrifugation (ranging from 0 to 80% sucrose) at 92,444 × g for 16 h at 4°C. Recombinant GFP widely dispersed along the gradient, showing a profile that was not coincident with that of the total fluorescence (Fig. 1a). In contrast to what would be expected for a monodispersal conformational model, such a lack of matching indicates a variable functional status within the soluble protein population. This functional heterogeneity was further confirmed by Western blot quantitative analysis of GFP as described previously (8) to determine the specific fluorescence, which peaked as three independent protein fractions (Fig. 1b). The GFP population enclosed in peak number 1 was further purified by size exclusion chromatography using fast protein liquid chromatography equipment, resulting in a highly (84%) pure and fluorescent population. Therefore, according to the conventional concept of soluble protein characteristics, the protein species found therein would be conformationally homogeneous and functional, having reached the native conformation. However, when we analyzed this particular population by transmission electron microscopy we observed a spectrum of soluble aggregates ranging from particulate material to fibril-like structures, with both categories demonstrating immunoreactivity with anti-GFP antibodies (Fig. 2a) as inclusion bodies themselves (Fig. 2b). Moreover, the Fourier transform infrared spectroscopy (FTIR) pattern of these protein species was compared with that of GFP inclusion bodies. The insoluble aggregates were observed to be rich in intermolecular β-sheets, with a minor occurrence of native-like α-helix and/or unfolded stretches (Fig. 1c). However, the soluble GFP did show a wider set of conformational types in which α-helix and intramolecular native-like β-sheets were prominent. The significant occurrence of these protein forms, indicative of native conformation, accounts for the higher specific emission level found in the soluble protein species within peak 1 (and also peak 2; Fig. 1b) compared with that of inclusion bodies (shown in the inset of Fig. 2b). In addition, such native-like folding patterns in soluble material were accompanied by unfolded and intermolecular β-sheet species. The smoother peaks in the FTIR plot of soluble protein (resulting from the contribution of several bands between 1,650 and 1,630 cm−1) compared to those of the more defined inclusion body FTIR spectra and the higher number of structural patterns again stressed the high structural heterogeneity of soluble protein species. Polypeptides aggregated as inclusion bodies (Fig. 2b) would then represent a particular and narrow subpopulation among the total recombinant protein species which, upon clustering through intermolecular interactions, deposit as insoluble material. Still being conformationally diverse, they would be significantly more homogeneous regarding their folding state than their soluble counterparts.
FIG. 1.
(a) Distribution of the soluble version of the aggregation-prone mGFP (triangles) and fluorescence emission (circles) along a sucrose gradient. (b) Specific fluorescence emission of soluble GFP populations. Arrows indicate discrete peaks of highly fluorescent protein subfractions (with GFP purity values given as percentages), and the inset shows values for the average specific fluorescence of soluble GFP in the whole soluble cell fraction. (c) FTIR spectra (normalized at the tyrosine peak by use of GRAMS/AI spectroscopic software, version 7) of isolated GFP inclusion bodies (dashed line) and the purified GFP population (from peak 1) (continuous line). Vertical lines indicate the relevant peaks corresponding to α-helix (α), unfolded stretches (U), native intramolecular β-sheet (β intra), and intermolecular cross β-sheet (β inter).
FIG. 2.
(a) Transmission electron microscopy analysis of fast protein liquid chromatography-purified GFP from peak 1, showing fibril-like and particulate microaggregates (top). At the bottom, the results of immunodetection of GFP on the same samples are shown. (b) Immunolabeling of GFP in inclusion bodies, on cryosections of GFP-producing E. coli cells. In the inset figures, the specific fluorescence values of GFP determined on isolated inclusion bodies are indicated. Bars, 0.2 μm.
In summary, the soluble fraction of a recombinant GFP produced in E. coli is composed of diverse protein populations with distinct specific fluorescence characteristics, with the most pure and fluorescent subfraction (peak 1) still being formed by a spectrum of protein forms and microaggregates. All those protein species, and the less fluorescent variants of peaks 2 and 3, generate, as a set, an average specific emission that defines the quality of the recombinant protein in the soluble cell fraction (note the average specific fluorescence of soluble GFP given in the inset of Fig. 1b). The protein “quality” of model mGFP, and presumably of other recombinant polypeptides, could then be statistically observed as the relative abundance of the most active protein species, which would be closer to the canonical native conformation. Adjusting either the production conditions or the genetic composition of the cell quality control system (either by knocking down or by overexpressing chaperone or protease genes) would alter protein quality by unbalancing the prevalence of active (native or native-like) and inactive (misfolded) protein species in the cell context.
Acknowledgments
We appreciate the financial support to our research on recombinant protein production through grants BIO2007-61194 and BIO2005-23732-E (MEC) and grant 2005SGR-00956 (AGAUR). We are also grateful for the financial support provided by the CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN, promoted by ISCIII), Spain. M.M.-A., N.G.-M., and E.G.-F. are recipients of fellowships from MEC, Spain.
We thank Salvador Bartolomé and Alejandro Sànchez Chardi for helpful technical assistance.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Aris, A., and A. Villaverde. 2000. Molecular organization of protein-DNA complexes for cell-targeted DNA delivery. Biochem. Biophys. Res. Commun. 278:455-461. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx, F., and M. Mujacic. 2004. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22:1399-1408. [DOI] [PubMed] [Google Scholar]
- 3.Carrio, M., N. Gonzalez-Montalban, A. Vera, A. Villaverde, and S. Ventura. 2005. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 347:1025-1037. [DOI] [PubMed] [Google Scholar]
- 4.de Marco, A., and A. Schroedel. 2005. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doglia, S. M., D. Ami, A. Natalello, P. Gatti-Lafranconi, and M. Lotti. 2008. Fourier transform infrared spectroscopy analysis of the conformational quality of recombinant proteins within inclusion bodies. Biotechnol. J. 3:193-201. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Fruitos, E., A. Aris, and A. Villaverde. 2007. Localization of functional polypeptides in bacterial inclusion bodies. Appl. Environ. Microbiol. 73:289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Fruitos, E., N. Gonzalez-Montalban, M. Morell, A. Vera, R. M. Ferraz, A. Aris, S. Ventura, and A. Villaverde. 2005. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Fact. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Fruitos, E., M. Martinez-Alonso, N. Gonzalez-Montalban, M. Valli, D. Mattanovich, and A. Villaverde. 2007. Divergent genetic control of protein solubility and conformational quality in Escherichia coli. J. Mol. Biol. 374:195-205. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Montalban, N., E. Garcia-Fruitos, S. Ventura, A. Aris, and A. Villaverde. 2006. The chaperone DnaK controls the fractioning of functional protein between soluble and insoluble cell fractions in inclusion body-forming cells. Microb. Cell Fact. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Montalban, N., E. Garcia-Fruitos, and A. Villaverde. 2007. Recombinant protein solubility—does more mean better? Nat. Biotechnol. 25:718-720. [DOI] [PubMed] [Google Scholar]
- 11.Nahálka, J. 2008. Physiological aggregation of maltodextrin phosphorylase from Pyrococcus furiosus and its application in a process of batch starch degradation to α-d-glucose-1-phosphate. J. Ind. Microbiol. Biotechnol. 35:219-223. [DOI] [PubMed] [Google Scholar]
- 12.Nahalka, J., I. Dib, and B. Nidetzky. 2008. Encapsulation of Trigonopsis variabilis d-amino acid oxidase and fast comparison of the operational stabilities of free and immobilized preparations of the enzyme. Biotechnol. Bioeng. 99:251-260. [DOI] [PubMed] [Google Scholar]
- 13.Nahálka, J., P. Gemeiner, M. Bucko, and P. G. Wang. 2006. Bioenergy beads: a tool for regeneration of ATP/NTP in biocatalytic synthesis. Artif. Cells Blood Substit. Immobil. Biotechnol. 34:515-521. [DOI] [PubMed] [Google Scholar]
- 14.Nahálka, J., A. Vikartovska, and E. Hrabarova. 2008. A crosslinked inclusion body process for sialic acid synthesis. J. Biotechnol. 134:146-153. [DOI] [PubMed] [Google Scholar]
- 15.Navrátil, M., P. Gemeiner, J. Klein, E. Sturdík, A. Malovíková, J. Nahálka, A. Vikartovská, Z. Dömény, and D. Smogrovicová. 2002. Properties of hydrogel materials used for entrapment of microbial cells in production of fermented beverages. Artif. Cells Blood Substit. Immobil. Biotechnol. 30:199-218. [DOI] [PubMed] [Google Scholar]
- 16.Sachdev, D., and J. M. Chirgwin. 1999. Properties of soluble fusions between mammalian aspartic proteinases and bacterial maltose-binding protein. J. Protein Chem. 18:127-136. [DOI] [PubMed] [Google Scholar]
- 17.Sørensen, H. P., and K. K. Mortensen. 2005. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura, S., and A. Villaverde. 2006. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 24:179-185. [DOI] [PubMed] [Google Scholar]
- 19.Vera, A., N. Gonzalez-Montalban, A. Aris, and A. Villaverde. 2007. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol. Bioeng. 96:1101-1106. [DOI] [PubMed] [Google Scholar]