Abstract
Genomic data combined with reverse genetic approaches have contributed to the characterization of major virulence factors of Vibrio species; however, these studies have targeted primarily human pathogens. Here, we investigate virulence factors in the oyster pathogen Vibrio splendidus LGP32 and show that toxicity is correlated to the presence of a metalloprotease and its corresponding vsm gene. Comparative genomics showed that an avirulent strain closely related to LGP32 lacked the metalloprotease. The toxicity of LGP32 metalloprotease was confirmed by exposing mollusk and mouse fibroblastic cell lines to extracellular products (ECPs) of the wild type (wt) and a vsm deletion mutant (Δvsm mutant). The ECPs of the wt induced a strong cytopathic effect whose severity was cell type dependent, while those of the Δvsm mutant were much less toxic, and exposure to purified protein demonstrated the direct toxicity of the Vsm metalloprotease. Finally, to investigate Vsm molecular targets, a proteomic analysis of the ECPs of both LGP32 and the Δvsm mutant was performed, revealing a number of differentially expressed and/or processed proteins. One of these, the VSA1062 metalloprotease, was found to have significant identity to the immune inhibitor A precursor, a virulence factor of Bacillus thuringiensis. Deletion mutants corresponding to several of the major proteins were constructed by allelic exchange, and the ECPs of these mutants proved to be toxic to both cell cultures and animals. Taken together, these data demonstrate that Vsm is the major toxicity factor in the ECPs of V. splendidus.
Vibrionaceae are a predominant family of gram-negative bacteria found in aquatic environments (30). Bacteria within this family demonstrate a high degree of genetic diversity and are able to colonize very different types of niches. They live freely as planktonic forms in the water column or are associated in biofilms or with host organisms as pathogenic, commensal, or mutualistic bacteria. To date, eight genome sequences from Vibrionaceae have been made available: those of Vibrio cholerae strains N16961 and 0395, V. parahaemolyticus RIMD2210633, V. vulnificus strains YJ016 and CMCP6, V. fischeri ES114, V. harveyi ATCC BAA-1116, and Photobacterium profundum SS9 (3, 10, 19, 24, 32). More recently, we have completed the sequencing of the genome of V. splendidus strain LGP32, an oyster (Crassostrea gigas) pathogen (F. Le Roux, M. Zouine, N. Chakroun, J. Binesse, D. Saulnier, C. Goarant, C. Bouchier, N. Zidane, L. Ma, C. Rusniok, C. Buchrieser, C. Médigue, M. F. Polz, and D. Mazel, presented at the Second Conference on the Biology of Vibrios, Paris, France, 28 November to 1 December 2007).
Genomics data combined with reverse genetics have largely contributed to the characterization of the main virulence factors in human pathogens: e.g., cholera toxin (CT) and the toxin-coregulated pilus in V. cholerae, the thermostable direct thermolysin and the type III secretion system in V. parahaemolyticus, and the capsular polysaccharide in V. vulnificus (for a comprehensive review, see The Biology of Vibrios [29]). However, epidemiological studies have clearly established that virulence is most often multifactorial. For instance, although CT is the most important factor in the enteric disease cholera, CT-deficient strains of V. cholerae still elicit mild to severe diarrhea in humans, indicating that other factors are likely to contribute to the pathogenesis of the disease (5, 11, 27, 28). Furthermore, the relative contribution of a given gene product may vary with the strain, host, and type of infection. In such a scheme, accessory virulence factors such as hemolysins, lipases, and proteases may facilitate virulence.
Extracellular metalloproteases have a number of properties that suggest their involvement in pathogenesis, including their cytotoxicity, tissue-destructive activities, and inhibitory effect on phagocytosis (9). Proteases may similarly interact with other virulence factors to potentiate their expression and/or effects on the host. For instance, the hemagglutinin/protease of V. cholerae cleaves and activates the cholera enterotoxin (2), and it has been shown previously that the enterotoxic hemolysin is extracellularly processed into mature hemolysin after cleavage by various proteolytic enzymes, including a metalloprotease (22). Recently, we have shown that Vsm, a metalloprotease of V. splendidus strain LGP32, is an essential determinant of lethality when extracellular products (ECPs) are injected into oysters (7, 15).
Like several other Vibrio species, V. splendidus has been associated with mortalities of marine invertebrates, causing major economic losses in aquaculture (for a review, see reference 14). Compared to those of human pathogens, the mechanisms of the strains involved in such invertebrate vibriosis are poorly understood. Such a lack of knowledge is in part a consequence of the absence of a standardized model for in vivo studies. Indeed, with no inbred mollusk lines, the genetic background of the experimental animals is heterogeneous. Because of these difficulties, ex vivo studies have become necessary to better characterize the several bacterial activities that appear to be involved in V. splendidus virulence. Virulence caused by factors such as the extracellular enzymes and stress proteins can be recognized by the loss of cell adherence, the induction of cell lysis, or apoptosis. A clam primary cell culture, despite consisting of a heterogeneous cell population, has been used previously to study the interaction between cells and a pathogenic Vibrio species (4, 13). Indeed, Labreuche et al. (12) showed that the level of lethality in animals treated with the ECPs of another Vibrio species, V. aestuarianus, was correlated ex vivo to the significant inhibition of the phagocytic capacity of hemocytes, as well as their ability to adhere to the substrate.
In the present study, the ECPs of LGP32, a virulent strain of V. splendidus, were investigated to identify the products responsible for the toxicity of this strain. We first showed that the absence of toxicity of an avirulent V. tasmaniensis strain closely related to LGP32, LMG20012T, toward oysters correlated with the absence of both a metalloprotease activity (designated Vsm) and its encoding gene. The toxicity of the ECPs of LGP32 and of a Δvsm strain, a metalloprotease deletion mutant of LGP32, and that of the Vsm purified protein toward whole oysters, Bge cells (a mollusk cell line), and NIH 3T3 cells (a mouse fibroblastic cell line) were tested. Our data show that the ECPs of LGP32 induced a strong cytopathic effect whose severity was cell type dependent. Indeed, the purified Vsm protein was sufficient to induce in vivo and ex vivo toxicity, while the ECPs of the Δvsm strain were much less toxic. In order to identify a Vsm target protein(s) in the ECPs, proteomic analyses of the ECPs of both LGP32 and the Δvsm mutant were performed. Interestingly, these studies revealed a number of differentially expressed and/or processed proteins, among which the VSA1062 metalloprotease was found to have significant identity to the immune inhibitor A precursor, a virulence factor of Bacillus thuringiensis. New deletion mutants corresponding to three differentially expressed and/or processed extracellular proteins, the VSA576, VS2864, and VSA1062 proteins, were constructed by allelic exchange. The ECPs of these mutants proved to be toxic both in cell culture and in animals, showing that none of the deleted genes was essential for the toxicity of LGP32 ECPs injected into oysters. Altogether, our data demonstrate that Vsm is the major determinant of toxicity in the ECPs of V. splendidus.
MATERIALS AND METHODS
Bacterial cultures.
The bacterial strains used in this study and their relevant properties are described in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) broth or, for strain π3813, Mueller-Hinton broth at 37°C. Vibrio strains were grown in LB broth-0.5 M NaCl or marine broth or on marine agar at 20°C. All media were from Difco. Chloramphenicol (12.5 mg liter−1), thymidine (0.3 mM), and diaminopimelate (0.3 mM) were added as supplements when necessary. The induction of ccdB expression under the control of the PBAD promoter was achieved by the addition of 0.2% l-arabinose to the growth medium, and conversely, this expression was repressed by the addition of 1% d-glucose.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Bacterial strains | ||
| π3813 | lacIqthi-1 supE44 endA1 recA1 hsdR17 gyrA462 zei298::Tn10 ΔthyA::(erm-pir-116) (Tcr Ermr) | 15 |
| β3914 | (F−) RP4-2-Tc::Mu ΔdapA::(erm-pir) gyrA462 zei-298::Tn10 (Kmr Emr Tcr) | 15 |
| LGP32 | V. splendidus | 6 |
| LMG20012T | V. tasmaniensis | 31 |
| Δvsm mutant | LGP32 Δvsm (M4 metalloprotease) | 15 |
| Δ1062 mutant | LGP32 Δ1062 (M6 metalloprotease) | This study |
| Δ2864 mutant | LGP32 Δ2864 | This study |
| Δ576 mutant | LGP32 Δ576 (unknown function) | This study |
| Δvsm-1062 mutant | LGP32 Δvsm Δ1062 | This study |
| Plasmids | ||
| pSW4426T | oriVR6KγoriTRP4araC-PBAD-ccdB (Spr Cmr) | 15 |
| pSWΔ1062T | pSW4426T Δ1062 (Spr Cmr) | This study |
| pSWΔ2864T | pSW4426T Δ2864 (Spr Cmr) | This study |
| pSWΔ576T | pSW4426T Δ576 (Spr Cmr) | This study |
PCR amplification.
PCR amplifications for plasmid construction were performed in 50-μl reaction mixtures containing Pfu DNA polymerase (Promega). For long-range PCR, we used the Herculase II fusion enzyme (Stratagene). Routine PCRs were performed in 50-μl reaction mixtures using the Bioline Taq polymerase according to the manufacturer's instructions. Conditions for amplification were as follows: 94°C for 3 min and 30 cycles of 94°C for 30 s, 10°C less than the melting temperature for 30 s, and 72°C for 60 s per kb.
Gene annotation and comparative genomics.
The whole genome sequence of V. splendidus LGP32, like the genome sequences of V. cholerae, V. parahaemolyticus, V. vulnificus, and V. fischeri, has been transferred into the annotation database MaGe (Magnifying Genomes), which allows dynamic and permanent updating of the genome annotation (http://www.genoscope.cns.fr/agc/mage). The MaGe system offers a set of graphical interfaces that allow relevant expert annotation of microbial genomes to be performed. One of the easily accessed features of this system is the ability to explore gene context by searching for conserved synteny. Putative orthologous relationships between two genomes are defined by the presence of gene couples satisfying the bidirectional best-hit criterion or an alignment threshold (a minimum of 30% sequence identity over 80% of the length of the smaller protein). These relationships are subsequently used to search for gene clusters, e.g., synteny groups, conserved among several bacterial genomes. From these comparisons, we defined a species-specific gene as a gene having no ortholog in the compared species (significant similarities were not detected).
Suicide vector construction.
Alleles carrying an internal deletion were generated in vitro using a two-step PCR construction method (20). Using genomic DNA of the strain LGP32, independent PCR amplifications of the regions encompassing the VSA1062, VS2864, and VSA576 open reading frames (ORFs) were performed using two primer pairs per ORF: 1062-1 and 1062-2 (yielding a 457-bp product) and 1062-3 and 1062-4 (yielding a 447-bp product), 2864-1 and 2864-2 (generating a 341-bp product) and 2864-3 and 2864-4 (giving a 341-bp product), and 576-1 and 576-2 (generating a 500-bp product) and 576-3 and 576-4 (resulting in a 465-bp product). Primer sequences are listed in Table S1 in the supplemental material. After gel purification, 100-ng aliquots of the two PCR products from each ORF were mixed, and a final PCR amplification was carried out using the more external primer pair: 1062-1 and 1062-4, 2864-1 and 2864-4, and 576-1 and 576-4. After gel purification, the PCR products, referred to as the Δ1062, Δ2864, and Δ576 alleles, were EcoRI digested and ligated into an EcoRI-linearized pSW4426T plasmid described previously (15).
Conjugation.
We have previously developed a suicide vector based on the pir-dependent R6K replicative origin, which can be transferred by RP4-based conjugation into V. splendidus and which also carries the plasmid F toxin gene (ccdB) under the control of the PBAD promoter (15). This genetic system allows the efficient counterselection of integrated plasmids in V. splendidus in the presence of arabinose. This approach thus permits efficient markerless allelic replacement in this species. The different conjugation experiments were performed according to the filter mating procedure using a donor/recipient ratio of 1/10 as described previously (1). Selection against the ΔdapA donor was achieved by plating onto a medium that was devoid of diaminopimelic acid and supplemented with 1% glucose and chloramphenicol. Antibiotic-resistant colonies were grown in LB broth-NaCl and spread onto plates containing 0.2% arabinose. Mutants were screened by PCR amplification as described previously (15) using the primer pairs 1062-5 and 1062-6, 2864-5 and 2864-6, and 576-5 and 576-6.
Preparation of the ECPs.
Bacterial ECPs were obtained by the cellophane overlay method as described by Liu (16). Briefly, 500 μl of an overnight culture of bacteria was spread onto a marine agar plate covered by a cellophane film. After 24 h of incubation at 20°C, bacteria were collected using 1 ml of cold 1× phosphate-buffered saline. After centrifugation for 10 min at 4,000 × g, the supernatant (ECPs) was passed through a 0.22-μm-pore-size filter. The protein concentrations of ECPs (ranging from 100 to 500 mg liter−1) were estimated using the Bradford method with Coomassie protein assay reagent according to the instructions of the manufacturer (Pierce Biotechnology, Rockford, IL). Proteins were separated on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) gel and were stained using Coomassie blue (Bio-Rad).
Protease activity measurement.
Protease activity was determined using azocasein (Sigma) as a substrate. Briefly, crude ECPs (250 μl) were added to 250 μl of azocasein (5 mg ml−1 in 50 mM Tris-HCl buffer, pH 8.0). The mixture was incubated at 20°C for 1 h. The undigested substrate was precipitated by adding 500 μl of 10% trichloroacetic acid to the reaction mixture and centrifuging at 12,000 × g and 4°C for 5 min. Supernatant (500 μl) was neutralized by the addition of an equal volume of 1 N NaOH. After mixing, the absorbance at 440 nm of triplicate samples was measured.
Purification of the metalloprotease.
ECPs of LGP32 were concentrated five times at 4°C by ultrafiltration (using a stirred cell system with a 10-kDa molecular mass cutoff; Pall). Fifteen milliliters of concentrated ECPs was applied to a Hiload 26/60 Superdex 75-pg column (GE Healthcare) equilibrated in 50 mM Tris-HCl, pH 7.5. Fractions of 3.5 ml were collected at a flow rate of 0.5 ml/min. Fractions containing Vsm were checked on an SDS-10% PAGE gel, quantified by the Bradford method, and tested for their protease activity as described previously.
Protein identification by two-dimensional (2D) electrophoresis and peptide mass fingerprinting.
The separation of extracellular proteins was performed as described previously (26); briefly, 350 μg of protein was loaded onto each strip. Isoelectric focusing was carried out using immobilized pH gradients (pH 4 to 7; Bio-Rad Laboratories, Hercules, CA) in a Protean II isoelectric focusing cell (Bio-Rad). Strips were subsequently loaded onto SDS-10% PAGE gels, and electrophoresis was performed in a Protean II xi cell (Bio-Rad). Experiments were conducted in triplicate. Proteins were visualized by staining with Bio-Safe Coomassie blue (Bio-Rad) and identified by peptide mass fingerprinting after trypsinolysis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) at the Plateau d'Analyses Protéomiques par Séquençcage et Spectrométrie de Masse (INRA, Jouy-en-Josas, France). Protein identification was achieved using the MS-Fit software (University of California—San Francisco Mass Spectrometry Facility; http://prospector.ucsf.edu). All searches were performed against the database of the V. splendidus LGP32 genome sequence (Le Roux et al., presented at the Second Conference on the Biology of Vibrios, Paris, France, 28 November to 1 December 2007).
Virulence assays.
Bacteria were grown under constant agitation at 20°C for 36 h in marine broth, harvested, and resuspended in sterile seawater (121°C for 15 min) at an optical density at 600 nm of 1, which corresponded to bacterial concentrations ranging from 109 to 2 × 109 CFU ml−1 as determined by conventional dilution plating on marine agar (data not shown). Oysters were intramuscularly injected with bacterial strains or ECPs as described previously (6, 12). After injection, the oysters were transferred into aquaria (15 to 20 oysters per aquarium of 2.5 liters) containing 20°C aerated seawater passed through a 5-μm-pore-size filter and were kept under static conditions and fed daily with a mixture of planktonic algae (Isochrysis galbana and Chaetoceros calcitrans). Each treatment was performed in duplicate, and mortality was recorded daily.
Cell culture and microscopy.
Bge cells from the tropical snail Biomphalaria glabrata (ATCC CRL-1494) were grown at 26°C on 22% Schneider medium complemented with lactalbumin hydrolysate (4.5 g liter−1), d-galactose (1.3 g liter−1), and 10% heat-inactivated fetal calf serum. NIH 3T3 cells were maintained at 37°C in a CO2 atmosphere on Dulbecco's modified Eagle medium with 10% heat-inactivated fetal calf serum. All cell culture reagents were from Invitrogen.
For microscopy, cells were seeded onto coverslips. Fixation was carried out with 3.7% paraformaldehyde in PHEM buffer (for 1 liter, add 18.14 g of PIPES, 6.5 g of HEPES, 3.8 g of EGTA, and 0.99 g of MgSO4 and adjust the pH to 7.0 with 10 M KOH). The observation of the formazan needle-like crystals present at the cell surface after MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] treatment was performed by diffraction interference contrast (DIC) microscopy. The immunostaining of the microtubule cytoskeleton was performed using a primary rabbit polyclonal anti-β-tubulin antibody and a Texas Red-conjugated secondary antibody (Molecular Probes). Actin staining was carried out using Texas Red-conjugated phalloidin. Cells were then mounted in Moviol mounting medium before analysis.
Cytotoxicity assay.
Nonconfluent cell cultures were obtained by seeding 65,000 Bge cells or 40,000 NIH cells in 300 μl of medium into each well of a 24-well plate and incubating the plates overnight under the respective culture conditions. ECP fractions with a final protein concentration of 10 μg ml−1 (amounts of which were normalized based on the protein content) were added to the cells and left in contact from 3 to 90 h before the addition of 33 μl of an MTT solution (5 mg ml−1) from the Vybrant MTT cell proliferation assay kit (Molecular Probes, The Netherlands). For quantitative assays, cells were further incubated for 4 h and were then lysed with a 0.5% SDS solution and incubated at 37°C for 16 h according to the recommendations of the assay kit manufacturer. The optical densities at 570 nm of triplicate samples were determined.
Nucleotide sequence accession numbers.
The nucleotide sequences of the VSA1062, VSA576, and VS2864 genes have been deposited in the GenBank database under the accession numbers EU349011, EU349012, and EU349013.
RESULTS
The avirulent LMG20012T vibrio is devoid of Vsm metalloprotease.
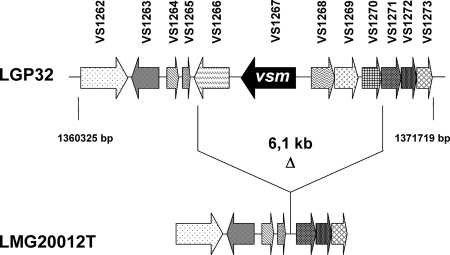
A preliminary screening to test for the virulence of live bacteria belonging to several strains closely related to V. splendidus indicated that the LMG20012T strain had essentially no virulence (i.e., bacterial injection gave results similar to those of the seawater treatment [data not shown]). Similarly, the injection of LGP32 and LMG20012T ECPs, instead of bacteria, into oysters (at 5 μg g−1 of flesh) resulted in 93.3 and 3.3% mortalities, respectively, by 3 days postinjection (Table 2). In addition, no protease activity could be detected in the LMG20012T ECPs by using the azocasein assay (Table 2), thus further suggesting a role for the Vsm metalloprotease in V. splendidus virulence (15). Indeed, PCR amplification of LMG20012T genomic DNA by using primers framing the vsm gene-containing region (11.4 kbp) produced a DNA fragment lacking a 6.1-kbp region that most notably carries vsm in V. splendidus LGP32 and four other ORFs encoding conserved proteins of unknown function (Fig. 1). Altogether, these results suggest that the absence of the vsm gene in strain LMG20012T and, thus, of its metalloprotease activity may partially explain the avirulence of this strain.
TABLE 2.
Oyster mortality 24 h postinjection with ECPs, purified Vsm protein, or sterile seawater and proteolytic activity
| Inoculum | Oyster mortality (%) (mean ± SD) | Proteolytic activity (protease units/mg of protein) |
|---|---|---|
| ECPs from: | ||
| LGP32 | 93.3 ± 9.4 | 50 |
| LMG20012T | 3.3 ± 4.7 | 0 |
| Vsm | 56.1 ± 13.3 | 25.6 |
| ECPs from: | ||
| Δvsm mutant | 16.7 ± 4.7 | 11 |
| Δ1062 mutant | 93.3 ± 9.4 | 46 |
| Δ2864 mutant | 86.7 | 52 |
| Δ576 mutant | 80 | 46 |
| Δvsm-1062 mutant | 3.3 ± 4.7 | 0 |
| Sterile seawater | 0 | 0 |
FIG. 1.
Genetic organization of the vsm gene region. The region flanking vsm, localized in chromosome 1 of V. splendidus LGP32, was PCR amplified in LMG20012T and sequenced. Corresponding genes found in both LMG20012T and LGP32 are represented by arrows with the same pattern. The absence in LMG20012T of a genomic region of 6.1 kb containing the vsm gene is indicated (Δ). ORFs were annotated as encoding the following products: VS1267, Vsm extracellular zinc metalloprotease; VS1262, tryptophanase; VS1263, putative helix-turn-helix-type transcriptional activator; VS1264, VS1265, VS1266, VS1269, VS1271, VS1272, and VS1273, conserved hypothetical proteins; VS1268 and VS1269, hypothetical proteins.
Vsm is lethal for oysters.
To purify the Vsm metalloprotease, the ECPs from LGP32 were subjected to gel filtration chromatography. The pool of fractions 39 to 41 produced a single band upon SDS-PAGE while displaying the highest protease activity (data not shown). As shown in Table 2, the injection of 10 μg of this purified Vsm protein per oyster induced 56% mortality within 24 h postchallenge, while the injection of LGP32 and Δvsm mutant ECPs induced 93 and 16% mortalities, respectively (Table 2). The fact that purified Vsm appeared to be slightly less toxic than crude LGP32 ECPs correlated with a decrease of the proteolytic activity of the purified Vsm (25 protease units/mg) compared to that of the LGP32 ECPs (50 protease units/mg) (Table 2), suggesting that the purification step altered the protease activity or that a cofactor, present in the crude ECPs, is required for full Vsm activity. Taken together, these results suggest that the metalloprotease Vsm is necessary and sufficient to induce oyster mortality.
Distinct proteins in the ECPs of the Δvsm strain and LGP32.
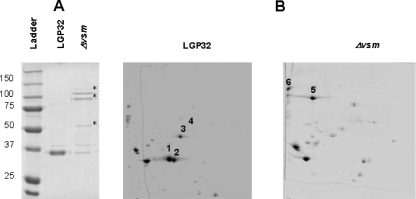
Because Vsm metalloprotease might act indirectly by cleaving another virulence factor(s) in the LGP32 ECPs, SDS-PAGE was carried out to compare the protein profiles of the ECPs of LGP32 and its previously described derivative Δvsm strain (15). SDS-PAGE analysis revealed a predominant band (approximately 35 to 37 kDa) in LGP32 ECPs that was missing in the ECPs of the Δvsm mutant (Fig. 2A). In addition, Δvsm mutant ECPs displayed two bands of weak intensity migrating at 100 kDa and one band at 55 kDa (Fig. 2A). 2D gel analyses of the ECPs of LGP32 and the Δvsm strain over a pI range from 4 to 7 revealed six major differential spots (Fig. 2B). MALDI-TOF MS analysis and peptide mass fingerprinting using the LGP32 database (Le Roux et al., presented at the Second Conference on the Biology of Vibrios, Paris, France, 28 November to 1 December 2007) allowed the identification of 56 of 126 spots with high matching scores (Table 2; also see the supplemental material). Only six spots were found to differ constantly between the ECPs of the two strains, and they corresponded to the Vsm metalloprotease (Fig. 2B, spots 1 to 3) and the VSA576 (Fig. 2B, spot 4), VSA1062 (Fig. 2B, spot 5), and VS2864 (Fig. 2B, spot 6) proteins. VSA576 and VS2864 ORFs encode predicted 73.7- and 94.6-kDa proteins of unknown function, with isoelectric points of 5.31 and 4.64, respectively. The VSA1062 gene encodes a 101-kDa metalloprotease that belongs to the M6 peptidase family, according to the MEROPS classification (http://merops.sanger.ac.uk). Interestingly, this protein shows significant identity (41%) to the immune inhibitor A precursor, a virulence factor encoded by the ina gene of B. thuringiensis (18). Thus, one of the proteins expressed in the Δvsm strain is a metalloprotease that may compensate for the missing Vsm.
FIG. 2.
ECP profiles for LGP32 and the Δvsm mutant. (A) ECP protein analysis on a Coomassie blue-stained SDS-10% PAGE gel. Bands corresponding to proteins differentially expressed and/or processed in the Δvsm mutant are indicated (*). (B) 2D analysis of the ECPs. The most significant differential spots are numbered 1 to 3 for Vsm, 4 for VSA576, 5 for VSA1062, and 6 for VS2864.
The differentially expressed proteins are not involved in virulence.
Deletion mutants corresponding to the differentially expressed and/or processed proteins, the VSA1062, VSA576, and VS2864 proteins, were obtained by a method we described previously (15). The metalloprotease activities of the ECPs from the Δ1062, Δ2864, and Δ576 mutants, determined using the azocasein assay, were found to be in the range of that of the wild-type strain ECPs (Table 2). In contrast, the ECPs of the Δvsm mutant displayed dramatically reduced protease activity (20% of the LGP32 ECP protease activity), while those of the Δvsm-1062 double mutant had essentially no activity. Twenty-four hours postinjection into the oyster adductor muscles (5 μg g−1 of flesh), the observed mortalities for the wild-type and the Δ1062 mutant ECPs were in the same range (∼93%). The mortalities for the Δ2864 and Δ576 mutant ECPs (86.7 and 80%, respectively) were slightly decreased compared to that for the wild type, while those for the Δvsm mutant ECPs (16.7%) and for the Δvsm-1062 double mutant ECPs (3.3%) were dramatically reduced (Table 2). In live oysters, the degree of experimental variation is high; therefore, we considered that a difference of 10% in the mortality rate was a minor or nonsignificant effect. In addition, and in agreement with the findings in our previous report (15), the mortality rates observed for the different mutants were the same as that observed for LGP32 when live bacteria instead of ECPs were injected into oysters (data not shown). The data demonstrate that these factors that are differentially expressed and/or processed in LGP32 and the Δvsm strain are not, or are only marginally, involved in virulence in the oyster. Moreover, these results suggest that Vsm toxicity is not mediated through these factors.
Virulent ECPs alter cell morphology.
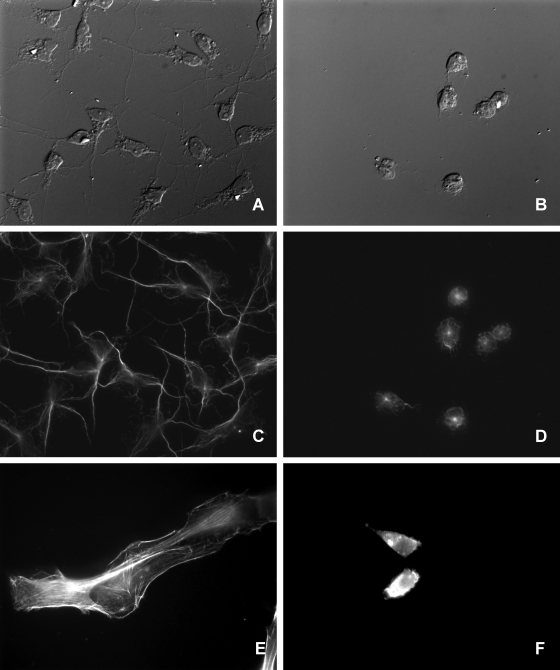
Nonconfluent Bge cells, a mollusk cell line, were exposed to ECPs at a final protein concentration of 10 μg ml−1. Observation by DIC microscopy revealed the retraction of cytoplasmic extensions and cell rounding within 3 h after ECP treatment, suggesting a cytopathic effect of LGP32 (Fig. 3A and B). Interestingly, this cytopathic effect was reproduced by using 10 μg of purified Vsm ml−1. In contrast, the ECPs of either LMG20012T or the Δvsm strain had no apparent effect, even after 24 or 48 h of exposure (data not shown). After 8 h, cells treated with LGP32 ECPs were floating mostly as groups of 15 to 20 closely associated rounded cells (Fig. 3B). The immunostaining of microtubules revealed strong alteration of the microtubule cytoskeleton (Fig. 3C and D), thus corroborating the morphology of the LGP32 ECP-treated cells. In contrast, cells incubated with LMG20012T or Δvsm strain ECPs did not display any alteration of their microtubule cytoskeletons (data not shown).
FIG. 3.
ECP alteration of cell morphology. Nonconfluent cell cultures were obtained by seeding coverslips with 65,000 Bge or 40,000 NIH cells in 300 μl of medium in 24-well plates and were grown overnight under the respective culture conditions. ECPs were added to the cells at a final protein concentration of 10 μg ml−1, and cells were further incubated for 6 h. (A and B) DIC microscopy images. ECP-treated Bge cells (B) showed cytoplasmic retraction and cell rounding compared to untreated Bge cells (A). (C to F) Fluorescence microscopy images. (C and D) The immunostaining of β-tubulin on Bge cells showed the retraction of the microtubule cytoskeletons of ECP-treated cells (D) compared to those of untreated cells (C). (E and F) NIH cells. (F) Actin labeling showed the disruption of the actin cytoskeletons of ECP-treated cells. (E) Untreated cells.
The experiment was then repeated with nonconfluent NIH 3T3 cells, a mouse fibroblast cell line. Cells were rounded as early as 1 h after treatment with the LGP32 ECPs or purified Vsm, while 2 h later, they were already floating in the culture medium (data not shown). Labeled phalloidin revealed intensely stained stress fibers in untreated NIH cells (Fig. 3E), while LGP32 ECP-treated cells displayed large speckles of aggregated actin in the retracted cytoplasm (Fig. 3F). Similar observations were made using the Δ1062, Δ576, and Δ2864 mutants, while the Δvsm and Δvsm-1062 strains did not induce any apparent modification of the cell morphology or actin cytoskeleton (data not shown). The ECPs of the virulent LGP32 vibrio, in contrast to those of the Δvsm mutant, induced changes in cell morphology and then detachment from the extracellular matrix, and these alterations occurred more rapidly and to a greater extent for mammalian NIH cells than for mollusk Bge cells. This activity has to be attributed to Vsm, as the purified protein is sufficient to induce cell rounding.
ECP alteration of viability is cell type dependent.
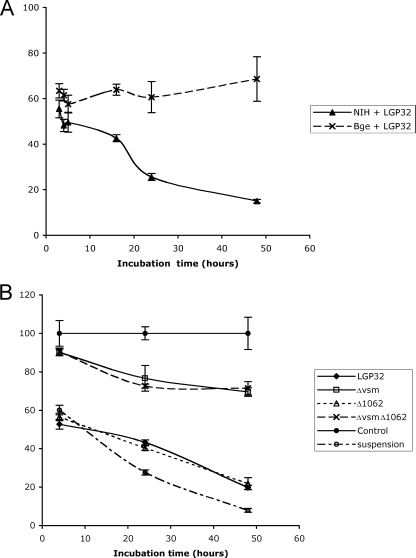
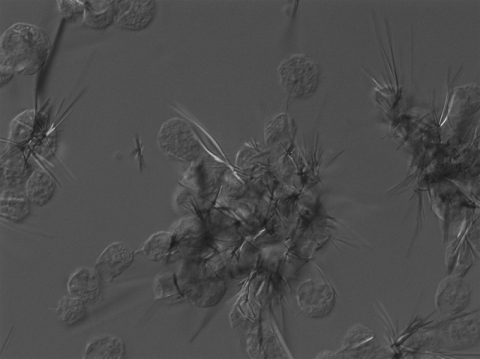
To determine whether the ECP treatment altered Bge cell physiology, cell viability was estimated using the MTT assay that requires MTT endocytosis, reduction and polymerization, and exocytosis as needle-like crystals (17). Indeed, this assay revealed a 27% decrease of the MTT reduction activity as early as 3 h after treatment with LGP32 ECPs (see Fig. 5A). Interestingly, kinetics showed no further alteration of the rate of MTT reduction when the LGP32 ECP treatment duration was increased to 48 h (see Fig. 5A). DIC microscopy analysis of ECP-treated cells revealed long and abundant needle-like crystals at the peripheries of Bge cells (Fig. 4), even when incubation with MTT was performed as late as 90 h post-ECP treatment (data not shown), a time when most cells were associated in groups and in suspension. When performed on NIH 3T3 cells, the MTT assay revealed a significant alteration of cell metabolism, with decreases of 45, 75, and 85% after exposure to LGP32 ECPs for 3, 24, and 48 h, respectively (Fig. 5A). In addition, the ECPs of the Δ576 and Δ2864 mutants (data not shown) and the Δ1062 mutant (Fig. 5B) were as cytotoxic for NIH cells as LGP32 ECPs, and the ECPs of the Δvsm strain, although devoid of Vsm protease, still induced a moderate degree of cytotoxicity (Fig. 5B). These data indicated that an additional cytotoxic factor(s) was present in the Δvsm mutant and, thus, was likely to be present in LGP32 ECPs. To confirm this hypothesis, the Δvsm-1062 double-deletion mutant was constructed as described above and its ECPs were shown by the azocasein assay to be devoid of metalloprotease activity (Table 2). Nevertheless, when assayed on NIH cells, the ECPs of the Δvsm-1062 double mutant strain were shown to be as cytotoxic as those of the Δvsm mutant (Fig. 5B), thus confirming that the putative VSA1062 metalloprotease carries little or no cytotoxicity. Again, similar results were obtained using purified Vsm instead of LGP32 ECPs (data not shown). Untreated NIH cells were then either seeded into 24-well plates or left in suspension for extensive periods of time, and viability at 3, 24, and 48 h was tested using the MTT assay. Our data, expressed as percentages of the attached cells' activity, indicate that the ability of unattached NIH cells to metabolize MTT decreased at a rate similar to that of LGP32 ECP-treated NIH cells (Fig. 5B). Our findings thus suggest that the difference in the rate of MTT reduction between ECP-treated Bge and NIH cells reflects the ability of Bge cells, but not NIH cells, to survive in suspension for extensive periods of time (our unpublished observation).
FIG. 5.
Results of the viability testing of ECP-treated cells. Cells were treated as described in the legend to Fig. 3 and were then incubated for 3 to 48 h before the addition of 33 μl of an MTT solution (5 mg ml−1) from the Vybrant MTT cell proliferation assay kit (Molecular Probes, The Netherlands). The optical densities at 570 nm of triplicate samples were determined. Cell viability is expressed as a mean percentage of the MTT reduction to formazan relative to the activity of untreated cells (defined as 100%) at each time point. (A) Kinetics of the cytotoxicity of LGP32 ECPs to Bge and NIH cells. The upper curve shows that despite the rounding of Bge cells after ECP treatment, the capacity to metabolize MTT was altered only slightly. In contrast, the ECPs induced a continuous decrease of MTT metabolism by NIH cells. (B) Kinetics of the cytotoxicity of mutant ECPs to NIH cells. Cells were incubated with equivalent amounts of ECPs normalized based on the content of total protein and were treated with MTT as described above. The Δvsm strain and the Δvsm-1062 double mutant displayed moderate cytotoxicity, indicating that VSA1062 is not a factor of virulence in the ECPs. Indeed, the ECPs of the Δ1062 strain and LGP32 were equally cytotoxic. Interestingly, the MTT reduction assay provided essentially the same percentages of viability for NIH cells maintained in suspension as for cells treated with virulent ECPs.
FIG. 4.
Rounded ECP-treated Bge cells with formazan crystals. Bge cells were seeded onto coverslips, treated with LGP32 ECPs for 24 h, and then incubated for 4 h with MTT and fixed with paraformaldehyde as mentioned above. DIC microscopy revealed groups of rounded and closely associated cells. These cells display long crystals of formazan, indicating active MTT metabolism despite the ECP treatment.
DISCUSSION
In this study, we investigated the virulence of V. splendidus LGP32, a strain that was isolated in 2004 from the hemolymph of oysters experiencing out breaks of summer mortality and which later proved to be pathogenic for oysters and clams (6). Taxonomic analyses failed to identify markers associated with V. splendidus pathogenicity (6, 7). Therefore, the virulence of the isolated strains had to be assessed by experimental challenge. Analysis of the data provided by this approach is hampered by the variability of the physiological state of the animals, which depends upon a continuously changing environment (including food, temperature, salinity, and pollutants), and by the heterogeneous genetic background (25). Hence, there is a need for the development of cellular and/or molecular tests to evaluate the virulence of a strain. Indeed, standardized in vitro assays are necessary to screen for distinct activities covered by the generic term virulence, such as adherence and cytolytic effects. However, marine invertebrate cell lines do not exist, and so far the experiments to examine interactions between cells and pathogenic vibrios rely on primary lines of hemocytes maintained under survival conditions (4, 8, 23). Nevertheless, the use of such a cell population has drawbacks. The hemocyte population is made of distinct cell types that remain to be fully characterized. Moreover, each animal provides a limited number of cells (about 106), and this strategy requires the pooling of hemocytes from different individuals with heterogeneous genetic backgrounds. Even more disturbing is the fact that the oyster hemolymph is naturally contaminated with bacteria that can confound the conclusions, especially if the experiment lasts long enough to allow bacterial proliferation. Interestingly, a recent report describes the use of a flounder gill cell line as a test for the cytotoxicity of the recombinant metalloprotease of V. anguillarum, a fish vibrio (33).
To investigate the role of the secreted metalloprotease of V. splendidus as well as those of other proteins present in the ECPs, new genetic tools were developed to establish knockout mutants of this species (15). ECPs of these mutants were then tested through animal challenges performed in parallel with cytotoxicity assays of cell lines.
We first tested the ECPs on Bge cells, the only mollusk cell line available to date. DIC microscopy revealed that the ECPs produced by the virulent LGP32 strain altered Bge cell morphology as soon as 3 h posttreatment and that later on, they induced cells to detach from the extracellular matrix. This cytotoxic effect was not observed with the ECPs of LMG20012T, a nonvirulent vibrio, or the ECPs of the Δvsm strain, a vsm deletion mutant, whereas the purified Vsm induced the detachment. These data demonstrated a main role for the Vsm metalloprotease in inducing the rounded cell shape. However, by the MTT reduction assay, only a moderate alteration of Bge cell metabolism was observed, even when the assay was performed as late as 90 h after the start of incubation with LGP32 ECPs or purified Vsm. These results indicated that despite the inhibition of cell anchorage, Vsm alters Bge cell viability and metabolic functions only slightly. It is thus apparent that cytoplasmic retraction and cell rounding as observed by DIC microscopy may not per se be sufficient to determine the virulence of an ECP or of a protein.
Because this observation might be cell type dependent, the LGP32 ECP cytotoxicity and the purified Vsm were also assessed using NIH 3T3 cells, a fibroblastic mouse cell line sensitive to extracellular matrix attachment. Our data show a strong correlation between the alteration of the NIH cell morphology, accompanied by a quick detachment of the plate floor, and the alteration of cell metabolism (Fig. 5). These dramatic changes were accompanied by a total destructuralization of the actin cytoskeleton, suggesting the nonviability of most of the cells. Indeed, the MTT assay revealed a strong alteration of NIH cell metabolism, resulting in a 75% inhibition 24 h after ECP treatment (Fig. 5B). This clear discrepancy between the responses of the two cell types suggests that the Vsm activity promotes the NIH cell death by inducing cell detachment. Indeed, we have shown that when NIH cells were left in suspension for extended periods of time, the MTT metabolism decreased as if the cells were ECP treated (Fig. 5B). On the other hand, Bge cells are thought to be of hematopoietic origin; hemocytes are circulating cells involved in the invertebrate immune defense, and their survival is not attachment dependent. Indeed, Bge cells maintained for 96 h in suspension were still metabolically active and capable of rapidly forming a monolayer (our unpublished observation). The striking rate of oyster mortality induced by LGP32 ECPs indicates that the Vsm metalloprotease is not likely to act on hemocytes but that instead it actually targets a most sensitive cell type(s), the alteration of which quickly leads to the death of the organism.
The second part of this study was aimed at identifying additional cytotoxic proteins by studying some of the ECP proteins that are differentially expressed in LGP32 and the Δvsm strain, the mutant lacking Vsm. 2D gel analysis of LGP32 and Δvsm strain ECPs revealed six spots representing differentially produced proteins that were identified using MALDI-TOF MS analysis and peptide mass fingerprinting. The Δvsm ECPs were found to contain a predicted protein of unknown function, the VS2864 protein, and more interestingly, the VSA1062 protein, an additional metalloprotease of the M6 family, while LGP32 contained an unknown protein, the VSA576 protein, in addition to exhibiting three spots corresponding to Vsm. The results of the azocasein assay showed that while the Δ576, Δ2864, and Δ1062 deletion mutants expressed levels of metalloprotease activity similar to that expressed by LGP32, the level of activity in the Δvsm strain was down to 20% of the level in the wild type and the Δvsm-1062 double mutant had no detectable activity (Table 2). Moreover, the Δ576, Δ2864, and Δ1062 deletion mutants proved to be as cytotoxic to NIH cells as LGP32, as shown by DIC microscopy and the MTT assay. It was tempting to propose that V. splendidus VSA1062 metalloprotease, which was detected in Δvsm mutant ECPs but not in LGP32 ECPs, might compensate for some of the functions fulfilled by the product of the deleted vsm gene. Indeed, Milton et al. (21) showed that V. anguillarum mutants lacking a functional metalloprotease carried two different proteases which were not detected in the wild-type strain, suggesting that these proteases might compensate for the loss of the empA protease. Strikingly, while the Δvsm-1062 double mutant displayed no detectable metalloprotease activity, the cytotoxicity of its ECPs was similar to that of the ECPs of the Δvsm strain (Fig. 5B). These data suggest that although the VSA1062 metalloprotease shows significant identity (41%) to the immune inhibitor A precursor (InHA), a virulence factor encoded by the ina gene of B. thuringiensis (18), it is not, or is only marginally, involved in the ECP toxicity. Instead, like InHA, which is involved in antibacterial peptide resistance, the VSA1062 protein may play a role in bacterial resistance against the oyster immune response, a possibility that will be the focus of future investigations. Altogether, genome analysis indicating no additional metalloprotease gene in V. splendidus (Le Roux et al., presented at the Second Conference on the Biology of Vibrios, Paris, France, 28 November to 1 December 2007) and the data presented in this report show that although Vsm is the major determinant of the toxicity of the ECPs, an additional factor(s) may have a significant contribution to the degree of virulence of this pathogen.
Conclusion.
In this study, we have shown that Vsm, a metalloprotease, is the major determinant of the toxicity of the ECPs from a virulent strain of V. splendidus. Our data, obtained by using two distinct cell lines and animal challenge, illustrate the multiple-criterion definition of virulence. The term cytotoxicity is used when referring to the alteration of cell morphology or of cell viability. Cell morphology assessment is certainly a quick and valid test for cytotoxicity, since the results were corroborated by those from animal challenges. Nevertheless, viability assays of Bge cells have shown that morphology can be durably altered without implying cell death. The mechanism of cytotoxicity of the metalloprotease Vsm remains to be explored, and the observed difference regarding mollusk and mammalian cells is of particular interest.
Supplementary Material
Acknowledgments
We are indebted to Nathalie Morin and Bernard Kloareg for helpful discussions during the course of this work. We acknowledge Patricia Anglade for technical assistance in proteomic analysis. We thank Edward Ruby, Cyrille Goarant, and Céline Loot for critical reading of the manuscript.
This study was carried out with financial assistance from the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS-URA 2171), the Institut Françcais de Recherche pour l'Exploitation de la Mer (IFREMER), and the Institut de Génomique Marine (Ministère de la Recherche contract no. 0425). Johan Binesse acknowledges a Ph.D. scholarship from the French Ministry of Research and Technology (MRT).
Footnotes
Published ahead of print on 3 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Biskri, L., M. Bouvier, A. M. Guerout, S. Boisnard, and D. Mazel. 2005. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 187:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booth, B. A., M. Boesman-Finkelstein, and R. A. Finkelstein. 1984. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect. Immun. 45:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choquet, G., P. Soudant, C. Lambert, J. L. Nicolas, and C. Paillard. 2003. Reduction of adhesion properties of Ruditapes philippinarum hemocytes exposed to Vibrio tapetis. Dis. Aquat. Organisms 57:109-116. [DOI] [PubMed] [Google Scholar]
- 5.Coster, T. S., K. P. Killeen, M. K. Waldor, D. T. Beattie, D. R. Spriggs, J. R. Kenner, A. Trofa, J. C. Sadoff, J. J. Mekalanos, and D. N. Taylor. 1995. Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet 345:949-952. [DOI] [PubMed] [Google Scholar]
- 6.Gay, M., F. C. Berthe, and F. Le Roux. 2004. Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Organisms 59:49-56. [DOI] [PubMed] [Google Scholar]
- 7.Gay, M., T. Renault, A. M. Pons, and F. Le Roux. 2004. Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis. Aquat. Organisms 62:65-74. [DOI] [PubMed] [Google Scholar]
- 8.Goarant, C., J. Herlin, R. Brizard, A. L. Marteau, C. Martin, and B. Martin. 2000. Toxic factors of Vibrio strains pathogenic to shrimp. Dis. Aquat. Organisms 40:101-107. [DOI] [PubMed] [Google Scholar]
- 9.Hase, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaper, J. B., H. Lockman, M. M. Baldini, and M. M. Levine. 1984. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidates. Nature 308:655-658. [DOI] [PubMed] [Google Scholar]
- 12.Labreuche, Y., P. Soudant, M. Gonçcalves, C. Lambert, and J. L. Nicolas. 2006. Effects of extracellular products from the pathogenic Vibrio aestuarianus strain 01/32 on lethality and cellular immune responses of the oyster Crassostrea gigas. Dev. Comp. Immunol. 30:367-379. [DOI] [PubMed] [Google Scholar]
- 13.Lane, E., and T. H. Birkbeck. 1999. Toxicity of bacteria towards hemocytes of Mytilus edulis. Aquat. Living Resour. 12:343-350. [Google Scholar]
- 14.Le Roux, F., and B. Austin. 2006. Vibrio splendidus, p. 285-296. In F. L. Thompson, B. Austin, and J. Swings (ed.), The vibrios. ASM Press, Washington, DC.
- 15.Le Roux, F., J. Binesse, D. Saulnier, and D. Mazel. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, P. V. 1957. Survey of hemolysin production among species of pseudomonads. J. Bacteriol. 74:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Y., D. A. Peterson, H. Kimura, and D. Schubert. 1997. Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J. Neurochem. 69:581-593. [DOI] [PubMed] [Google Scholar]
- 18.Lovgren, A., M. Zhang, A. Engstrom, G. Dalhammar, and R. Landen. 1990. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol. Microbiol. 4:2137-2146. [DOI] [PubMed] [Google Scholar]
- 19.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto-Mashimo, C., A. M. Guerout, and D. Mazel. 2004. A new family of conditional replicating plasmids and their cognate Escherichia coli host strains. Res. Microbiol. 155:455-461. [DOI] [PubMed] [Google Scholar]
- 21.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamune, K., K. Yamamoto, A. Naka, J. Matsuyama, T. Miwatani, and T. Honda. 1996. In vitro proteolytic processing and activation of the recombinant precursor of El Tor cytolysin/hemolysin (pro-HlyA) of Vibrio cholerae by soluble hemagglutinin/protease of V. cholerae, trypsin, and other proteases. Infect. Immun. 64:4655-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nottage, A. S., and T. H. Birkbeck. 1990. Interactions between different strains of Vibrio alginolyticus and hemolymph fractions from adult Mytilus edulis. J. Invertebr. Pathol. 56:15-19. [DOI] [PubMed] [Google Scholar]
- 24.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samain, J. F., L. Dégremont, P. Soletchnik, J. Haure, E. Bédier, M. Ropert, J. Moal, A. Huvet, H. Bacca, A. Van Wormhoudt, M. Delaporte, K. Costil, S. Pouvreau, C. Lambert, V. Boulo, P. Soudant, J. L. Nicolas, F. Le Roux, T. Renault, B. Gagnaire, F. Géret, I. Boutet, T. Burgeot, and P. Boudry. 2007. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection process. Aquaculture 268:227-243. [Google Scholar]
- 26.Sanchez, B., M. C. Champomier-Verges, P. Anglade, F. Baraige, C. G. de Los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2008. A preliminary analysis of Bifidobacterium longum exported proteins by two-dimensional electrophoresis. J. Mol. Microbiol. Biotechnol. 14:74-79. [DOI] [PubMed] [Google Scholar]
- 27.Tacket, C. O., G. Losonsky, J. P. Nataro, S. J. Cryz, R. Edelman, A. Fasano, J. Michalski, J. B. Kaper, and M. M. Levine. 1993. Safety and immunogenicity of live oral cholera vaccine candidate CVD 110, a ΔctxA Δzot Δace derivative of El Tor Ogawa Vibrio cholerae. J. Infect. Dis. 168:1536-1540. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, D. N., K. P. Killeen, D. C. Hack, J. R. Kenner, T. S. Coster, D. T. Beattie, J. Ezzell, T. Hyman, A. Trofa, and M. H. Sjogren. 1994. Development of a live, oral, attenuated vaccine against El Tor cholera. J. Infect. Dis. 170:1518-1523. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, F. L., B. Austin, and J. Swings. 2006. The biology of vibrios. ASM Press, Washington, DC.
- 30.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Mol. Biol. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, F. L., C. C. Thompson, and J. Swings. 2003. Vibrio tasmaniensis sp. nov., isolated from Atlantic salmon (Salmo salar L.). Syst. Appl. Microbiol. 26:65-69. [DOI] [PubMed] [Google Scholar]
- 32.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 33.Yang, H., J. Chen, G. Yang, X. H. Zhang, Y. Li, and M. Wang. 2007. Characterization and pathogenicity of the zinc metalloprotease empA of Vibrio anguillarum expressed in Escherichia coli. Curr. Microbiol. 54:244-248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.