Abstract
Vibrio parahaemolyticus is a food-borne pathogen that naturally inhabits both marine and estuarine environments. Free-living protozoa exist in similar aquatic environments and function to control bacterial numbers by grazing on free-living bacteria. Protozoa also play an important role in the survival and spread of some pathogenic species of bacteria. We investigated the interaction between the protozoan Acanthamoeba castellanii and the bacterium Vibrio parahaemolyticus. We found that Acanthamoeba castellanii does not prey on Vibrio parahaemolyticus but instead secretes a factor that promotes the survival of Vibrio parahaemolyticus in coculture. These studies suggest that protozoa may provide a survival advantage to an extracellular pathogen in the environment.
Vibrio parahaemolyticus is a gram-negative, incidental pathogen that causes food-borne gastroenteritis, septicemia, and wound infections. The most common source of infection is contact with undercooked or raw seafood, particularly oysters (9). The pathogenicity of V. parahaemolyticus has been attributed to multiple virulence factors, of which the most well studied are the thermostable direct hemolysin (TDH) and the similar TDH-related hemolysin (TRH) (42). These hemolysins are found in most clinical strains and have been associated with cytotoxicity, enterotoxicity, cardiotoxicity, and hemolytic activity (39, 42, 44). V. parahaemolyticus also possesses two distinct type III secretion systems (T3SS), which allow for the direct translocation of bacterial proteins into host cells (26). Bacteria use these translocated proteins, known as effectors, to manipulate the host to promote bacterial survival and/or virulence (13). Although the precise role that each T3SS plays in V. parahaemolyticus pathogenesis is not understood, it has been shown that the T3SS encoded on the large chromosome (T3SS1) contributes to cytotoxicity toward eukaryotic cells and that the T3SS encoded on the smaller chromosome (T3SS2) is responsible for enterotoxicity (7, 35, 36).
V. parahaemolyticus is commonly isolated from many environmental sources, including marine, estuarine, and coastal waters (20, 21). The prevalence of V. parahaemolyticus in the environment and incidence of infection have been linked to rising water temperatures caused by global warming (32). V. parahaemolyticus has been found to be associated with various plankton, copepods, and crustaceans; however, sampling from various coastal sites reveals that V. parahaemolyticus is present in sediment, as well as free-living in water (5, 8, 19-21). In fact, the number of V. parahaemolyticus organisms detected in water samples is greater than or equal to the number associated with plankton, supporting the proposal that free-living bacteria contain a large population of V. parahaemolyticus in the environment (5, 20).
In the environment, free-living bacteria coexist with protozoa. The interaction between these organisms is complex and can be parasitic, commensal, or mutualistic in nature. Acanthamoeba castellanii is a unicellular free-living protozoan that is found ubiquitously in the environment. This species of protozoa has been isolated from various sources, including estuaries, freshwater lakes, rivers, saltwater lakes, beaches, and sediment (22). Although pathogenic in its own right, many studies have highlighted the role of Acanthamoeba spp. as reservoirs and/or vectors of pathogenic bacteria (4, 14). The most well characterized example is that of the intracellular pathogen Legionella pneumophila, which has been shown to survive and replicate within both A. castellanii and A. polyphaga after phagocytosis (12, 23). Moreover, L. pneumophila utilizes the same mechanism to survive within both Acanthamoeba spp. and macrophages from higher eukaryotes (14). This suggests that the association of bacteria with protozoa has significant consequences for the evolution of bacteria (27). Recently, it was shown that Vibrio cholerae is able to survive in the presence of A. castellanii and can be detected within amoebae, suggesting that protozoa may serve as hosts for, and promote the spread of, waterborne pathogens in the environment (1, 2). In this study, we examined the interaction between A. castellanii and V. parahaemolyticus. As these two species occupy similar ecological niches, their interaction likely has significant biological and ecological consequences. In a laboratory coculture microcosm, A. castellanii is able to promote the survival of V. parahaemolyticus. Surprisingly, this interaction does not involve the intracellular survival of V. parahaemolyticus but rather is mediated by a factor secreted by A. castellanii. These studies provide the first insight into how the bacterial pathogen V. parahaemolyticus may evade phagocytosis and utilize a protozoan factor to promote its own survival in the environment.
MATERIALS AND METHODS
Microorganisms and culture conditions.
The Vibrio parahaemolyticus strains used in this study are listed in Table 1. To determine the presence/absence of the TDH, genomic DNA was isolated using the Qiagen DNeasy tissue kit according to the manufacturer's protocols. The presence or absence of tdh was detected by PCR as described previously (6). The strains POR1, POR2, POR3, LM5312, and LM5674 were maintained on minimal marine medium agar plates (5 mM K2SO4, 77 mM K2HPO4, 35 mM KH2PO4, 20 mM NH4Cl, 5 mM MgSO4·7H2O, 2% NaCl, 0.4% galactose, and 1.5% Bacto agar per liter) supplemented with 0.1% sodium pyruvate at 30°C. The strains KO2201 and KO2202 were maintained on marine Luria broth (MLB; 1% tryptone, 0.5% yeast extract, and 3% NaCl per liter) supplemented with 0.1% sodium pyruvate at 30°C. To establish cocultures, all V. parahaemolyticus strains were cultured in MLB at 30°C for 16 to 18 h with aeration. Escherichia coli K-12 was kindly provided by V. Sperandio (University of Texas Southwestern Medical Center) and was maintained in Luria broth (LB; 1% tryptone, 0.5% yeast extract, and 1% NaCl per liter) at 37°C. To establish cocultures, E. coli was cultured in LB at 37°C for 16 to 18 h with aeration.
TABLE 1.
V. parahaemolyticus strains used in this study
| Strain | Relevant genotype | Relevant phenotype | Source | Reference(s) |
|---|---|---|---|---|
| RIMD2210633 | Wild type | Pandemic isolate, Kanagawa phenomenon positive, TDH+, competent for secretion/translocation via T3SS1 and T3SS2 | Clinical isolate; T. Honda | 26 |
| POR1 | ΔtdhA ΔtdhS | Lacks TDH, derived from RIMD2210633 | Clinical isolate; T. Honda | 36 |
| POR2 | ΔtdhA ΔtdhS ΔvcrD1 | Deficient for secretion/translocation via the T3SS1, derived from POR1 | Clinical isolate; T. Honda | 34, 36 |
| POR3 | ΔtdhA ΔtdhS ΔvcrD2 | Deficient for secretion/translocation via T3SS2, derived from POR1 | Clinical isolate; T. Honda | 34, 36 |
| LM5312 | BB22, wild type | Wild-type opaque colony morphology, Kanagawa phenomenon positive, TDH+, competent for secretion/translocation via T3SS1 and T3SS2 | Clinical isolate; L. L. McCarter | 17, 31 |
| LM5674 | ΔopaR | Lacks quorum sensing regulator OpaR, translucent colony morphology, increased production of lateral flagella, derived from LM5312 | Clinical isolate; L. L. McCarter | 17, 31 |
| KO2201 | Unknown | Environmental isolate, Kanagawa phenomenon positive, TDH+ | Blue crab hemolymph; V. parahaemolyticus ATCC 27979 | |
| KO2202 | Unknown | Environmental isolate, Kanagawa phenomenon positive, TDH+ | Steamed crab; V. parahaemolyticus ATCC 35117 |
Anexic cultures of A. castellanii ATCC 30234 were obtained from the American Type Culture Collection and were maintained as monolayers in proteose peptone-yeast extract-glucose medium [PYG; ATCC medium 712; 2% proteose peptone, 0.1% yeast extract, 4 mM MgSO4·7H2O, 0.4 mM CaCl2, 0.05 mM Fe(NH4)2(SO4)2·6H2O, 2.5 mM Na2HPO4·7H2O, 2.5 mM KH2PO4, 0.1% sodium citrate dihydrate, and 0.1 M glucose, pH 6.5] in 75-cm2 tissue culture flasks, incubated at 25°C without aeration. Amoebae were routinely subcultured every 7 days.
Establishment of cocultures.
Logarithmic A. castellanii cultures were washed and the medium was changed 2 days prior to the start of the coculture. One day prior to coculture, the monolayer was suspended and amoebae were enumerated using a hemocytometer (Reichert). Amoebae were diluted and seeded at a density of 1 × 105 amoebae/ml in PYG in a 24-well tissue culture dish and incubated at 25°C for 24 h. The day the coculture was established (day 0), the medium was removed and replaced with either fresh PYG (cultures with amoebae alone) or PYG containing suspended bacteria at a multiplicity of infection of 10 (approximately 1 × 106 bacteria/ml) (cocultures). For each experiment, one well of amoebae was disrupted by vigorous pipetting and organisms were enumerated; the number of bacteria to be added to the coculture was adjusted accordingly. Cultures of bacteria alone at the same concentration in PYG were also established. Plates were centrifuged at 200 × g for 5 min to facilitate contact between amoebae and bacteria, followed by incubation at 25°C for up to 35 days.
Parachamber cocultures were established as described above with the following modifications. Logarithmic A. castellanii cultures were washed and the medium was changed 1 day prior to the start of the coculture. The day the coculture was established (day 0), the monolayer was suspended by tapping the flask, and the number of amoebae was determined. A transwell membrane with a 0.2-μm pore size (Nunc) was inserted into each well of the 24-well tissue culture dish. Amoebae were diluted in PYG to a density of approximately 2.5 × 105 amoebae/ml, and 400 μl of this suspension was added to the top chamber (1 × 105 total amoebae). V. parahaemolyticus POR1 was added to the bottom chamber at a density of 2.5 × 106 bacteria/ml in PYG, and 400 μl of this suspension was added to the bottom chamber (1 × 106 total bacteria). As controls, cultures were set up containing A. castellanii alone (top), V. parahaemolyticus alone (bottom), and A. castellanii and V. parahaemolyticus together in the bottom chamber.
Preconditioned A. castellanii medium was generated by collecting medium from cultures of A. castellanii alone, grown for 1, 4, and 7 days. The medium was centrifuged at 800 × g for 10 min, filtered with a 0.22-μm filter, and used to establish a coculture with POR1 as described above.
Analysis of coculture.
Cocultures were monitored visually every day; at various time points, the total number of amoebae was determined by first disrupting each well by vigorous pipetting and counting aliquots with a hemocytometer. The ratio of live to dead amoebae was determined by staining aliquots of the coculture with trypan blue for 10 min (18). At each time point, the number of extracellular bacteria was enumerated by plating serial dilutions on minimal marine medium or MLB supplemented with 0.1% sodium pyruvate (V. parahaemolyticus) or LB (E. coli) as appropriate. To detect the presence of intracellular bacteria, cocultured A. castellanii organisms were gently washed and treated with 200 μg/ml gentamicin in phosphate-buffered saline for 2 hours at room temperature to kill all extracellular bacteria. The cells were gently washed again and lysed in 0.1% Triton X-100 in phosphate-buffered saline for 15 min at room temperature to release intracellular bacteria. Serial dilutions of this suspension were plated as described above. We established that the gentamicin treatment resulted in efficient killing of bacteria without affecting the amoebae and that the Triton X-100 treatment resulted in efficient lysis of A. castellanii without affecting the bacteria (data not shown). Parachamber cocultures were visually monitored for up to 21 days and analyzed as described above. Preconditioned-medium cultures were visually monitored for up to 10 days and analyzed as described above.
RESULTS
Survival of V. parahaemolyticus in the presence of A. castellanii.
We initially chose to carry out our experiments with POR1, a strain of V. parahaemolyticus that lacks the TDH toxin but is competent for secretion by both T3SSs (34). When this strain was cultured in the A. castellanii-specific medium, PYG, the number of CFU/ml decreased sharply over the first 3 days and viable bacteria became undetectable by day 4 (Fig. 1A). We did observe variation between experiments and found that viable bacteria become undetectable between days 4 and 8 when cultured in PYG (data not shown). This is in contrast to what was found when we cultured E. coli K-12 in the same medium; viable bacteria can be detected for up to 31 days at approximately 1 × 108 CFU/ml in PYG (Fig. 1B).
FIG. 1.
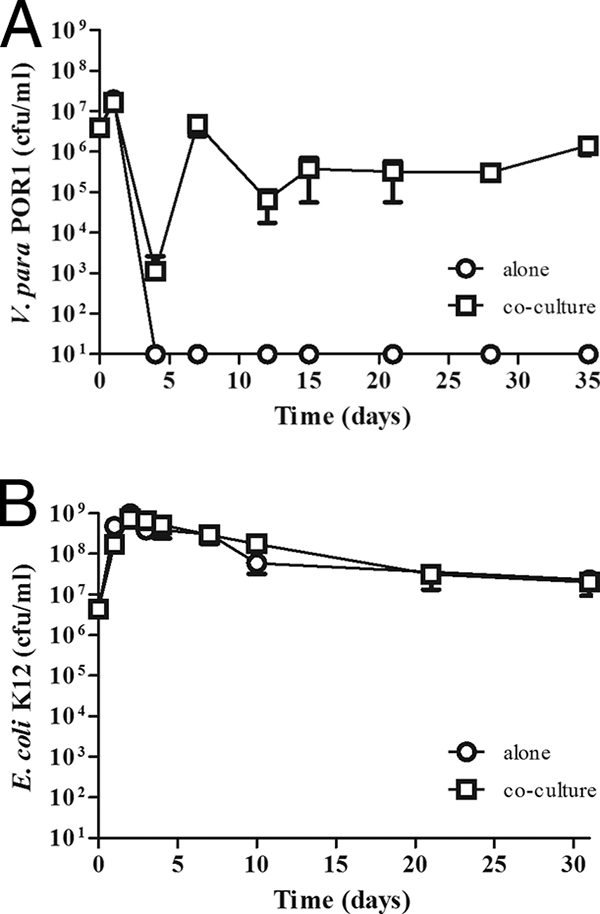
A. castellanii promotes the survival of V. parahaemolyticus. V. parahaemolyticus (V. para) POR1 (A) and E. coli K-12 (B) were cultured in the absence (open circles) or presence (open squares) of A. castellanii for 35 (A) and 31 (B) days. The number of bacteria was determined by plating serial dilutions on media selective for V. parahaemolyticus or E. coli as described in Materials and Methods. Results are reported as the number of bacteria recovered per ml; the limit of detection is 10 CFU. Data points represent the means ± standard deviations (SD) from triplicate cultures and are representative of at least three independent experiments.
When V. parahaemolyticus POR1 was cultured in the presence of A. castellanii, the number of viable bacteria also initially declined during the first 3 days (Fig. 1A). However, after this decline, POR1 replicated and was able to survive and persist for up to 35 days (Fig. 1A and Table 2). E. coli K-12 also survives for up to 31 days in the presence of A. castellanii (Fig. 1B). However, this is likely due to the ability of E. coli K-12 to survive in the culture medium alone, as the numbers of recovered bacteria are similar in the absence (2.30 × 107 ± 0.20 × 107 CFU/ml) and presence (2.10 × 107 ± 1.10 × 107 CFU/ml) of amoebae at day 31 (Fig. 1B).
TABLE 2.
Coculture of various V. parahaemolyticus strains with A. castellanii
| Strain | No. of bacteria recovered (CFU/ml) 10 days after coculturea
|
Mean no. of viable A. castellanii organisms (amoebae/ml) (106) ± SD | ||
|---|---|---|---|---|
| PYG alone | Cocultured with A. castellanii
|
|||
| Extracellular | Intracellular | |||
| None | NA | NA | NA | 3.95 ± 0.90 |
| RIMD2210633 | ND | 1.89 (±0.74) × 106 | ND | 3.93 ± 0.34 |
| POR1 | ND | 2.19 (±1.44) × 106 | ND | 2.59 ± 0.30 |
| POR2 | ND | 1.58 (±0.35) × 106 | ND | 3.69 ± 0.59 |
| POR3 | ND | 1.44 (±0.08) × 106 | ND | 3.88 ± 0.28 |
| LM5312 | ND | 1.91 (±1.26) × 105 | ND | 4.25 ± 0.22 |
| LM5674 | ND | 3.96 (±1.88) × 104 | ND | 3.83 ± 0.54 |
| KO2201 | ND | 2.84 (±1.01) × 107 | ND | 4.87 ± 1.40 |
| KO2202 | ND | 9.97 (±1.40) × 106 | ND | 3.40 ± 0.11 |
NA, not applicable; ND, none detected.
The ability of some bacteria to survive in the presence of protozoa such as A. castellanii has been attributed to the ability of the bacteria to survive within amoebae (14). To examine whether the survival of POR1 was due to the ability of the bacteria to gain entry into and survive within amoebae, the number of intracellular POR1 organisms was examined by gentamicin protection assays. Unlike what has been described for V. cholerae, L. pneumophila, and other bacteria, V. parahaemolyticus does not reside within A. castellanii, as we were unable to recover any intracellular bacteria (1, 2, 12, 23) (Table 2). It should be noted that V. parahaemolyticus has been described to enter into a viable but nonculturable (VBNC) state under some long-term-stress conditions (40, 41). As the addition of sodium pyruvate to culture media has been shown to enhance the recovery of bacteria that have entered the VBNC state, the medium was supplemented with 0.1% sodium pyruvate (33). Therefore, our inability to recover intracellular bacteria is not due to their entry into a VBNC state. Rather, V. parahaemolyticus is able to resist phagocytosis by A. castellanii and employ an extracellular strategy of survival.
In an attempt to identify the factors that contribute to this phenotype, we obtained and tested multiple V. parahaemolyticus strains in our coculture; the strains, their origins, and their relative genotypes and phenotypes are listed in Table 1. The V. parahaemolyticus strain POR1 lacks the TDH; thus, we predict that this virulence factor does not contribute to the survival of the bacteria in the context of our coculture. However, to rule out any involvement of the TDH, we tested the POR1 parent strain, RIMD2210633, which encodes the TDH. To examine the role of the two T3SSs, we tested strains of V. parahaemolyticus, POR2 and POR3, containing an in-frame deletion of vcrD1 and vcrD2, respectively, which renders them incompetent for secretion by T3SS1 and T3SS2, respectively (Table 1). We also examined an additional clinical isolate, LM5312, and two environmental isolates, KO2201 and KO2202, all of which encode the TDH (Table 1). KO2201 and KO2202, obtained from the ATCC, were isolated from blue crab hemolymph and steamed crab, respectively. Another strain, LM5674, is derived from LM5312 but lacks the opaR gene, which encodes a transcriptional regulator of the LuxR quorum-sensing regulator family (31). Strains lacking opaR form translucent colonies and lack the production of the capsule polysaccharide (11). LM5674 also exhibits altered biofilm formation and a hyperswarmer phenotype, which correlates with the overexpression of genes encoding the lateral flagellar system of V. parahaemolyticus (10, 17). Similar to what was found for POR1, all of the strains were unable to survive in PYG and none were found to reside within A. castellanii (Table 2). Moreover, all of the strains tested were able to survive in the presence of A. castellanii for at least 10 days (Table 2). This is consistent with the hypothesis that A. castellanii promotes the survival of V. parahaemolyticus irrespective of the presence of the TDH, the T3SSs, the quorum-sensing regulator OpaR, or the source of isolation.
Bacterial survival is mediated by a factor secreted by A. castellanii.
The survival of V. parahaemolyticus in the presence of A. castellanii does not require the bacteria to gain entrance into A. castellanii. To determine whether cell contact is necessary for survival, we examined the ability of POR1 to survive in a parachamber with A. castellanii. In these experiments, a transwell membrane with a 0.2-μm pore size is used to physically separate the bacteria and A. castellanii. In the parachamber, the amoebae and bacteria are cultured in the top and bottom chambers, respectively. In control cultures, both amoebae and bacteria are added to the bottom chamber. As shown in Fig. 2, POR1 survives equally well when physically separated from A. castellanii or when grown in a coculture. At day 1, 1.46 × 108 ± 0.64 × 108 CFU were recovered from the parachamber, whereas 0.76 × 108 ± 0.26 × 108 CFU were recovered from the coculture. As observed previously, the number of recovered bacteria decreases slightly by day 7 in both the parachamber and the coculture (1.60 × 106 ± 1.01 × 106 CFU and 1.58 × 106 ± 0.29 × 106 CFU, respectively) and remains relatively constant over the next 14 days (0.41 × 106 ± 0.23 × 106 CFU and 3.34 × 106 ± 1.46 × 106 CFU at day 14 and 0.51 × 106 ± 0.22 × 106 CFU and 4.70 × 106 ± 2.65 × 106 CFU at day 21 in the parachamber and coculture, respectively). In contrast, bacteria grown in the absence of A. castellanii are unable to survive and the bacterial population dies between days 7 and 14 (Fig. 2). Thus, V. parahaemolyticus is able to utilize a factor secreted by A. castellanii to promote its survival independently of direct contact with the amoebae. In addition, the numbers of bacteria recovered are similar in both the parachamber and the coculture, further supporting the model that V. parahaemolyticus resists phagocytosis.
FIG. 2.
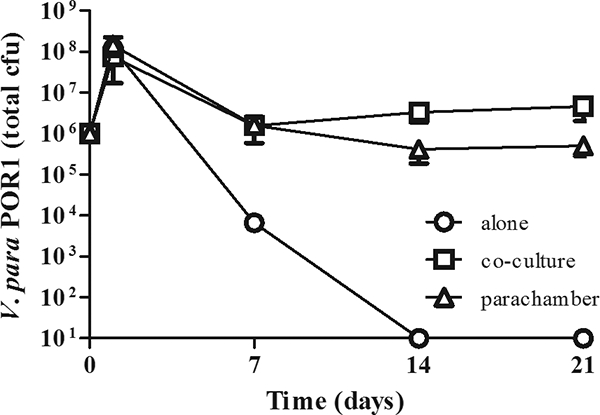
Survival of V. parahaemolyticus is mediated by a factor secreted by A. castellanii. V. parahaemolyticus (V. para) POR1 was cultured in the presence of A. castellanii for 21 days but separated from amoebae by a 0.2-μm-pore-size membrane (triangles). As a control, bacteria were cultured in PYG alone (circles) and in the presence of A. castellanii (squares) as in the legend to Fig. 1. The number of bacteria was determined as described for Fig. 1. Results are reported as numbers of total bacteria; the limit of detection is 10 CFU. Data points represent the means ± SD from triplicate cultures and are representative of at least three independent experiments.
The results from the parachamber experiment indicated that a factor secreted by the amoebae was responsible for the survival of the bacteria. We then tested whether media from a culture of A. castellanii alone could recapitulate the same survival effect. Media from 1-, 4-, and 7-day-old cultures of A. castellanii alone were used to generate preconditioned media and establish cocultures. However, as shown in Fig. 3, we found that media preconditioned from a culture of A. castellanii alone does not support the growth of V. parahaemolyticus. This suggests that the amoebae may require the presence of the bacteria to elicit production of the factor. Alternatively, the survival factor produced by the amoebae is either rapidly metabolized or requires continuous production.
FIG. 3.
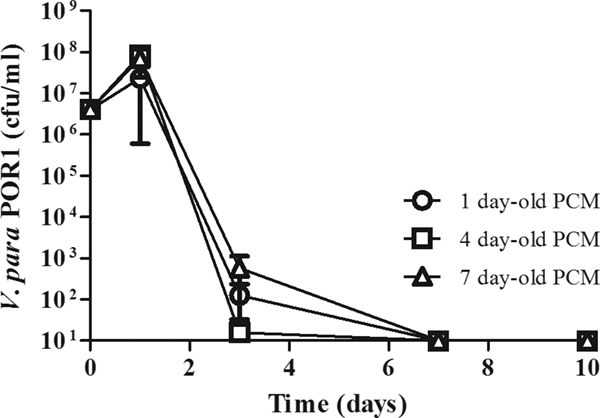
Medium conditioned from A. castellanii alone does not promote the survival of V. parahaemolyticus. V. parahaemolyticus (V. para) POR1 was cultured for 10 days in the presence of media conditioned from 1 (circles)-, 4 (squares)-, and 7 (triangles)-day-old cultures of A. castellanii alone. PCM, preconditioned medium. The number of bacteria was determined as described in the legend to Fig. 1. Results are reported as numbers of bacteria recovered per ml; the limit of detection is 10 CFU. Data points represent the means ± SD from triplicate cultures and are representative of at least three independent experiments.
V. parahaemolyticus is not cytotoxic toward A. castellanii.
V. parahaemolyticus has been shown to be cytotoxic toward many cell types, and this toxicity has been attributed to the presence of the T3SS1 (7, 25, 34). As cytotoxicity may be a mechanism by which V. parahaemolyticus resists phagocytosis and promotes its own survival, we determined the number of viable A. castellanii organisms in the coculture. Over 10 days, the number of viable amoebae in PYG alone increases to 3.95 × 106 ± 0.90 × 106 amoebae/ml (Table 2). When cocultured with RIMD2210633, POR1, POR3, LM5312, or LM5674, the number of viable amoebae after 10 days is similar to the number of amoebae in PYG alone (Table 2). Furthermore, we do not see an increase in the number of viable amoebae when they are cocultured with POR2, a strain lacking the T3SS1 (3.69 × 106 ± 0.59 × 106 amoebae/ml) (34, 35). Therefore, V. parahaemolyticus is not cytotoxic toward A. castellanii. The morphology of A. castellanii in the presence and absence of bacteria was examined by light and confocal microscopy. No significant alterations in cell morphology were observed (data not shown), suggesting that V. parahaemolyticus resists predation by A. castellanii through a mechanism other than direct killing of the amoebae.
DISCUSSION
The ability of V. parahaemolyticus to survive as a free-living bacterium in the environment requires that it coexist with other organisms, such as protozoa. In this study, we utilized a coculture microcosm to investigate the interaction between V. parahaemolyticus and the model protozoan A. castellanii. We observe that V. parahaemolyticus, unlike some species of bacteria, is able to avoid predation and survive in the presence of A. castellanii. Furthermore, the mechanism used by V. parahaemolyticus involves extracellular survival and is dependent on a factor produced and secreted by the protozoan. These studies provide the first insight into how these two species may coexist in the environment.
The ability of A. castellanii to promote the survival of V. parahaemolyticus does not require direct contact between amoebae and the bacteria. We found that V. parahaemolyticus POR1 survived equally well when directly cocultured with A. castellanii or when physically separated by a membrane. Thus, survival is mediated by a diffusible factor produced by the amoebae. However, media preconditioned from cultures of A. castellanii alone were unable to support the survival of the bacteria. This indicates that the bacteria may require continuous production of the factor or that the factor may be rapidly metabolized. If the factor is metabolized, this would support the idea that A. castellanii provides a nutritional source for V. parahaemolyticus that satisfies a requirement not fulfilled by the media. This alternative food source may come in the form of dying amoeba or metabolic end products, produced by the amoebae as they grow. This type of cross-feeding between species is known as syntrophy and is common among organisms in a symbiotic relationship (24, 37). Furthermore, it has been suggested that Mycobacterium avium survives in the presence of A. polyphaga by feeding off by-products secreted by the amoebae (43). However, we found that amoeba lysate could not promote the survival of V. parahaemolyticus POR1 (data not shown). Again, this may be due to the rapid metabolism of the factor or the requirement for continuous production. Alternatively, the ability of V. parahaemolyticus to survive in the presence of A. castellanii may be more complex and involve an active molecular conversation between the two organisms. In fact, the inability of preconditioned media to support survival suggests that production of the survival factor by the amoebae may require the presence of the bacteria. It is becoming increasingly clear that bacteria are capable of interspecies and interkingdom communication and that these processes are important for the evolution and survival of the bacteria (16). As such, the ability to sense and respond to protozoa in the environment would benefit the bacteria, and it may contribute to the persistence of V. parahaemolyticus as a free-living organism.
Interestingly, V. parahaemolyticus appears to exist in a nonproliferative state in the presence of the amoebae, as the number of viable bacteria does not increase over more than 30 days in coculture. This nonproliferative/nondividing state is in some ways reminiscent of the VBNC state described for various Vibrio species, including V. parahaemolyticus (40, 41). In both cases, the bacteria are held in a static, nonreplicative state in which they appear to be dormant. This state may be a consequence of limited nutrients late in the coculture. However, if this was the case, we would expect to observe a decrease in the number of bacteria as nutrients become more and more limiting. The fact that we do not see this trend suggests that this state may have more-significant biological relevance. Furthermore, if this state is representative of free-living bacteria in the environment, entry into this state may further facilitate the persistence of bacteria in the environment.
The ability to survive in the presence of amoebae has been reported for other bacteria, such as V. cholerae; Abd and colleagues reported that the V. cholerae O1 and El Tor strains can survive in the presence, but not the absence, of A. castellanii for up to 14 days (1). However, the persistence of V. cholerae in the presence of A. castellanii is likely due to the ability of V. cholerae to survive within this protozoan (1, 2). In contrast, we were unable to recover intracellular V. parahaemolyticus under any conditions. Furthermore, the number of bacteria recovered from the coculture did not change when A. castellanii was physically separated from V. parahaemolyticus. If A. castellanii was able to internalize bacteria, even at a low level, we would expect that the number of viable bacteria recovered would be much higher in the parachamber than in the coculture. Thus, it appears that V. parahaemolyticus not only survives in the presence of A. castellanii but actively resists predation by the protozoa.
Many species of bacteria have evolved clever mechanisms to resist predation by protozoa (29). Comamonas acidovorans, Flectobacillus spp., and other bacteria resist engulfment through the formation of long filaments, greater than 10 μm in length (15). Other bacteria, such as Pseudomonas aeruginosa, Janthinobacterium lividum, and Chromobacterium violaceum, target the protozoa with toxins that cause rapid cell lysis (3, 29, 30, 38). Still other species increase their swimming speed to avoid grazing or form biofilms that are resistant to predation (28, 29). We did not observe elongation of the bacteria during coculture or lysis of the amoebae by the bacteria. Furthermore, we observe that the ability to avoid phagocytosis is common for all V. parahaemolyticus strains studied. Thus, the mechanism utilized by V. parahaemolyticus is independent of the TDH, T3SS machinery, and quorum sensing and is unrelated to the source of isolation. Interestingly, we observed that the strain lacking OpaR, LM5674, was recovered in lower numbers after 10 days of coculture with A. castellanii than all other V. parahaemolyticus strains tested (Table 2). Thus, while survival and resistance to phagocytosis are not absolutely dependent on OpaR, optimum survival may require OpaR or an OpaR-regulated pathway. Future work will focus on understanding the molecular mechanism used by V. parahaemolyticus to resist phagocytosis; this will provide further insight into the survival of this pathogen in the environment.
Overall, these observations have an impact on our understanding of the ability of V. parahaemolyticus to persist and spread in the environment. While V. parahaemolyticus is a major health and economic issue in Southeast Asia, problems associated with V. parahaemolyticus infections in the United States are believed to be largely underdiagnosed and may represent a major health risk (9). Therefore, understanding the mechanisms used by V. parahaemolyticus for survival and persistence is essential to control and prevent infection. The ability of amoebae to promote the survival of V. parahaemolyticus in coculture may reveal a mechanism by which this pathogen persists in the environment. Future studies will reveal the nature of the molecular relationship between V. parahaemolyticus and A. castellanii and will focus on the use of this knowledge for the development of tools to prevent the survival and spread of the bacterium in the environment.
Acknowledgments
We thank Linda McCarter, Takeshi Honda, and Tetsuya Iida for their generosity in supplying the V. parahaemolyticus strains and Vanessa Sperandio for supplying the E. coli K-12 strain. We thank Michael Norgard, John MacMillan, and all of the members of the Orth lab for their support, helpful discussions, and critical reading of the manuscript.
K.O. and M.A.L.-A. are supported by the NIH-AID (grant RO1-AI056404) and the Welch Research Foundation (grant I-1561). K.O. is a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease and a W. W. Caruth, Jr., Biomedical Scholar.
Footnotes
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Abd, H., A. Saeed, A. Weintraub, G. B. Nair, and G. Sandstrom. 2007. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60:33-39. [DOI] [PubMed] [Google Scholar]
- 2.Abd, H., A. Weintraub, and G. Sandstrom. 2005. Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ. Microbiol. 7:1003-1008. [DOI] [PubMed] [Google Scholar]
- 3.Abd, H., B. Wretlind, A. Saeed, E. Idsund, K. Hultenby, and G. Sandstrom. 2008. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J. Eukaryot. Microbiol. 55:235-243. [DOI] [PubMed] [Google Scholar]
- 4.Auran, J. D., M. B. Starr, and F. A. Jakobiec. 1987. Acanthamoeba keratitis. A review of the literature. Cornea 6:2-26. [PubMed] [Google Scholar]
- 5.Baffone, W., R. Tarsi, L. Pane, R. Campana, B. Repetto, G. L. Mariottini, and C. Pruzzo. 2006. Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea (Italy) and study of their pathogenicity-associated properties. Environ. Microbiol. 8:1299-1305. [DOI] [PubMed] [Google Scholar]
- 6.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. Vickery, D. D. Jones, and C. A. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 7.Burdette, D. L., M. L. Yarbrough, A. Orvedahl, C. J. Gilpin, and K. Orth. 2008. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc. Natl. Acad. Sci. USA 105:12497-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covazzi Harriague, A., M. D. Brino, M. Zampini, G. Albertelli, C. Pruzzo, and C. Misic. 2008. Vibrios in association with sedimentary crustaceans in three beaches of the northern Adriatic Sea (Italy). Mar. Pollut. Bull. 56:574-579. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 10.Enos-Berlage, J. L., Z. T. Guvener, C. E. Keenan, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55:1160-1182. [DOI] [PubMed] [Google Scholar]
- 11.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 13.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 14.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, M. W., E. R. Moore, and M. G. Hofle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes, D. T., and V. Sperandio. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaques, S., and L. L. McCarter. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 188:2625-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong, H. J., E. S. Jang, B. I. Han, K. H. Lee, M. S. Ock, H. H. Kong, D. I. Chung, S. Y. Seol, D. T. Cho, and H. S. Yu. 2007. Acanthamoeba: could it be an environmental host of Shigella? Exp. Parasitol. 115:181-186. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko, T., and R. R. Colwell. 1975. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl. Microbiol. 29:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko, T., and R. R. Colwell. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko, T., and R. R. Colwell. 1975. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl. Microbiol. 30:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan, N. A. 2006. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol. Rev. 30:564-595. [DOI] [PubMed] [Google Scholar]
- 23.Kilvington, S., and J. Price. 1990. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 24.Kooijman, S. A., P. Auger, J. C. Poggiale, and B. W. Kooi. 2003. Quantitative steps in symbiogenesis and the evolution of homeostasis. Biol. Rev. Camb. Philos. Soc. 78:435-463. [DOI] [PubMed] [Google Scholar]
- 25.Liverman, A. D., H. C. Cheng, J. E. Trosky, D. W. Leung, M. L. Yarbrough, D. L. Burdette, M. K. Rosen, and K. Orth. 2007. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. USA 104:17117-17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 27.Marciano-Cabral, F., and G. Cabral. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16:273-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matz, C., T. Bergfeld, S. A. Rice, and S. Kjelleberg. 2004. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6:218-226. [DOI] [PubMed] [Google Scholar]
- 29.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 30.Matz, C., A. M. Moreno, M. Alhede, M. Manefield, A. R. Hauser, M. Givskov, and S. Kjelleberg. 2008. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarter, L. L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin, J. B., A. DePaola, C. A. Bopp, K. A. Martinek, N. P. Napolilli, C. G. Allison, S. L. Murray, E. C. Thompson, M. M. Bird, and J. P. Middaugh. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353:1463-1470. [DOI] [PubMed] [Google Scholar]
- 33.Mizunoe, Y., S. N. Wai, T. Ishikawa, A. Takade, and S. Yoshida. 2000. Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol. Lett. 186:115-120. [DOI] [PubMed] [Google Scholar]
- 34.Ono, T., K. S. Park, M. Ueta, T. Iida, and T. Honda. 2006. Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, T. Iida, and T. Honda. 2004. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48:313-318. [DOI] [PubMed] [Google Scholar]
- 36.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer, T., and S. Bonhoeffer. 2004. Evolution of cross-feeding in microbial populations. Am. Nat. 163:E126-E135. [DOI] [PubMed] [Google Scholar]
- 38.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raimondi, F., J. P. Kao, C. Fiorentini, A. Fabbri, G. Donelli, N. Gasparini, A. Rubino, and A. Fasano. 2000. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect. Immun. 68:3180-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, S. A., D. McDougald, and S. Kjelleberg. 2000. Vibrio vulnificus: a physiological and genetic approach to the viable but nonculturable response. J. Infect. Chemother. 6:115-120. [DOI] [PubMed] [Google Scholar]
- 41.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirai, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang, G., T. Iida, H. Inoue, M. Yutsudo, K. Yamamoto, and T. Honda. 1997. A mutant cell line resistant to Vibrio parahaemolyticus thermostable direct hemolysin (TDH): its potential in identification of putative receptor for TDH. Biochim. Biophys. Acta 1360:277-282. [DOI] [PubMed] [Google Scholar]


