Abstract
The virulence factor internalin A (InlA) facilitates the uptake of Listeria monocytogenes by epithelial cells that express the human isoform of E-cadherin. Previous studies identified naturally occurring premature stop codon (PMSC) mutations in inlA and demonstrated that these mutations are responsible for virulence attenuation. We assembled >1,700 L. monocytogenes isolates from diverse sources representing 90 EcoRI ribotypes. A subset of this isolate collection was selected based on ribotype frequency and characterized by a Caco-2 cell invasion assay. The sequencing of inlA genes from isolates with attenuated invasion capacities revealed three novel inlA PMSCs which had not been identified previously among U.S. isolates. Since ribotypes include isolates with and without inlA PMSCs, we developed a multiplex single-nucleotide polymorphism (SNP) genotyping assay to detect isolates with virulence-attenuating PMSC mutations in inlA. The SNP genotyping assay detects all inlA PMSC mutations that have been reported worldwide and verified in this study to date by the extension of unlabeled primers with fluorescently labeled dideoxynucleoside triphosphates. We implemented the SNP genotyping assay to characterize human clinical and food isolates representing common ribotypes associated with novel inlA PMSC mutations. PMSCs in inlA were significantly (ribotypes DUP-1039C and DUP-1045B; P < 0.001) or marginally (ribotype DUP-1062D; P = 0.11) more common among food isolates than human clinical isolates. SNP genotyping revealed a fourth novel PMSC mutation among U.S. L. monocytogenes isolates, which was observed previously among isolates from France and Portugal. This SNP genotyping assay may be implemented by regulatory agencies and the food industry to differentiate L. monocytogenes isolates carrying virulence-attenuating PMSC mutations in inlA from strains representing the most significant health risk.
Listeria monocytogenes is a facultative intracellular pathogen that is the etiologic agent of listeriosis, a severe invasive disease in humans (35). In 1999, Mead and coworkers estimated that 2,500 human listeriosis cases occur each year in the United States, resulting in nearly 500 deaths (22). People with definite immunocompromising circumstances are predominantly inclined to succumb to invasive listeriosis, the clinical manifestations of which may include life-threatening conditions such as septicemia, meningitis, or encephalitis or spontaneous abortion in pregnant women (36). While human clinical listeriosis cases are relatively rare, they usually lead to hospitalization (85 to 90%) and often result in death (20 to 30%), making listeriosis a substantial public health problem (22).
More than 90% of human listeriosis cases are caused by a few specific L. monocytogenes serotypes (i.e., 1/2a, 1/2b, and 4b), with serotype 4b strains being held responsible for the majority of listeriosis epidemics (15, 21). Several different molecular subtyping approaches (e.g., EcoRI ribotyping and pulsed-field gel electrophoresis) have been used to group L. monocytogenes isolates into two major genetic lineages (I and II) and a third minor lineage (III) that correlate with serotype classification (see, e.g., references 25, 37, and 38). Molecular epidemiology studies demonstrated that lineage I is overrepresented by isolates from humans with clinical listeriosis (11, 37). In addition, two highly clonal L. monocytogenes molecular subtypes within lineage I (termed epidemic clones I and II) that belong to serotype 4b have been linked to most listeriosis epidemics (7, 15) and have frequently been isolated in sporadic listeriosis cases (11). While lineage II includes isolates that cause human disease, this lineage also includes molecular subtypes that are isolated most commonly from food and have rarely or never been associated with human listeriosis cases (11).
The ability of L. monocytogenes to traverse multiple host barriers is a key characteristic that allows this pathogen to establish a systemic infection. L. monocytogenes invades nonprofessional phagocytic cells by expressing a family of proteins known as the internalins. While several internalins have been identified, InlA and InlB are the most well characterized internalins involved in the invasion of host cells (1). InlA is an 800-amino-acid (aa) protein, encoded by the key virulence-associated gene inlA, which facilitates the entry of L. monocytogenes into nonprofessional phagocytic cells expressing the human isoform of E-cadherin (19). The interaction between InlA and E-cadherin is a critical first step in crossing the intestinal barrier during the initial stages of an L. monocytogenes infection (19). The importance of this interaction has been demonstrated previously by the significantly attenuated virulence of inlA null mutants observed in cell culture and animal infection experiments (9, 18).
Previous studies described multiple distinct single-nucleotide polymorphisms (SNPs) in inlA leading to premature stop codons (PMSCs) upstream of the region encoding the C-terminal LPXTG membrane-anchoring motif (6, 12, 14, 26, 28, 29, 30, 33). These PMSC mutations in inlA led to the expression of a truncated form of InlA that was secreted rather than anchored to the bacterial cell wall (14, 26, 28, 33) or terminated InlA production (29). Another study recently completed by our group showed that PMSC mutations in inlA appear to be fully responsible for attenuated virulence, as demonstrated by characterizing sets of paired isogenic mutants with and without PMSCs in inlA by cell culture invasion assays and guinea pig infection experiments (23). The characterization of large representative L. monocytogenes isolate collections in the United States and France revealed that isolates carrying a PMSC mutation in inlA represent >30% of food isolates but <2% of human clinical isolates (13, 26).
Taken together, the findings of previous studies suggest that L. monocytogenes isolates found in food include (i) a subpopulation of subtypes that are responsible for the majority of human disease (i.e., epidemic clones I and II) and (ii) a significant subpopulation of subtypes that carry virulence-attenuating PMSC mutations in inlA. Although the underlying genetic mechanisms that explain the association between epidemic clone strains and the majority of human disease have yet to be elucidated, mutations leading to a PMSC in inlA have been shown to be responsible for attenuated mammalian virulence (23). Thus, SNPs leading to a PMSC in inlA serve as suitable genetic markers to differentiate isolates with virulence-attenuating mutations in inlA from fully virulent L. monocytogenes isolates that encode full-length InlA. Although a recent study by Ducey et al. (5) developed a subtyping assay based on SNP sites identified through multilocus sequence typing, the assay is restricted to characterizing L. monocytogenes lineage I isolates and includes only one SNP associated with attenuated virulence. The present study was thus conducted to (i) identify additional mutations leading to PMSCs in inlA genes from a large representative collection of genetically diverse L. monocytogenes isolates from the United States and (ii) develop a multiplex SNP genotyping assay to differentiate L. monocytogenes isolates that carry virulence-attenuating PMSC mutations in inlA from virulent isolates that encode full-length InlA. The SNP genotyping assay was designed to detect all PMSC mutations in inlA that have been described previously (6, 12, 14, 26, 28, 29, 33), along with those discovered through this study. However, we were unable to verify the presence of two inlA PMSC mutations through inlA sequencing and SNP genotyping analyses of multiple DNA samples provided by a research group in France (33). Also, another research group from France described one additional putative inlA PMSC mutation in a single L. monocytogenes isolate from France after the present study was submitted for publication (30). In future work, we will attempt to confirm this newly described inlA PMSC mutation and incorporate this marker into the SNP genotyping assay developed in this study.
MATERIALS AND METHODS
Bacterial isolates for high-throughput invasion assay.
We assembled a large collection of L. monocytogenes isolates to represent the genetic diversity of this pathogen and different sources of isolation. While L. monocytogenes isolates from a variety of sources (e.g., humans with clinical infections, food, and various environments) were included in this isolate set, our efforts were focused on the well-characterized set of human clinical and food isolates described by Gray et al. (11). In addition to the isolates described by Gray et al. (11), we selected L. monocytogenes isolates from various environments, including urban and pristine environments (34), farm environments (27), and food-processing plant environments (17). Together, the isolates from these studies constitute a combined total of 1,731 L. monocytogenes isolates representing 90 unique EcoRI ribotypes (11, 17, 27, 34). Based on the frequency of ribotype occurrence, we then selected a representative subset of L. monocytogenes isolates (n = 203), from which isolates belonging to ribotypes previously associated with PMSC mutations in inlA reported by Nightingale et al. (26) were excluded. Specifically, we selected all isolates belonging to ribotypes that occurred in 5 or fewer isolates, 3 isolates per ribotype for ribotypes occurring at a frequency of between 6 and 50 isolates, and 4 isolates per ribotype for ribotypes occurring in >50 isolates. All isolates included in this subset are described in detail in Table S1 in the supplemental material.
High-throughput Caco-2 cell invasion assay screening.
The Caco-2 cell invasion phenotypes of the subset of 203 L. monocytogenes isolates described above were determined using a 24-well plate Caco-2 cell invasion assay designed to screen L. monocytogenes isolates for attenuated invasion. Briefly, L. monocytogenes cultures were grown in brain heart infusion broth (Becton Dickson, Sparks, MD) at 30°C for 18 h without shaking. Semiconfluent Caco-2 cells were infected with approximately 2 × 107 L. monocytogenes cells per well. After infection for 30 min at 37°C, nonadherent bacteria were removed by washing the wells three times with phosphate-buffered saline and then adding medium containing 150 μg of gentamicin/ml to kill extracellular bacteria. At 1.5 h postinfection, wells were washed three times with phosphate-buffered saline to remove traces of antibiotics before the lysing of host cells with cold deionized water. Intracellular L. monocytogenes cells were enumerated by plating appropriate serial dilutions onto brain heart infusion plates in duplicate. A standard laboratory control strain (10403S) (2) was included in each assay, and the invasion efficiency for this strain was set to 100%, while the invasion efficiencies of other L. monocytogenes isolates were expressed as percentages of that of intracellular 10403S. A second or third independent assay was performed to confirm the reproducibility of attenuated invasion efficiency (see Table S1 in the supplemental material).
DNA sequencing.
Full-length inlA sequencing was performed for L. monocytogenes isolates that demonstrated reproducibly attenuated invasion of Caco-2 cells (i.e., an average invasion efficiency of <50% of that of the laboratory control strain) (see Table S1 in the supplemental material). The preparation of the DNA template, PCR amplification, and the purification of PCR products were performed as detailed previously (8, 29), and DNA concentrations in purified PCR products were determined using a spectrophotometer (ND-1000; NanoDrop Technologies). DNA sequencing was performed at Cornell University's Biological Resource Center (Ithaca, NY) or Colorado State University's Proteomics and Metabolomics Facility (Fort Collins, CO) using previously described PCR primers and internal sequencing primers (29), BigDye Terminator chemistry, and AmpliTaq-FS DNA polymerase. Sequencing reaction mixtures were electrophoresed on an ABI PRISM 3730 or 3100 DNA analyzer. Nucleotide sequences were assembled and aligned with SeqMan and MegAlign software (DNAStar; Lasergene, Madison, WI), respectively. The sequencing of targeted inlA regions was also performed to confirm the presence of PMSC mutation types 4 through 7 and 12 in up to five isolates shown by SNP genotyping to carry those mutations. As we have demonstrated that a mutation leading to a PMSC in sigB is also associated with attenuated Caco-2 cell invasion, the sigB genes from two L. monocytogenes isolates that showed significantly attenuated Caco-2 cell invasion that could not be explained by mutations in inlA were sequenced using primers and conditions detailed in our earlier study (25). All sequences not associated with novel inlA PMSC mutations are available through the PathogenTracker database (www.pathogentracker.net).
Extension primer design for SNP genotyping assay.
Partial inlA sequences from L. monocytogenes isolates corresponding to each unique inlA PMSC mutation identified to date among isolates from the United States (26, 29; this study) and other countries (6, 12, 14, 28, 33), along with the inlA sequence from a wild-type L. monocytogenes strain, EGD-e (10), which carries an inlA allelic type encoding full-length InlA (10), were used to design extension primers for SNP genotyping (Table 1). Extension primers were designed to position the 3′-terminal nucleotide of each primer 1 base 5′ of the targeted SNP. An alignment of 40 full inlA sequences from a set of diverse L. monocytogenes isolates (29) was used to identify mismatches in extension primer sequences. Extension primers were designed to anneal to either the sense or the antisense strand in an attempt to avoid mismatches to the greatest extent possible (Table 2). When mismatches were unavoidable, the incorporation of degenerate nucleotides was found to greatly improve the annealing of extension primers to the template and, thus, the extension product peak intensity when samples were electrophoresed. A noncomplementary tail was added to the 5′ end of extension primers so that all primers differed by at least 6 nucleotides in length in order to facilitate the detection of multiple SNPs in a single reaction. A poly(G) tail was incorporated onto the 5′ end of extension primer US-G to permit the better separation of fluorescently labeled dideoxynucleoside triphosphate (ddNTP) extension products from primers US-F and US-G (Table 2). Due the length of extension primer US-G, this primer also required purification by polyacrylamide gel electrophoresis.
TABLE 1.
L. monocytogenes isolates used to develop an SNP genotyping assay to differentiate isolates with and without a PMSC in inlA
| Isolatea | Amino acid positionb corresponding to inlA PMSC (mutation type) | Reference |
|---|---|---|
| FSL F2-539 (wild type) | No inlA PMSC | 10 |
| Natural isolates | ||
| FSL F2-563 | 606 (1) | 26 |
| FSL R2-074 | 656 (2) | 26 |
| FSL F2-515 | 700 (3) | 26 |
| FSL F2-640 | 9 (4) | 29 |
| FSL R2-080 | 189 (5) | This study |
| FSL T1-045 | 492 (6) | This study |
| FSL T1-061 | 562 (7) | This study |
| NV8 | 460 (8) | 33 |
| NV7 | 519 (9) | 33 |
| NV4 | 677 (10) | 33 |
| NV5 | 685 (11) | 33 |
| L028 | 576 (12) | 14 |
| 36-25-1 | 527 (13) | 12 |
The isolate described as the wild type was a laboratory control strain (EGD-e) that carried full-length inlA, whereas isolates described as natural were isolated from food samples and carried naturally occurring mutations that led to a PMSC in inlA. We did not detect the presence of mutations leading to a PMSC in inlA from isolates NV4 and NV7, which were previously reported to harbor PMSC mutations 10 and 9, respectively (33), by using inlA sequencing or the SNP genotyping assay, even after obtaining multiple DNA samples.
Amino acid numbering is relative to the sequence of the wild-type control strain EGD-e.
TABLE 2.
Description of extension primers incorporated into the SNP genotyping assay developed to differentiate L. monocytogenes isolates with and without a mutation leading to a PMSC in inlA
| Primer name | Primer concnd (μM) | Targeted annealing strand | Multiplex reaction no. | PMSC mutation type | Product size (bp)a | Mutant allelic type (dye color)b | Wild-type allelic type (dye color)b | Primer sequence (5′ to 3′)c |
|---|---|---|---|---|---|---|---|---|
| US-A | 0.4 | Antisense | 1 | 3 | 31-32 | G (blue) | C (black) | GCA AAT GAY ATT ACG CTG TA |
| US-B | 0.2 | Antisense | 1 | 4 | 34-35 | C (black) | A (green) | [GCC] GGA GTG TAT ATA GTG AGA ARA AA |
| US-C | 0.2 | Antisense | 1 | 1 | 39-40 | A (green) | T (red) | [CTG CCT G]GG TTA TAC TTT CAA AGG CTG GTA |
| US-D | 0.2 | Antisense | 1 | 2 | 44-45 | T (red) | C (black) | [GAC CGA CCA AGC TCG] CAA CGA CTC AAG CAG TAG ACT AT |
| US-E | 0.2 | Antisense | 1 | 5 | 49-51 | T (red) | C (black) | [GAG ATC GGC AGA GCG GCG AGT A]GC TTT CAG GTT TAA CTA GTC TA |
| US-F | 2.0 | Antisense | 1 | 6 | 54-55 | T (red) | C (black) | [CGT ACA GAC TGG CGA TCG GCA TGC TAC T]AA CAT TTA GYG GAA CYG TGA CR |
| US-G | 0.2 | Sense | 1 | 7 | 62-63 | A (green) | G (blue) | [GGG GGG GTC GTC CTG GCT CAA TCT CGG CCT GGC TAG CT]C TGT GTA GCT GTT AAT ACT AAA TT |
| FR-A | 0.2 | Antisense | 2 | 8 | 30-31 | A (green) | G (blue) | ACA CAG AGC CTG ATA TAA CAT G |
| FR-B | 0.2 | Antisense | 2 | 9 | 33-34 | A (green) | G (blue) | [TTG AT]G CAA AGA AAC AAC CAA AGA AGT G |
| FR-C | 0.2 | Antisense | 2 | 10 | 39-40 | T (red) | A (green) | [AGC AGC AGC TAC G]AA WCA ACG ACT CAA RCA GTA G |
| FR-D | 0.2 | Antisense | 2 | 11 | 44-45 | A (green) | G (blue) | [CCA GCG AAT CAT GCC A]AC GAA AAA ACA GAT GGG AAA AAA T |
| FR-E | 1.0 | Antisense | 2 | 12 | 50-51 | T (red) | A (green) | [CTG CGC ACG ATC GAT GGC TCG ATC T]AA ACM GGY GGA ACT AAM TGG R |
| JP-A | 0.2 | Antisense | 2 | 13 | 57-58 | T (red) | A (green) | [GCA TCC GTC CAC TTC GAT CAA TCG CGC CGC TC]C TGA ACC AGC TAA GCC YGT A |
The sizes of the electrophoresed extension products will differ from the sizes expected based on primer lengths due to the influence of the weight of each particular fluorescent dye on the mobility shifting of the DNA fragments.
Colors indicate which fluorescent dye was incorporated into each ddNTP.
A noncomplementary tail (indicated by brackets and boldface type) was added to the 5′ end of extension primers to adjust the length to a distinct size in order to facilitate the detection of multiple SNPs in a single reaction. Degenerate nucleotides, indicated as follows, were incorporated into some extension primer sequences to accommodate polymorphic sites in targeted annealing regions: W, A + T; R, A + G; Y, C + T; and M, A + C.
Final primer concentration in SNaPshot extension reaction.
SNP genotyping.
The amplification of the full-length inlA gene was completed as described above (29). The preparation of the PCR template for primer extension and SNP genotyping was performed using the ABI PRISM SNaPshot multiplex kit according to the instructions of the manufacturer (Applied Biosystems, Foster City, CA). Amplicons were purified by treatment with shrimp alkaline phosphatase (Fermentas, Glen Burnie, MD) and exonuclease I (Fermentas). Purified inlA PCR products were subsequently used as DNA templates for single-base-pair extension by fluorescently labeled ddNTPs. Two separate multiplex reactions were constructed, including (i) reaction 1 with seven extension primers to detect all SNPs previously reported or shown here to result in an inlA PMSC among L. monocytogenes isolates from the United States and (ii) reaction 2 with six extension primers to detect SNPs leading to an inlA PMSC observed among isolates from countries other than the United States (Table 2). Primer concentrations in both reaction mixtures were adjusted to optimize the multiplex detection of peaks from all single-base extensions within each reaction (Table 2). Each extension reaction was completed using SNaPshot ready reaction mix, a purified full-length inlA gene PCR product, and pooled primers for reaction 1 or 2. Single-base-pair extension was performed in a thermal cycler, and positive and negative controls were prepared and included on each reaction plate. Following extension reactions, samples were treated with calf intestinal phosphatase (New England Biolabs, Ipswich, MA), combined with a GeneScan-120 LIZ size standard (Applied Biosystems) and Hi-Di (deionized) formamide (Applied Biosystems), and subsequently denatured. The amounts of the extension product and size standard in each reaction mixture were adjusted as necessary to optimize peak intensity. Lastly, samples were analyzed by capillary electrophoresis on an ABI 3100 DNA analyzer. Data analysis was performed using GeneScan version 3.7 software (Applied Biosystems), and specific bins and panels were constructed to distinguish wild-type and mutant allelic types in each reaction.
Implementation of the multiplex SNP genotyping assay to characterize a larger panel of L. monocytogenes isolates.
PathogenTracker is a World Wide Web-based database that contains epidemiological and molecular subtype data for more than 6,000 L. monocytogenes isolates (7). We queried PathogenTracker to determine the distribution of EcoRI ribotypes newly associated with inlA PMSC mutations among human clinical and food isolates (Table 3). We assessed the ability of the SNP genotyping assay (including reactions 1 and 2) to characterize a larger panel of isolates than that described above by determining the prevalence of inlA PMSC mutations among common ribotypes newly associated with these mutations. All ribotype DUP-1029A isolates described in PathogenTracker (n = 2) demonstrated attenuated Caco-2 cell invasion and were shown to carry PMSC mutation type 5 by inlA sequencing as described above. Ribotype DUP-1041A, DUP-1048A, and DUP-1056A isolates were not analyzed, as isolates representing these subtypes were rare in the entire database (i.e., data on ≤10 natural isolates of these ribotypes were available). Up to 30 isolates from (i) humans with clinical listeriosis and (ii) food products were selected to represent common ribotypes newly associated with inlA PMSCs, including DUP-1039C, DUP-1045B, and DUP-1062D. Altogether, the SNP genotyping assay was used to determine the prevalence of inlA PMSC mutations in 123 L. monocytogenes isolates from humans with listeriosis and from food ( Table 4).
TABLE 3.
Distribution of L. monocytogenes ribotypes associated with inlA PMSC mutation types identified in this study among isolates in the PathogenTracker database
| EcoRI ribotypea | Mutation type(s) | No. (%) of isolates in PathogenTrackerb obtained from:
|
Total no. (%) of isolates | ||
|---|---|---|---|---|---|
| Humansc | Foodd | Other | |||
| DUP-1029A | 5 | 0 (0.00) | 2 (0.11) | 0 (0.00) | 2 (0.02) |
| DUP-1039C | 7, 4, and 12 | 56 (3.96) | 115 (6.08)* | 275 (5.06) | 446 (5.10) |
| DUP-1041A | 4 | 0 (0.00) | 3 (0.16) | 0 (0.00) | 3 (0.03) |
| DUP-1045B | 4 | 20 (1.41) | 26 (1.37) | 61 (1.12) | 107 (1.22) |
| DUP-1048A | 7 | 0 (0.00) | 0 (0.00) | 3 (0.05) | 3 (0.03) |
| DUP-1056A | 4 | 2 (0.14) | 5 (0.26) | 3 (0.05) | 10 (0.11) |
| DUP-1062D | 5 and 6 | 4 (0.28) | 38 (2.01)* | 15 (0.28) | 57 (0.65) |
| Total in PathogenTracker | 1,415 | 1,892 | 5,439 | 8,746 | |
All isolates were of lineage II, assigned based on ribotypes as described previously (38).
Numbers indicate the numbers of isolates present in PathogenTracker on 28 March 2008; detailed source information on isolates can be obtained at www.pathogentracker.net.
The majority of human isolates (>90%) were obtained from patients in North America with invasive listeriosis.
The majority of food isolates (>95%) were obtained from North America. Asterisks indicate the significant overrepresentation (P < 0.01) of L. monocytogenes isolates belonging to a given ribotype.
TABLE 4.
Prevalence of PMSCs in inlA among five EcoRI ribotypes associated with inlA PMSC mutations identified in this study
| Ribotypea and inlA PMSC mutation type | Molecular serogroup(s) (no. of isolates characterized)b | No. of inlA PMSC mutations (no. of isolates screened) among isolates obtained fromc:
|
Total | % Prevalence of inlA PMSCs among all isolates belonging to ribotype | |
|---|---|---|---|---|---|
| Humans | Food | ||||
| DUP-1039C | |||||
| 7 | 1 (7) | 0 (29) | 7 (28) | ||
| 4 | 3 (3) | 4 (29) | 10 (28) | ||
| 12 | 3 (2) | 1 (29) | 1 (28) | ||
| Total | 1 and 3 | 5 (29) | 18 (28)** | 23 (57) | 40.4 |
| DUP-1045B | 1 (10) | 1 (17) | 12 (17)** | 13 (34) | 38.2 |
| DUP-1062D | |||||
| 6 | 1 (8) | 0 (4) | 11 (28) | ||
| 5 | 1 (2) | 0 (4) | 2 (28) | ||
| Total | 1 | 0 (4) | 13 (28)* | 13 (32) | 40.7 |
DUP-1029A isolates were not characterized by the SNP genotyping assay because all DUP-1029A isolates available were shown by DNA sequencing to carry an inlA PMSC. Isolates belonging to ribotypes DUP-1041A, DUP-1048A, and DUP-1056A were not characterized by the SNP genotyping assay, as these ribotypes were very rare among isolates in the PathogenTracker database. Ribotype DUP-1039C isolates were associated with three PMSC mutations corresponding to separate serotypes. Ribotype DUP-1062D isolates were associated with two PMSC mutations that corresponded to the same serotype.
Serogroup 1 isolates were of serotype 1/2a or 3a, and serogroup 3 isolates were of serotype 1/2c or 3c. Up to 10 isolates belonging to each common ribotype newly associated with an inlA PMSC were characterized by molecular serotyping as described by Doumith et al. (4).
A total of 123 isolates, including 57 DUP-1039C isolates, 34 DUP-1045B isolates, and 32 DUP-1062D isolates, were screened by reactions 1 and 2 of the SNP genotyping assay. Asterisks indicate significant (**, P < 0.001) or marginal (*, P = 0.11) overrepresentation of L. monocytogenes isolates carrying an inlA PMSC. More than one inlA PMSC was present in some ribotypes, and statistical analysis was based on the detection of any PMSC in isolates belonging to each ribotype.
Molecular serotyping.
A multiplex PCR assay was performed as described by Doumith et al. (4) to determine the molecular serogroups of select isolates belonging to common ribotypes newly associated with inlA PMSC mutation types 4 through 7 and 12. Each of the previously described multiplex PCR profile groups (groups 1 to 4) (4) includes L. monocytogenes isolates belonging to more than one serotype, with group 1 comprising serotypes 1/2a and 3a, group 2 comprising serotypes 1/2b, 3b, and 7, group 3 comprising serotypes 1/2c and 3c, and group 4 comprising serotypes 4b, 4d, and 4e, as noted previously (4). L. monocytogenes isolates classified as serotypes 1/2a, 1/2b, 1/2c, and 4b by conventional serotyping were included as controls in each multiplex PCR assay. L. monocytogenes isolates were assigned to a serogroup based on the observed multiplex PCR profile for each isolate.
Statistical analysis.
All statistical analyses were performed using Statistical Analysis Systems (SAS) software (version 9.1; SAS Institute, Cary, NC), and P values of <0.05 were considered to be statistically significant in all cases. A one-sample t test, as implemented using the t test procedure in SAS, was used to compare the average invasion efficiency of each L. monocytogenes isolate to that of the standard laboratory control strain 10403S, which was set at 100%. A chi-square test of independence or Fisher's exact test (as appropriate) was implemented using the SAS frequency procedure to describe the distribution of common EcoRI ribotypes associated with inlA PMSC mutation types 4 through 7 and 12 among human clinical and food isolates listed in PathogenTracker. Fisher's exact test was used to analyze the distribution of L. monocytogenes isolates carrying an inlA PMSC among human clinical and food isolates belonging to commonly occurring ribotypes newly associated with inlA PMSCs.
Nucleotide sequence accession numbers.
Sequences for isolates identified as carrying novel inlA PMSC mutations were deposited in GenBank under accession no. EU699430 to EU699434.
RESULTS AND DISCUSSION
We assembled an expansive L. monocytogenes isolate collection to screen for the presence of additional PMSC mutations in inlA among isolates from the United States. Caco-2 cell invasion assays and the sequencing of inlA from isolates with attenuated invasion phenotypes revealed three novel PMSC mutations in inlA (i.e., mutation types 5 through 7) that had not been identified previously among L. monocytogenes isolates from the United States. We then developed a multiplex SNP genotyping assay that was designed to detect mutations leading to PMSCs in inlA that have been reported worldwide and verified in this work to date. We assessed the ability of the SNP genotyping assay to characterize a larger panel of isolates through the implementation of this assay to determine the prevalence of inlA PMSCs among human clinical and food isolates belonging to common ribotypes newly associated with these mutations. SNP genotyping revealed a fourth novel inlA PMSC mutation among L. monocytogenes isolates from the United States which had been discovered previously among isolates from France and Portugal (6, 14). Consistent with the findings of our previous studies, results from the present study showed that ribotypes include isolates with and without PMSC mutations in inlA and that the inlA PMSC mutations identified in the present work were common among food isolates but rare among isolates from listeriosis patients (26). The multiplex SNP genotyping assay developed in this study can be utilized to differentiate virulence-attenuated isolates carrying PMSC mutations in inlA from fully virulent L. monocytogenes isolates encoding full-length InlA.
Combined Caco-2 cell invasion screening and targeted inlA sequencing lead to the identification of novel PMSCs in inlA among L. monocytogenes isolates from the United States.
Previous studies identified four mutations resulting in a PMSC in inlA (termed mutation types 1 through 4) among L. monocytogenes isolates from the United States (26, 29). In the present study, we assembled a collection of >1,700 L. monocytogenes isolates representing 90 unique EcoRI ribotypes from diverse sources to screen for additional PMSC mutations in inlA among isolates from the United States. A subset (n = 203) of the large isolate collection was then selected to represent each unique ribotype based on the frequency of occurrence, and the Caco-2 cell invasion phenotypes of this subset of isolates were determined (see Table S1 in the supplemental material). Results from Caco-2 cell invasion assays revealed that 24 of 203 isolates showed a reproducibly attenuated invasion phenotype, defined by an average normalized invasion efficiency of <50% (Fig. 1). The average invasion efficiencies for these 24 isolates ranged from 0.07 to 49.6% when normalized to that of the laboratory control strain 10403S, which was set at 100% (Fig. 1). Fifteen of these 24 isolates demonstrated significantly attenuated invasion efficiencies (P < 0.05; one-sample t test) compared to that of the standard laboratory control strain 10403S. Since previous studies showed that an attenuated Caco-2 cell invasion phenotype may be attributed to the presence of a mutation leading to a PMSC in inlA (6, 26, 28, 29, 33), full-length inlA sequencing was performed to screen for the presence of inlA PMSCs in all 24 isolates demonstrating attenuated Caco-2 cell invasion.
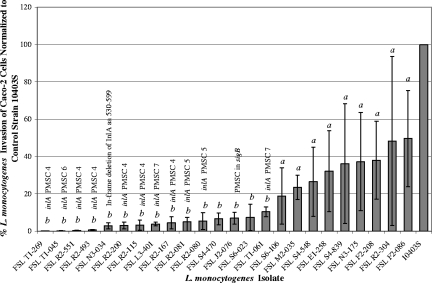
FIG. 1.
Caco-2 cell invasion efficiencies of 24 L. monocytogenes isolates, from the subset of 203 isolates characterized by the Caco-2 cell invasion assay described herein, that demonstrated reproducibly attenuated invasion (see Table S1 in the supplemental material). The invasion efficiencies of L. monocytogenes isolates were normalized to that of a standard laboratory control strain, 10403S (the level of invasion by this strain was set at 100%), which was included in each invasion assay. Columns represent the average invasion efficiency observed for each isolate, and error bars indicate the minimum and maximum invasion efficiencies observed for each isolate. The presence of PMSC types 4 through 7 is noted above the names of the isolates carrying each respective mutation. A one-sample t test was applied to compare the invasion efficiency of each isolate to that of 10403S. Different letters indicate the presence or absence of statistical significance, defined by P of <0.05. Specifically, isolates denoted by a did not show significantly reduced invasion (P > 0.05), whereas isolates denoted by b showed significantly reduced efficiencies of invasion (P < 0.05) of Caco-2 cells compared to that of 10403S. All L. monocytogenes isolates harboring PMSC mutations in inlA had significantly reduced invasion (P < 0.02) of Caco-2 cells. FSL J2-076 displayed attenuated invasion of Caco-2 cells that was associated with the presence of a PMSC in sigB (31) rather than a PMSC in inlA.
The virulence-associated gene inlA was successfully amplified from all 24 L. monocytogenes isolates demonstrating attenuated invasion, and analyses of inlA sequence data revealed the presence of three novel PMSC mutations (mutation types 5 through 7) that had not been identified previously among isolates from the United States (Fig. 2). The mutation leading to inlA PMSC type 6 was previously observed among L. monocytogenes isolates from France (28) and Portugal (6). Also, a previously described 5′-end frameshift mutation resulting in inlA PMSC type 4 (29) was detected in six isolates belonging to ribotypes not previously associated with this mutation (Fig. 1). All 11 isolates shown to carry a PMSC mutation in inlA demonstrated average invasion efficiencies that were <10% of that observed for 10403S (P < 0.05) (Fig. 1). Four isolates demonstrated significantly attenuated invasion of Caco-2 cells that could not be explained by the presence of a PMSC mutation in inlA. However, one of these isolates (FSL N3-034) contained a large in-frame deletion of InlA aa 530 to 599, which may explain the reduced invasion efficiency of this isolate. The generation of isogenic strains with and without this deletion mutation by allelic exchange mutagenesis and the further characterization of these isogenic strains by Caco-2 cell invasion assays will be necessary to determine if this deletion mutation is responsible for reduced invasion efficiency. Attenuated Caco-2 cell invasion by a rare lineage III isolate (FSL J2-076) was not associated with a PMSC mutation in inlA but can likely be attributed to the presence of a previously identified PMSC mutation in sigB (30), an alternative sigma factor gene shown to contribute to the invasion of Caco-2 cells through the regulation of InlA expression (16). Two isolates (FSL S4-480 and FSL S6-023) demonstrated significantly reduced invasion of Caco-2 cells that could not be explained by mutations leading to a PMSC in inlA or sigB. Further work is needed to probe the underlying genetic mechanisms associated with the observed attenuated invasion phenotype of these two isolates.
FIG. 2.
Full-length InlA (A) and locations of PMSC mutations in InlA as identified previously by research groups in France (D, E, F, H, J, M, and N), Japan (G), Portugal (B, E, and J), and the United States (B, C, E, I, J, K, L, and O). The map of full-length InlA (A) represents the sequence for L. monocytogenes strain EGD-e. N, N-terminal end; S, signal sequence; LRR, leucine-rich repeat; IR, intergenic repeat; MA, membrane anchor; C, C-terminal end. Numbers below the EGD-e full-length InlA sequence represent amino acid positions. Numbers to the right of sequence diagrams B through O represent the amino acid position of each respective PMSC mutation relative to the sequence of EGE-e. References cited are as follows: Glaser et al., 2001, reference 10; Orsi et al., 2007, reference 29; Felicio et al., 2007, reference 6; Rousseaux et al., 2004, reference 33; Olier et al., 2002, reference 28; Handa-Miya et al., 2007, reference 12; Ragon et al., 2008, reference 30; Jonquieres et al., 1998, reference 14; and Nightingale et al., 2005, reference 26.
Along with other previously described inlA PMSC mutations (6, 12, 14, 26, 28, 29, 30, 33), the novel inlA PMSC mutations among L. monocytogenes isolates from the United States identified here through inlA sequencing (PMSC mutation types 5 through 7) are located upstream of the region encoding the C-terminal LPXTG membrane-anchoring motif (Fig. 2). Specifically, inlA sequencing revealed that two isolates (FSL R2-080 and FSL R2-081) carried a nonsense mutation (mutation type 5) at nucleotide 565 (relative to the EGD-e sequence), where a cytosine was converted to a thymidine, creating a PMSC (TAG) at codon 189. A second nonsense mutation in inlA (mutation type 6), which also converted a cytosine to a thymidine, was discovered in FSL T1-045 at nucleotide 1474 and resulted in a PMSC (TAG) at codon position 492. Two isolates (i.e., FSL L3-401 and FSL T1-061) carried a third nonsense mutation in inlA (mutation type 7) at nucleotide 1684, where a cytosine was replaced by a thymidine, resulting in a PMSC (TAA) at codon position 562 (Fig. 2). Six isolates contained a 5′-end frameshift mutation in a homopolymeric tract of adenine residues (PMSC mutation type 4) that was observed previously among L. monocytogenes isolates from the United States (29). Several distinct mutations leading to a PMSC in inlA and, thus, the production of a truncated form of InlA have been accumulated worldwide (Fig. 2). Interestingly, inlA PMSC mutations observed to date among L. monocytogenes isolates from the United States (with the exception of mutation types 6 and 12) differ from the mutations observed among isolates from other countries (6, 12, 14, 26, 28, 29, 30, 32), supporting the idea that mutations leading to loss of a full-length InlA arose independently.
L. monocytogenes isolates from the United States carrying inlA PMSC mutation types 5 through 7 belonged to four EcoRI ribotypes (Fig. 2). Specifically, inlA PMSC mutation type 5 was observed among isolates belonging to ribotypes DUP-1029A and DUP-1062D, PMSC mutation type 6 was present among ribotype DUP-1062D isolates, and PMSC mutation type 7 was found among ribotype DUP-1039C and DUP-1048A isolates (Fig. 2). In addition, targeted inlA sequencing revealed that PMSC mutation type 4, which was found previously only among ribotype DUP-1039C isolates (29), is also carried in ribotype DUP-1041A, DUP-1045B, and DUP-1056A isolates (Fig. 2), indicating hypermutability in the 5′-end homopolymeric tract of adenine residues where this mutation occurs (29). All ribotypes associated with inlA PMSC mutations newly identified among L. monocytogenes isolates from the United States in this study belonged to genetic lineage II, and our previous studies showed that a significant proportion of lineage II isolates carry PMSC mutations in inlA, providing evidence for the underrepresentation of lineage II subtypes in human clinical cases (11, 26). PathogenTracker queries revealed that while some ribotypes associated with newly identified inlA PMSCs were very rare among isolates in the entire database (i.e., ribotypes DUP-1029A, DUP-1041A, DUP-1048A, and DUP-1056A), others were common enough to allow statistical analysis to describe their distribution in human clinical and food isolate categories. Specifically, ribotypes DUP-1039C and DUP-1062D were isolated from food products significantly more commonly (P < 0.01) than from humans with listeriosis (Table 3). In a previous study, common L. monocytogenes EcoRI ribotypes associated with PMSCs in inlA (i.e., DUP-1039C, DUP-1045B, DUP-1052A, DUP-1062A, and DUP-1062D) accounted for 57% of 502 L. monocytogenes isolates obtained from >30,000 ready-to-eat food samples (11). While certain ribotypes (e.g., DUP-1062A) have been shown previously to exclusively be comprised of isolates with PMSC mutations in inlA, other ribotypes include isolates with and without these mutations (26). Thus, we developed an SNP genotyping assay to differentiate L. monocytogenes isolates with any of the currently identified and confirmed inlA PMSCs from isolates encoding full-length InlA at the nucleotide level.
A multiplex high-throughput SNP genotyping assay simultaneously detects PMSC mutations in inlA.
While epidemic clone strains I and II have been linked to the majority of listeriosis outbreaks worldwide (15) and are common in sporadic listeriosis cases (11), a significant proportion of L. monocytogenes isolates from foods carry a PMSC mutation in inlA that has been shown to be responsible for attenuated mammalian virulence (11, 13, 23, 26). SNPs leading to a PMSC in inlA thus represent suitable genetic markers to identify L. monocytogenes isolates with limited human virulence due to these mutations. We developed a multiplex SNP genotyping assay using the SNaPshot multiplex kit to detect all 13 mutations leading to PMSCs in inlA that had been reported worldwide and verified in this work to date (6, 12, 14, 26, 28, 29, 32), including those newly identified in this study. The SNaPshot multiplex kit relies on single-base-pair extension, or “minisequencing” chemistry, in which Taq polymerase extends each unlabeled extension primer with a single fluorescently labeled ddNTP to differentiate inlA allelic types resulting from the individual targeted SNPs (i.e., mutant allelic types carrying a PMSC mutation and wild-type allelic types encoding full-length InlA).
The SNP genotyping assay developed in this study successfully differentiated 11 of the 13 mutant inlA allelic types carrying an inlA PMSC relative to the wild-type inlA allele, which encodes full-length InlA (Fig. 3). We did not detect the presence of mutations leading to a PMSC in inlA in isolates NV4 and NV7, which were previously reported to harbor PMSC mutations 10 and 9, respectively (33), by using the SNP genotyping assay or inlA sequencing, even after obtaining multiple DNA samples. Peaks for all seven extension products in reaction 1 and all six extension products in reaction 2 were detected for all isolates used to develop this assay. Bins and panels created in GeneScan (version 3.7; Applied Biosystems) permitted automated differentiation between wild-type and mutant inlA allelic types based on peak color and size (Fig. 3). As more sequence data become available, it is reasonably likely that additional SNPs leading to a PMSC in inlA will be discovered. One of the major reasons we chose the SNaPshot multiplex kit is the ease through which the assay developed in this study can be modified to accommodate the detection of additional genetic markers by simply designing new extension primers and combining them into another multiplex reaction mixture.
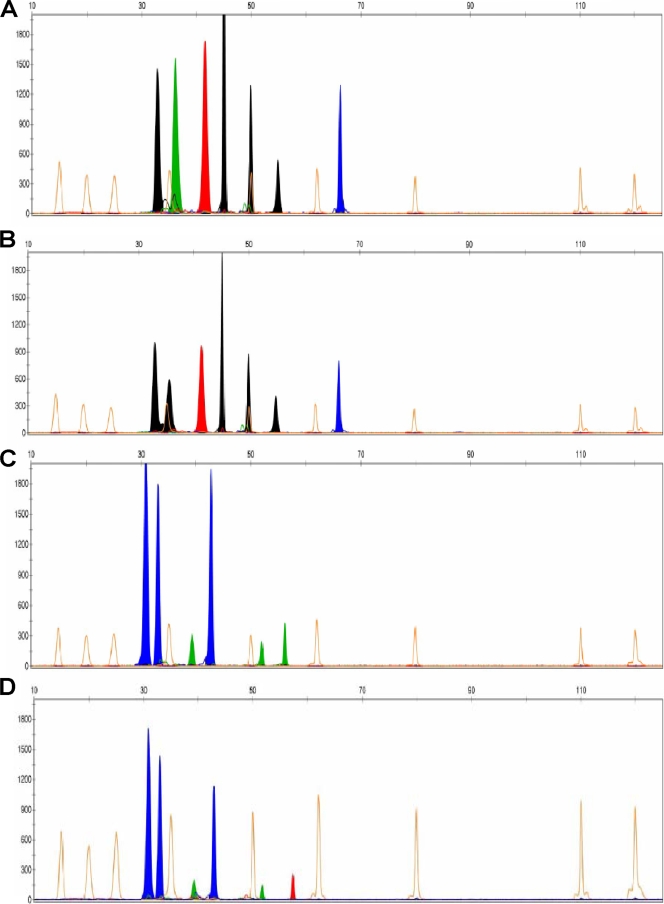
FIG. 3.
Electropherograms for multiplex SNaPshot reactions analyzed by GeneScan software. The upper x axis represents the sizes of extension product peaks relative to the GeneScan-120 LIZ size standard (Applied Biosystems), which was included in each SNaPshot reaction. The y axis represents the relative peak intensity. Orange peaks represent the GeneScan-120 LIZ size standard. Black peaks represent an extension product in which a fluorescently labeled ddCTP was incorporated onto the 3′ end of the extension primer. Similarly, green peaks represent the incorporation of ddATPs, red peaks represent the incorporation of ddTTPs, and blue peaks represent the incorporation of ddGTPs. Electropherogram A illustrates the multiplex SNP genotyping peak profile from reaction 1 for wild-type L. monocytogenes EGD-e. Electropherogram B represents the reaction 1 peak profile for an L. monocytogenes isolate carrying inlA PMSC mutation type 4. Electropherogram C shows the SNP genotyping peak profile from reaction 2 for wild-type L. monocytogenes EGD-e. Electropherogram D depicts the reaction 2 peak profile for an L. monocytogenes isolate carrying PMSC mutation type 13.
While previous studies described the presence of virulence-attenuating mutations in other key L. monocytogenes virulence-associated genes (i.e., actA, hly, and inlB), only a single isolate or a few isolates were reported to carry these mutations (3, 20, 24, 32). Mutations leading to the truncation of virulence factors other than InlA were not included in the SNP genotyping assay developed in this work because these mutations do not appear to have been accumulated at the population level and have not been shown to be responsible for attenuated virulence through the characterization of isogenic mutants in appropriate animal infection models. Although another recent study developed an SNP typing assay for L. monocytogenes, this assay was designed to characterize exclusively lineage I isolates and includes only a single genetic marker to differentiate isolates with attenuated virulence due to a PMSC mutation in inlA (5). Each SNP included in the SNP genotyping assay developed in this study leads to a PMSC in inlA, and this assay thus has the ability to differentiate virulence-attenuated L. monocytogenes isolates from epidemic clones and other strains encoding full-length InlA, which have been responsible for the majority of human illnesses and deaths.
Since ribotypes associated with a PMSC mutation in inlA were previously reported to include isolates with and without each respective PMSC in inlA, we utilized the SNP genotyping assay developed in this study to determine the prevalence of PMSC mutations in inlA among isolates belonging to common ribotypes newly associated with these mutations. Specifically, we characterized 123 human clinical and food isolates belonging to ribotypes DUP-1039C, DUP-1045B, and DUP-1062D to assess the ability of the SNP genotyping assay to characterize a larger panel of isolates. All 123 L. monocytogenes isolates were typeable by the SNP genotyping assay, and peaks representing all seven extension primers in reaction 1 and all six extension primers in reaction 2 were detected for all isolates. The sequencing of targeted regions of inlA was performed to confirm the presence of PMSC mutation types 4 through 7 and 12 for up to five isolates shown by SNP genotyping to carry each respective mutation. Results from inlA sequencing validated the ability of the SNP genotyping assay to detect inlA PMSC mutations.
Interestingly, the SNP genotyping of this set of 123 U.S. L. monocytogenes isolates revealed the presence of a fourth inlA PMSC (PMSC mutation type 12) novel among isolates from the United States in two ribotype DUP-1039C isolates (Table 4); PMSC mutation type 12 had previously been observed only among isolates from France and Portugal (Fig. 2). Consistent with the findings of our previous study (26), results from the SNP genotyping assay developed in the present study showed that ribotypes DUP-1039C, DUP-1045B, and DUP-1062D include isolates with and without mutations leading to a PMSC in inlA (Table 4). Cumulatively, 40.4, 38.2, and 40.7% of L. monocytogenes isolates representing ribotypes DUP-1039C, DUP-1045B, and DUP-1062D, respectively, carried a PMSC mutation in inlA (Table 4). Fisher's exact test showed that among isolates representing ribotypes DUP-1039C (P < 0.001), DUP-1045B (P < 0.001), and DUP-1062D (P = 0.11), the inlA PMSC mutation types were either significantly or marginally more common among food isolates than among isolates from humans with listeriosis. Along with the results of previous molecular epidemiology studies based on large representative L. monocytogenes isolate collections (see, e.g., references 13 and 26), our data contribute to an emerging body of evidence that L. monocytogenes isolates carrying a PMSC mutation in inlA are commonly present in food but appear to cause disease only on very rare occasions.
Interestingly, results from this study also demonstrated that a given ribotype may be associated with multiple distinct mutations resulting in a PMSC in inlA. For example, ribotype DUP-1039C L. monocytogenes isolates from the United States carry inlA PMSC mutation types 4, 7, and 12, while ribotype DUP-1062D isolates from the United States carry inlA PMSC mutation types 5 and 6 (Table 4). Select isolates belonging to ribotypes newly associated with a PMSC in inlA and subsequently shown by the SNP genotyping assay to carry a PMSC in inlA were grouped into a molecular serogroup by using a previously described multiplex PCR (4). All isolates clustered into a molecular serogroup associated with serotypes 1/2a and 3a (group 1) or 1/2c and 3c (group 3) (Table 4). Ribotype DUP-1039C, which carries more than one inlA PMSC, included isolates belonging to more than one serogroup (Table 4). These results are consistent with those of previous studies demonstrating that inlA PMSC mutations are observed most commonly among serotype 1/2a and 1/2c isolates (6, 13, 26). In addition, our previous study showed that inlA PMSC mutations may occasionally be found among serotype 1/2b isolates (26).
Conclusions.
A study recently completed by our group showed that mutations leading to a PMSC in inlA are responsible for attenuated mammalian virulence (23). Other previous molecular epidemiology studies reported that a significant proportion of L. monocytogenes strains isolated from food products carry PMSC mutations in inlA (11, 13, 26). Taken together, the results of these studies indicate that there is a significant subpopulation of L. monocytogenes strains in the food supply with a limited ability to cause human disease. We therefore developed a multiplex SNP genotyping assay that can rapidly detect SNPs leading to PMSCs in inlA, which were verified in the present study, to distinguish virulence-attenuated L. monocytogenes isolates from strains that have been linked to the majority of epidemic and sporadic listeriosis cases. The combined experimental approaches employed in the present study revealed the presence of four inlA PMSC mutations that had not been identified previously among L. monocytogenes isolates from the United States. The multiplex SNP genotyping assay developed in this work permits the rapid detection of virulence-attenuated L. monocytogenes isolates carrying a PMSC in inlA and provides a DNA sequence-based molecular subtyping tool that will be particularly useful for routine surveillance and outbreak investigations. Ultimately, the utilization of this assay for these purposes may lead to the inclusion of strain-specific virulence parameters into future risk assessments and facilitate the future revision of science-based regulations regarding the presence of L. monocytogenes strains with defined virulence characteristics in food.
Supplementary Material
Acknowledgments
The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number no. 2005-35201-16266.
We are indebted to Martin Wiedmann from the Department of Food Science at Cornell University for helpful discussions and for providing L. monocytogenes isolates to develop and validate the assay. We thank Claire Broton for inlA sequencing of invasion-attenuated isolates. We also thank Pascal Piveteau from the Laboratoire de Microbiologie for the generous gift of DNA from strains NV4, NV5, NV7, and NV8 and Bon Kimura from Tokyo University of Marine Science and Technology for kindly providing DNA from strain 36-25-1. We are also grateful for technical support and for assistance with the GeneScan software provided by David Gingrich and Jason Rivest (Proteomics and Metabolomics Facility, Colorado State University).
Footnotes
Published ahead of print on 3 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156-1166. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 3.Chakraborty, T., F. Ebel, J. Wehland, J. Dufrenne, and S. Notermans. 1994. Naturally occurring virulence-attenuated isolates of Listeria monocytogenes capable of inducing long term protection against infection by virulent strains of homologous and heterologous serotypes. FEMS Immunol. Med. Microbiol. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Doumith, M., C. Cazalet, N. Simones, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducey, T. F., B. Page, T. Usgaard, M. K. Borucki, K. Pupedis, and T. J. Ward. 2007. A single-nucleotide-polymorphism-based multilocus genotyping assay for subtyping lineage I isolates of Listeria monocytogenes. Appl. Environ. Microbiol. 73:133-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felicio, M. T., T. Hogg, P. Gibbs, P. Teixeira, and M. Wiedmann. 2007. Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fugett, E., E. Fortes, C. Nnoka, and M. Wiedmann. 2006. International Life Sciences Institute North American Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929-2938. [DOI] [PubMed] [Google Scholar]
- 8.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 9.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusnoik, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simones, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 11.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handa-Miya, S., B. Kimura, H. Takahashi, M. Sato, T. Ishikawa, K. Igarashi, and T. Fugii. 2007. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117:312-318. [DOI] [PubMed] [Google Scholar]
- 13.Jacquet, C., M. Doumith, J. I. Gordon, P. M. V. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 14.Jonquieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes L028 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lappi, V. R., J. Thimothe, K. K. Nightingale, K. Gall, V. N. Scott, and M. Wiedmann. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J. Food Prot. 67:2500-2514. [DOI] [PubMed] [Google Scholar]
- 18.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1724. [DOI] [PubMed] [Google Scholar]
- 19.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leimeister-Wachter, M., W. Goebel, and T. Chakraborty. 1989. Mutations affecting hemolysin production in Listeria monocytogenes located outside the listeriolysin gene. FEMS Microbiol. Lett. 53:23-29. [DOI] [PubMed] [Google Scholar]
- 21.McLauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 22.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nightingale, K. K., R. A. Ivy, A. J. Ho, E. D. Fortes, B. L. Njaa, R. M. Peters, and M. Wiedmann. 12 September 2008. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. doi: 10.1128/AEM.00997-08. [DOI] [PMC free article] [PubMed]
- 24.Nightingale, K. K., S. R. Milillo, R. A. Ivy, A. J. Ho, H. F. Oliver, and M. Wiedmann. 2007. Listeria monocytogenes F2365 is characterized by an elevated frequency of truncated genes, including inlB, and by unique phenotypic characteristics that limit its appeal to represent human outbreak-associated strains. J. Food Prot. 70:482-488. [DOI] [PubMed] [Google Scholar]
- 25.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes from human and animal cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes stubypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 29.Orsi, R. H., D. Ripoll, M. Yeung, K. K. Nightingale, and M. Wiedmann. 2007. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology 153:2666-2678. [DOI] [PubMed] [Google Scholar]
- 30.Ragon, M., T. Wirth, F. Hollandt, R. Lavenir, M. Lecuit, A. Le Monnier, and S. Brisse. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, A., K. K. Nightingale, G. Jeffers, E. Fortes, J. M. Kongo, and M. Wiedmann. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685-693. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, A., Y. Chan, and M. Wiedmann. 2005. Definition of genetically distinct attenuation mechanisms in naturally virulence-attenuated Listeria monocytogenes by comparative cell culture and molecular characterization. Appl. Environ. Microbiol. 71:3900-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousseaux, S., M. Olier, J. P. Lamaitre, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauders, B. D., M. Z. Durak, E. Fortes, K. Windham, Y Schukken, A. J. Lembo, Jr., B. Akey, K. K. Nightingale, and M. Wiedmann. 2006. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 69:93-105. [DOI] [PubMed] [Google Scholar]
- 35.Schlech, W. F., III. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Charkraborty, G. Dominguez-Bernal, W. Goebel, B. Zorn-Gonzalez, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 15:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.