Abstract
Several Bacillus and Paenibacillus species were isolated from Fe and Mn oxide minerals precipitating at a deep subsurface oxic-anoxic interface at Henderson Molybdenum Mine, Empire, CO. The isolates were investigated for their Mn(II)-oxidizing potential and interrogated for possession of the mnxG gene, a gene that codes for a putative Mn(II)-oxidizing enzyme in Bacillus species. Seven of eight Bacillus species were capable of Mn(II) oxidation; however, the mnxG gene was detected in only one isolate. Using sequences of known Bacillus species both with and without amplifiable mnxG genes and Henderson Mine isolates, the 16S rRNA and mnxG gene phylogenies were compared to determine if 16S rRNA sequences could be used to predict the presence or absence of an amplifiable mnxG gene within the genomes of the isolates. We discovered a strong correspondence between 16S rRNA sequence similarity and the presence/absence of an amplifiable mnxG gene in the isolates. The data revealed a complex phylogenetic distribution of the mnxG gene in which vertical inheritance and gene loss influence the distribution of the gene among the Bacillus species included in this study. Comparisons of 16S rRNA and functional gene phylogenies can be used as a tool to aid in unraveling the history and dispersal of the mnxG gene within the Bacillus clade.
The comparison of 16S rRNA and functional gene phylogenies for a common set of taxa is a useful tool for attempting to understand the evolutionary history of genomes. Similarities in tree topologies are interpreted as representing the vertical inheritance of genes (23, 27, 28), while incongruities can signify the occurrence of lateral gene transfer events (14), simultaneous evolution of genes with similar functions in different organisms (28), divergence of genes from a common origin (5), or gene loss (20). If positive correlations can be made between 16S rRNA gene sequences and the presence or absence of functional genes, then 16S rRNA surveys of environmental samples can be used to understand the functional potential of the community.
The microbial oxidation of manganese (Mn) influences a number of biogeochemical processes, since Mn(III/IV) oxides are chemically reactive and can (i) sequester numerous trace metals through sorption processes, (ii) serve as terminal electron acceptors for microbial respiration, and (iii) directly oxidize complex organic matter (7, 8, 30, 33, 35). Thus, to unravel several linked chemical cycles, it is important to understand the evolution of microbial Mn(II) oxidation capabilities and the microorganisms and genes involved.
In Bacillus species, mnxG, a putative multicopper oxidase, acts to catalyze the oxidation first of Mn(II) to Mn(III) and then of Mn(III) to Mn(IV) (6, 39). mnxG is localized in the exosporium, a layer that occurs outside of the spore coat in some Bacillus species (11, 12, 38). Mn-oxidizing Bacillus strains have been obtained from coastal surface sediments near San Diego, CA; in sediments and particulate plumes at deep-sea hydrothermal vents in the Guaymas Basin in the Gulf of California (6, 13, 24); and now at the Henderson Molybdenum Mine in Empire, CO.
The Henderson Mine was proposed as a site for a Deep Underground Science and Engineering Laboratory (DUSEL) in 2003 and has only recently been investigated for its potential to harbor unique microbial life in the deep subsurface (29). Anoxic, CO2- and metal-rich brines are released into aerated tunnels through boreholes at depths of >1 mile below the surface. Millimolar concentrations of Mn were detected in the fluids, and Mn oxide minerals were precipitating around the mouths of the boreholes. Eight Bacillus sp. isolates capable of Mn(II) oxidation were isolated from the mineral precipitates, while the mnxG gene was amplified from only one of these Mn-oxidizing isolates.
The inability to amplify mnxG from the other isolates led us to question if these isolates were more phylogenetically distinct from the mnxG-positive isolate than they were from each other and, if so, if 16S rRNA sequences could be used to predict the presence or absence of an amplifiable mnxG gene within the genomes of the Henderson Mine isolates. We hypothesized that the isolates would be dispersed throughout the Bacillus clade and predicted that the mnxG-positive isolate, HM06-02, would cluster with other mnxG-positive Bacillus isolates. We also predicted that their placement within the 16S rRNA phylogeny would relate to the presence or absence of mnxG within the genome of the isolates. We discovered a strong relationship between 16S rRNA sequence similarity and the presence or absence of an amplifiable mnxG gene in the Henderson Mine isolates. Additionally, the data revealed a complex phylogenetic distribution of the mnxG gene that we can use as a tool to aid in unraveling the history and dispersal of the mnxG gene within the Bacillus clade.
MATERIALS AND METHODS
Site description and sample collection.
Mn-oxidizing bacteria were isolated from deep-subsurface minerals collected from the Henderson Molybdenum Mine in Empire, CO, in March 2006. Sampling was conducted ∼3,000 ft below the surface, where a series of boreholes were drilled into granite in March 2005 to release deep-seated fluids. At the time of sampling, the draining fluids were ∼45°C and the ambient air temperature was ∼24°C. Secondary minerals, including Fe oxides, Mn oxides, and Ca sulfates, precipitated around the mouth of each borehole. Sterile Falcon tubes were filled with mineral precipitates and stored on ice (∼8 h) until return to the laboratory.
Isolation and identification of Mn(II)-oxidizing bacteria.
Samples were diluted 100-fold with water from the site that had been filtered through a sterile 0.2-μm polycarbonate filter. Samples were plated onto solid medium containing organic carbon and amended with 1 mM soluble Mn(II) and incubated in the dark at 25°C or 50°C. Growth occurred on Leptothrix medium, a chemically defined freshwater medium for the isolation of Mn-oxidizing bacteria (25), and K medium, a seawater-based rich medium containing yeast extract and peptone as carbon sources (34) (Table 1). Single colonies were transferred until they were morphologically uniform (∼6 generations), and then isolates were grown in liquid medium for DNA extraction (DNeasy blood and tissue kit; Qiagen).
TABLE 1.
Organisms isolated from the Henderson molybdenum mine
| Isolate | Accession no. | Sample source (depth [ft], location, type of precipitate) | Isolation mediumb | Isolation temp (°C) | Temp (°C) optimum(a) | Mn oxidationa | Amplifiable mnxG | Closest relative (% identity) |
|---|---|---|---|---|---|---|---|---|
| HM06-01 | EU004563 | 7,025, borehole 4 | K-asw | 50 | Not tested | ++ | No | Bacillus firmus B563 (100) |
| HM06-02 | EU004564 | 7,025, fracture, Mn oxides | K-asw | 25 | 37 | + | Yes | Bacillus sp. strain PL-12 (99) |
| HM06-02 mnxG | EU004573 | mnxG Bacillus sp. strain PL-16 (98) | ||||||
| HM06-03 | EU004565 | 7,025, borehole 1, Fe oxides | Leptothrix | 50 | Not tested | − | No | Paenibacillus sp. strain Smarlab 3301786 (99) |
| HM06-04 | EU004566 | 7,025, fracture, Mn oxides | K-asw | 25 | 37 | +++ | No | Bacillus sp. strain SD521 (99) |
| HM06-05 | EU004567 | 7,025, borehole 1, Fe oxides | Leptothrix | 50 | Not tested | − | No | Paenibacillus sp. strain Smarlab 3301786 (99) |
| HM06-06 | EU004568 | 7,025, borehole 3, Fe oxides | Leptothrix | 50 | Not detected | ++ | No | Bacillus sp. strain B-3 (99) |
| HM06-07 | EU004569 | 7,025, borehole 3, Fe oxides | Leptothrix | 50 | Not tested | − | No | Bacillus sp. strain B-3 (100) |
| HM06-08 | EU004570 | 7,025, borehole 4, Mn oxides | K-asw | 50 | 25 | ++ | No | Bacillus firmus B563 (100) |
| HM06-09 | EU004571 | 7,025, borehole 4, Mn oxides | K-asw | 50 | Not tested | ++ | No | Bacillus firmus B563 (100) |
| HM06-10 | EU004572 | 7,025, fracture, Mn oxides | K-asw | 25 | 25 and 37 | + | No | Bacillus sp. strain 2b-2 (98) |
| HM06-11 | EU525854 | 7,025, borehole 4, Mn oxides | Leptothrix | 50 | Not tested | ++ | No | Georgenia sp. strain 3A-1 (95) |
+, weak; ++, medium; +++, strong; −, none.
asw, artificial seawater.
PCR amplification and sequencing of 16S rRNA.
The 16S rRNA gene for each isolate was amplified by PCR. The total reaction mixture volume was 50-μl: 1 μl template DNA, 50 pmol of the forward (27F, 5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse (1492R, 5′-GGYTACCTTGTTACGACTT-3′) bacterial primers, 500 pmol deoxynucleoside triphosphates, reaction buffer, and 0.5 units of Taq. Thirty-five cycles were conducted in an Eppendorf thermal cycler (Westbury, NY) following a protocol of 95°C for 30 s, 50°C for 30 s, 72°C for 1 min, followed by 7 min at 72°C. Isolates that did not grow in liquid medium were amplified by adding a colony directly to a PCR and lysing the cells by holding them for 10 min at 94°C prior to starting the cycling routine. The PCR products were cleaned by using a QIAquick PCR purification kit (Qiagen). All sequencing was performed by SeqWright DNA Technology Services (Houston, TX). Approximately 1,400 bp of sequence was obtained by performing three sequencing reactions with 3′ primers (27F, 1074R, and 1492R) and assembling the data by using Sequencher 4.2, Gene Codes Corporation (Ann Arbor, MI). The full-length 16S rRNA sequences were searched against the GenBank database by using BLAST (1), and the closest sequenced relatives were identified.
Determination of Mn(II)-oxidizing capabilities and amplification of mnxG sequences.
To evaluate their Mn(II) oxidation capability, isolates were grown on plates amended with soluble Mn(II). After 1 week, the experimentally determined amount of time needed for sporulation to occur, dark colonies, potentially encrusted with brown Mn(IV) oxides, were tested for the presence of oxidized Mn by using a colorimetric leucoberbelin blue (LBB) assay (19). All Bacillus sp. isolates were initially tested for the presence of the mnxG gene by amplification with degenerate mnxG primers previously designed by Dick et al. (6), but only unrelated products were generated. More-specific primers, mnxGIF, 5′-ACGCATGTCTTTCACTATCATGTTCAT-3′, and mnxGIR, 5′-AAATAAGTGGTCATGGAAGAACCATGC-3′, previously designed by Francis et al. (13), yielded more-appropriate products. The PCR protocol was the following: 30 cycles of 94°C for 30 s, 45°C for 30 s, and 60°C for 1 min, followed by 1 cycle of 72°C for 15 min (13). PCR products were either directly sequenced by SeqWright DNA Technology Services, Houston, TX, or were cloned into chemically competent Escherichia coli cells by using a TOPO-TA cloning kit (Invitrogen). Clones containing the appropriate ∼800-bp PCR product were purified and sequenced by using the primer M13F. A BLAST search (1) of each sequence was conducted to determine if the sequences represented an mnxG gene.
Phylogenetic analysis. (i) Sequence attainment.
Of the 11 organisms isolated, eight were Bacillus, two were Paenibacillus sp., and one was a Georgenia sp. A total of 57 16S rRNA sequences were used in this study and include 24 Bacillus species not known to possess an mnxG gene, 23 sequences from mnxG-positive Bacillus organisms, the 7 sequences of mnxG-negative Henderson Mine Bacillus isolates, 1 sequence from the mnxG-positive Henderson Mine isolate, and the 2 sequences of the Henderson Mine isolates most similar to Paenibacillus species. 16S rRNA sequences were obtained by conducting nucleotide-nucleotide BLAST searches of Bacillus sp. strain SG-1 (1). Bacillus sp. strain SG-1 is a model organism used for the study of biotic manganese oxidation and possesses the mnxG gene (12, 24, 33, 35). The possession of an mnxG gene was determined by conducting a protein BLAST search using the protein sequence of the mnxG gene of Bacillus sp. strain SG-1. All Bacillus organisms identified in the mnxG search were included in the analysis to represent Mn-oxidizing members of the genus Bacillus. The protein product and nucleotide sequences of the mnxG gene in these Bacillus species were acquired from GenBank. BLAST searches were conducted on the 16S rRNA sequences of the isolates, and sequences of known Bacillus organisms with high identity to the Henderson Mine isolates (HM06-01 to -10) were included in the analysis. Many of the sequences were also used in previous Mn oxidation studies (6, 13).
(ii) Sequence alignment.
16S rRNA sequences were aligned using ClustalX (37) with default settings as follows: gap opening, 10.00; gap extension, 0.20; delay divergent sequences, 30%; DNA transition weight, 0.50; and negative matrix, OFF. An alignment of the mnxG protein sequence was constructed in ClustalX by using the following parameters: gap opening, 10.00; gap extension, 0.20; delay divergent sequences, 30%; protein weight matrix, Gonnet series; protein matrix, IUB; residue-specific penalties, ON; hydrophilic penalties, ON; hydrophilic residues, GPSNDQEKR; gap separation distance, 4; and end gap separation, OFF. GB02-29 was not included in the mnxG gene alignment because a protein sequence was not available for this organism. The nucleotide sequence of the mnxG gene of Henderson Mine isolate HM06-02 was translated into a protein sequence by using the translation tool ExPASy (http://www.expasy.ch/tools/dna.html) and included in the alignment.
(iii) Construction of trees.
A Bayesian analysis of the aligned 16S rRNA genes from all organisms was conducted using MrBayes (17). One million generations of an MCMC run were conducted with the following parameters: nst, 6; rates, gamma; sample frequency, 100; and nchains, 4. A total of 10,000 trees were constructed; the first 1,000 trees (equal to the first 10% or 100,000 generations of the analysis) were attributed to the burn-in period and excluded from further analyses. A 50% majority consensus tree was built in PAUP (version 4.0b10) (31) by using the results of the Bayesian analysis.
PAUP (version 4.0b10) (31) was used to construct a maximum parsimony and 50% majority rule bootstrap tree by using the protein sequence of the mnxG gene.
(iv) Character mapping.
Stochastic character mapping implemented with SIMMAP (2) allowed the estimation of the number of gains and losses of mnxG genes from the presence or absence data. The analysis was performed on 1,000 Bayesian 16S rRNA trees. We tested the hypothesis of no association between phylogeny and the presence or absence of mnxG by comparing the number of gains and losses for the observed data with the number of gains and losses after randomizing the presence or absence data across taxa 30 times, yielding a total of 30,000 mappings of gains and losses. The probability that this hypothesis is true was determined by following the method of Jones and Martin (18).
Nucleotide sequence accession numbers.
The partial 16S rRNA sequences of strains HM06-01 to HM06-11 were submitted to GenBank under accession numbers EU004563 to EU004572 and EU525854 (Table 1). The single mnxG gene identified was submitted to GenBank under accession number EU004573.
RESULTS
Growth and Mn(II) oxidation properties of isolates.
The maximum growth temperatures were determined for several isolates (Table 1). Of the 11 isolates from Henderson Mine, isolates HM06-02, -04, and -10 were grown at 25°C, while isolates HM06-05 to -09 and -11 were grown at 50°C. All isolates were examined microscopically and were rod-shaped spore formers. The results of the LBB colorimetric assay revealed that 8 of the 11 isolates exhibited Mn(II) oxidation (Table 1). Mn(II) oxidation varied from weak to intermediate to strong; this characterization was dependent on the rapidity of appearance and brightness of the blue color that indicated the oxidation of LBB by Mn(III/IV) oxides.
mnxG from a Henderson Mine Bacillus isolate is deeply nested within mnxG gene phylogeny.
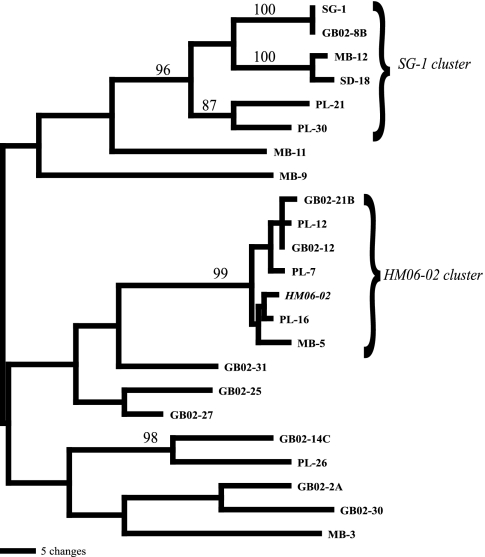
We successfully amplified and sequenced a single mnxG gene from isolate HM06-02. The gene, labeled HM06-02 mnxG, is most closely related to sequences recovered from marine sediments from Point Loma, CA, and is deeply nested within a well-supported clade (bootstrap value = 99) of mnxG genes sampled from four widely different localities: Point Loma, Guaymas Basin, Mission Bay, and Henderson Mine (6, 11, 13; this work) (Fig. 1).
FIG. 1.
Maximum parsimony bootstrap 50% majority rule consensus tree based on mnxG amino acid sequences. Bootstrap values are indicated at selected nodes and are based on 100 bootstrap replicates. The SG-1 cluster is labeled and is consistent across both the 16S rRNA and mnxG gene trees. The HM06-02 cluster is also labeled.
Phylogenetic placement of bacteria isolated from Henderson Mine.
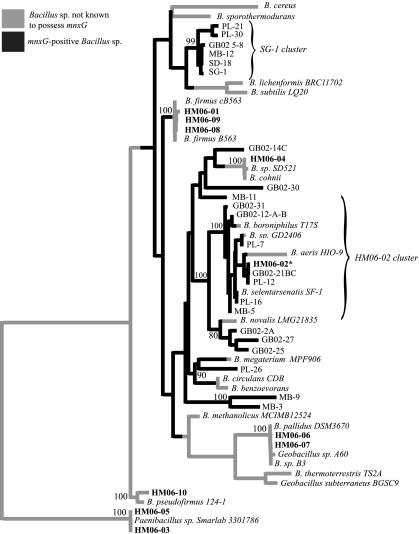
Seven unique 16S rRNA sequences were characterized from the 11 isolates. The sequence from isolate HM06-11 was most closely identical to Georgenia sp. strain 3A-1, and two others (HM06-03 and -05) were identical in sequence to an isolate of Paenibacillus (strain Smarlab 3301786). The other sequences grouped within a clade consisting of species and isolates of Bacillus and Geobacillus (Fig. 2). The discovery of Bacillus species in the mineral samples was not surprising, as the cloning and sequencing of environmental DNA extracted from adjacent borehole fluids detected Bacillus sp. isolates with sequences 94 to 96% similar to those of the isolates in our study (29). All of the isolates from which we were unable to amplify an mnxG gene grouped with high posterior probabilities (posterior probability = 1.0) within clades that lacked the mnxG gene, whereas the one isolate possessing an amplifiable mnxG gene was deeply nested within a well-supported clade (posterior probability = 1.0) consisting of other species that have the mnxG gene (Fig. 2). The results corroborate those of our functional assays: the phylogeny of the isolated microorganisms predicted the presence and absence of the mnxG gene.
FIG. 2.
Fifty-percent majority rule consensus phylogenetic tree based on 16S rRNA sequences of Bacillus species. Henderson Mine isolates are in boldface type, and the single asterisk indicates the mnxG-positive Bacillus organism isolated in this study. Consensus values are indicated at select nodes and are based on 9,000 Bayesian trees. The SG-1 and the HM06-02 clusters are labeled.
Few gains and frequent losses of mnxG.
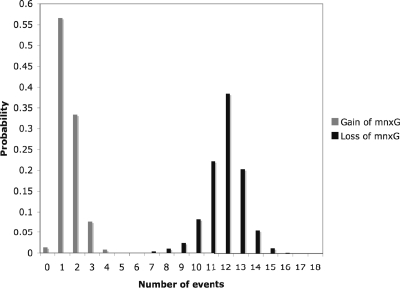
Stochastic character mapping was used to infer the history of mnxG gain and loss based on the presence and absence of the mnxG gene in taxa and the trees generated from Bayesian analysis of the 16S rRNA data. Across the many simulations of character evolution on 2,000 Bayesian trees, there were few gains of mnxG and many losses (Fig. 2 shows a typical example). The most-probable explanation of the data requires inferring 1 gain and 12 losses of mnxG (Fig. 3). A single origin of mnxG had the highest probability (0.566), yet the hypothesis for two and three independent gains of mnxG was not refuted (probabilities of 0.334 and 0.077, respectively). A broader range of number of gene losses was observed, with most of the distribution bounded by 10 and 14 losses.
FIG. 3.
Histogram depicting the probabilities of numbers of gains and losses of the mnxG gene needed to explain the observed data.
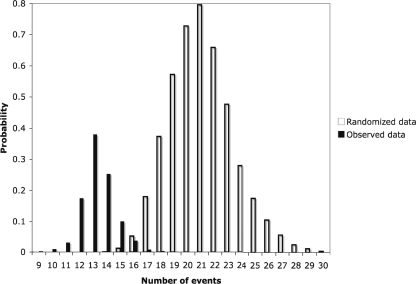
Thirteen character state changes (gains plus losses) best explained the data. This was less than expected if the character state changes were random and there was no relationship between the presence and absence of the mnxG gene and the 16S rRNA phylogeny of sampled isolates (Fig. 4); this indicates that vertical inheritance best explains the presence or absence of mnxG in Bacillus isolates.
FIG. 4.
Histogram comparing the number of changes needed within the trees constructed by Bayesian analysis to explain the distribution of the mnxG gene in the observed data versus the randomized data. Note that the distribution of observed values falls to the left of the distribution of random values; this indicates that the null hypothesis can be rejected.
DISCUSSION
Our results imply that phylogenetic placement on the basis of 16S rRNA sequence similarity predicts whether Bacillus species possess an mnxG gene. Furthermore, there is a complex pattern of distribution of the mnxG gene within the Bacillus clade revealed by the 16S rRNA relationships between the taxa. This distribution is best explained by a combination of vertical inheritance and gene loss within the Bacillus clade.
In the case of the Henderson Mine isolates, the results of 16S rRNA consensus tree analyses illustrate that the mnxG-positive isolate HM06-02 is most closely related to other mnxG-positive Bacillus organisms, while the mnxG-negative isolates were most closely related to other Bacillus organisms not known to possess an mnxG gene (Fig. 1). The evidence of phylogenetic conservation of the mnxG gene was further supported by the occurrence of an SG-1 cluster on both the 16S rRNA and mnxG trees (Fig. 1 and 2).
The phylogenetic inheritance of the mnxG gene within the Bacillus clade is further supported by stochastic character-mapping analyses. The most-probable number of gains and losses of the mnxG gene for the data set (13) is less than the number of changes (21) expected if the gene is randomly distributed on the phylogenetic trees (Fig. 4). These results indicate that phylogenetic inheritance plays a dominant role in the pattern of distribution of the gene. However, the multiple occurrences of gene loss suggest that there is a dynamic aspect to the evolution of the mnxG gene within the Bacillus clade.
The results of stochastic character-mapping analyses provided evidence for the multifaceted evolutionary process of mnxG within the Bacillus clade. Gene loss may explain relationships between taxa as seen on the 16S rRNA tree (Fig. 2), such as within the HM06-02 cluster or within the cluster of four isolates most closely related to GB02-2A, of which Bacillus novalis LMG21835 is the only isolate that lacks the gene (Fig. 2). The loss of genes is not uncommon and may be enhanced if the adaptation that the gene codes for is not used by the organism (4, 9, 16, 20, 22, 26). In Bacillus species, mnxG is only located in the exosporium of metabolically dormant spores (10). This may, in part, explain the frequency of loss of the gene, as the capability to oxidize Mn may not be necessary to the survival of metabolically dormant spores.
Despite the potential for the simultaneous occurrence of vertical inheritance and gene loss, it is still possible to use 16S rRNA-based phylogeny to predict to a first order the presence or absence of an amplifiable mnxG gene within the genome of a Bacillus organism.
The results of LBB tests for the presence of Mn(III/IV) oxides reveal that seven of eight Henderson Mine Bacillus isolates are capable of oxidizing manganese (Table 1), yet the mnxG gene was only amplified in one isolate, HM06-02. We present two lines of evidence supporting the observation that the mnxG gene is absent from the genomes of isolates HM06-01 and HM06-03 to -10: (i) the mnxG gene was not amplifiable, and (ii) the mnxG-negative organisms fall within other Bacillus clusters that are also not known to possess an mnxG gene. These data are in contrast to data from Guaymas Basin, where the authors were able to amplify an mnxG gene in all of their Mn(II)-oxidizing Bacillus species (6). There are two possible explanations for the lack of an amplifiable mnxG gene in Mn(II)-oxidizing Bacillus isolates from the Henderson Mine: (i) the mnxG-negative Bacillus sp. isolates may possess an entirely different gene that is responsible for the oxidation of manganese, or (ii) the primers designed to amplify mnxG may target only a subset of mnxG genes possessed by Bacillus sp. organisms. Henderson Mine Bacillus species are not the only examples of this paradox. Bacillus pumilus isolates capable of vigorously oxidizing manganese have been isolated from Mn crusts on basaltic rocks (A. S. Templeton and L. E. Mayhew et al., unpublished data), and yet mnxG could not be amplified from the genomic DNA in tests done in our and other labs (e.g., G. J. Dick, personal communication [regarding results of amplification using primers and conditions found in reference 6]). This was further corroborated by 16S rRNA phylogenies where B. pumilus does not fall within clusters of Mn(II)-oxidizing Bacillus species (6). Fortunately, the genome of Bacillus pumilus SAFR-032 has recently been completed (15). Protein sequences of all the putative manganese multicopper oxidases, mnxG (Bacillus sp. strain SG-1), cumA (Pseudomonas putida MT-2), and mofA (Leptothrix discophora SS-1), were searched against the Bacillus pumilus SAFR-032 genome by using BLAST. No genes homologous to mnxG were evident from the BLAST search; thus, it appears that mnxG was not amplified because it is not present in the genome. However, both cumA and mofA were most similar to cotA (24% and 27% identity, respectively), an outer spore coat protein of B. pumilus, supporting the idea that perhaps Bacillus sp. organisms possess multiple genes whose products have the functional capability of Mn(II) oxidation.
Conclusions.
16S rRNA phylogenies were determined to be a tool for predicting the presence or absence of a functional gene. The results of Bayesian analyses and character mapping indicate that 16S rRNA relationships between Bacillus organisms can be used to predict the presence or absence of the mnxG gene within the genome of a Bacillus organism. The one mnxG-positive Henderson Mine isolate falls in a cluster consisting predominantly of other mnxG-positive organisms. The mnxG-negative Henderson Mine isolates fall within clusters built solely of other mnxG-negative Bacillus organisms. These observations are supported by the high consensus values obtained from the Bayesian analyses. These conclusions agree with those found by other researchers investigating the relationship of 16S rRNA phylogenies to functional gene phylogenies (21, 23, 27, 28).
Gene loss was found to influence the occurrence of functional genes within the genomes of a subset of Bacillus species. A greater number of losses than gains have occurred (12 versus 1). In previous research, gene loss has been shown to influence the appearance of microbial genomes (4, 9). This research indicates that to fully understand the distribution of the mnxG gene within the genus Bacillus, the processes of phylogenetic inheritance and genomic remodeling through gene loss need to be considered.
Acknowledgments
We thank the two anonymous reviewers for constructive criticisms of earlier drafts of the manuscript. We also thank the Climax Molybdenum Company for access to the Henderson mine during the S2 phase of DUSEL proposal preparation. We also thank Jason Sahl for helpful discussions.
Funding for this work was provided by National Science Foundation grant MCB-0623815 (A. S. Templeton) and graduate fellowships awarded to L. E. Mayhew and E. D. Swanner by the Department of Geological Sciences, University of Colorado at Boulder.
Footnotes
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollback, J. 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinform. 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 5.Dar, S. A., L. Yao, U. van Dongen, J. G. Kuenen, and G. Muyzer. 2007. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 73:594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick, G. J., Y. E. Lee, and B. M. Tebo. 2006. Manganese(II)-oxidizing Bacillus spores in Guaymas Basin hydrothermal sediments and plumes. Appl. Environ. Microbiol. 72:3184-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, D., Y. M. Nelson, L. W. Lion, M. L. Shuler, and W. C. Ghiorse. 2000. Adsorption of Pb and Cd onto metal oxides and organic material in natural surface coatings as determined by selective extractions: new evidence for the importance of Mn and Fe oxides. Water Res. 34:427-436. [Google Scholar]
- 8.Ehrlich, H. L. 2002. Geomicrobiology, 4th ed. Marcel Dekker, Inc., New York, NY.
- 9.Feldgarden, M., N. Byrd, and F. M. Cohan. 2003. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology 149:3565-3573. [DOI] [PubMed] [Google Scholar]
- 10.Francis, C. A., K. L. Casciotti, and B. M. Tebo. 2002. Localization of Mn(II)-oxidizing activity and the putative multicopper oxidase, mnxG, to the exosporium of the marine Bacillus sp. strain SG-1. Arch. Microbiol. 178:450-456. [DOI] [PubMed] [Google Scholar]
- 11.Francis, C. A., E. M. Co, and B. M. Tebo. 2001. Enzymatic manganese(II) oxidation by a marine alpha-proteobacterium. Appl. Environ. Microbiol. 67:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, C. A., and B. M. Tebo. 2001. CumA multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl. Environ. Microbiol. 67:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis, C. A., and B. M. Tebo. 2002. Enzymatic manganese(II) oxidation by metabolically dormant spores of diverse Bacillus species. Appl. Environ. Microbiol. 68:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gersham, T. L. T., P. P. Sheridan, M. E. Watwood, Y. Fujita, and F. S. Colwell. 2007. Design and validation of ureC-based primers for groundwater detection of urea-hydrolizing bacteria. Geomicrobiology J. 24:353-364. [Google Scholar]
- 15.Gioia, J., S. Yerrapragada, X. Qin, H. Jiang, O. C. Igboeli, D. Muzny, S. Dugan-Rocha, Y. Ding, A. Hawes, W. Liu, L. Perez, C. Kovar, H. Dinh, S. Lee, L. Nazareth, P. Blyth, M. Holder, C. Buhay, M. R. Tirumalai, Y. Liu, I. Dasgupta, L. Bokhetache, M. Fujita, F. Karouia, P. Eswara Moorthy, J. Siefert, A. Uzman, P. Buzumbo, A. Verma, H. Zwiya, B. D. McWilliams, A. Olowu, K. D. Clinkenbeard, D. Newcombe, L. Golebiewski, J. F. Petrosino, W. L. Nicholson, G. E. Fox, K. Venkateswaran, S. K. Highlander, and G. M. Weinstock. 2007. Paradoxical DNA repair and peroxide resistance gene conservation in Bacillus pumilus SAFR-032. PLoS ONE 2:E928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao, W., and G. B. Golding. 2006. The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inferences of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 18.Jones, R. T., and A. P. Martin. 2006. Testing for differentiation of microbial communities using phylogenetic methods: accounting for uncertainty of phylogenetic inference and character state mapping. Microb. Ecol. 52:408-417. [DOI] [PubMed] [Google Scholar]
- 19.Krumbein, W. E., and H. J. Altmann. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgol. wiss. Meeresunters 25:347-356. [Google Scholar]
- 20.Kunin, V., and C. A. Ouzounis. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13:1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuske, C. R., S. M. Barns, C. C. Grow, L. Merrill, and J. Dunbar. 2006. Environmental survey for four pathogenic bacteria and closely related species using phylogenetic and functional genes. J. Forensic Sci. 51:548-558. [DOI] [PubMed] [Google Scholar]
- 22.Marri, P. R., W. Hao, and G. B. Golding. 2006. Gene gain and gene loss in Streptococcus: is it driven by habitat? Mol. Biol. Evol. 23:2379-2391. [DOI] [PubMed] [Google Scholar]
- 23.Martens, T., L. Gram, H. P. Grossart, D. Kessler, R. Muller, M. Simon, S. C. Wenzel, and T. Brinkhoff. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 24.Nealson, K. H., and J. Ford. 1980. Surface enhancement of bacterial manganese oxidation: implications for aquatic environments. Geomicrobiology J. 2:21-37. [Google Scholar]
- 25.Nelson, Y. M., L. W. Lion, W. C. Ghiorse, and M. L. Shuler. 1999. Production of biogenic Mn oxides by Leptothrix discophora SS-1 in a chemically defined growth medium and evaluation of their Pb adsorption characteristics. Appl. Environ. Microbiol. 65:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournie, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 27.Petri, R., and J. F. Imhoff. 2000. The relationship of nitrate-reducing bacteria on the basis of narH gene sequences and comparison of narH and 16S rDNA-based phylogeny. Syst. Appl. Microbiol. 23:47-57. [DOI] [PubMed] [Google Scholar]
- 28.Petri, R., L. Podgorsek, and J. F. Imhoff. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol. Lett. 197:171-178. [DOI] [PubMed] [Google Scholar]
- 29.Sahl, J. W., R. Schmidt, E. D. Swanner, K. W. Mandernack, A. S. Templeton, T. L. Kieft, R. L. Smith, W. E. Sanford, R. L. Callaghan, J. B. Mitton, and J. R. Spear. 2008. Subsurface microbial diversity in deep-granitic-fracture water in Colorado. Appl. Environ. Microbiol. 74:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunda, W. G., and D. J. Kieber. 1994. Oxidation of humic substances by manganese oxides yields low-molecular weight organic substrates. Nature 367:62-64. [Google Scholar]
- 31.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, MA.
- 32.Reference deleted.
- 33.Tebo, B. M., J. R. Bargar, B. G. Clement, G. J. Dick, K. J. Murray, D. Parker, R. Verity, and S. M. Webb. 2004. Biogenic manganese oxides: properties and mechanisms of formation. Ann. Rev. Earth Planetary Sci. 32:287-328. [Google Scholar]
- 34.Tebo, B. M., B. G. Clement, and G. J. Dick. 2007. Biotransformations of manganese, p. 1223-1238. In C. J. Hurst, R. L. Crawford, J. L. Garland, D. A. Lipson, A. L. Mills, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC.
- 35.Tebo, B. M., H. A. Johnson, J. K. McCarthy, and A. S. Templeton. 2005. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 13:421-428. [DOI] [PubMed] [Google Scholar]
- 36.Reference deleted.
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Waasbergen, L. G., M. Hildebrand, and B. M. Tebo. 1996. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J. Bacteriol. 178:3517-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb, S. M., B. M. Tebo, and J. R. Bargar. 2005. Structural characterization of biogenic Mn oxides produced in seawater by the marine Bacillus sp. strain SG-1. Am. Mineral. 90:1342-1357. [Google Scholar]
- 40.Reference deleted.