Abstract
Escherichia coli isolates containing the following extended-spectrum beta-lactamases have been detected in 11 of 57 fecal samples (19.3%) in Berlengas Island seagulls: TEM-52 (eight isolates), CTX-M-1 (one isolate), CTX-M-14a (one isolate), and CTX-M-32 (one isolate). Most of the extended-spectrum beta-lactamase-positive isolates harbored class 1 or class 2 integrons, which included different antibiotic resistance gene cassettes.
The emergence and wide dissemination of extended-spectrum beta-lactamases (ESBLs) among clinical Escherichia coli isolates in hospitals in recent years are of great concern and represent a problem for the treatment of infectious diseases (19). It has also been reported that E. coli isolates containing ESBLs, mostly of the CTX-M class, are frequently detected in community patients (4) and have also been found in food-producing animals and household pets (2, 3, 5, 8, 11, 14, 17, 24). Moreover, a previous report identified ESBLs in fecal E. coli isolates of wild animals (9), mainly in birds of prey, but seagulls were not included in that study. ESBLs seem to be widely distributed in bacteria of different ecosystems, although more information is needed, especially for wild ecosystems. The purpose of our study was to analyze the carriage of ESBL-containing E. coli isolates in fecal samples of Berlengas Island seagulls and also to characterize the type of ESBLs and the phylogenetic groups of isolates. Berlengas Island is part of the Berlengas Natural Reserve, located 5.7 miles from the Portuguese coast, and it belongs to the National Network of Protected Areas. Fishermen inhabited the island in the past, but currently nobody lives there year round, although some tourists visit the island and a few people stay for vacations. Diverse species of seagulls make their nests on this island; in the last few years the population of seagulls has increased significantly and is considered a true plague (18).
Fifty-seven fresh seagull fecal droppings were obtained in different areas of the Berlengas Island during September 2007 and were tested for the presence of ESBL-containing E. coli isolates. Fecal samples were seeded in Levine agar plates supplemented with cefotaxime (CTX; 2 μg/ml), and colonies with typical E. coli morphology were selected and identified by classical biochemical methods and by the API 20E system (bioMérieux, La Balme Les Grottes, France). Susceptibility of the recovered E. coli isolates to 16 antibiotics (ampicillin, amoxicillin plus clavulanic acid, cefoxitin, CTX, ceftazidime, aztreonam, imipenem, gentamicin, amikacin, tobramycin, streptomycin, nalidixic acid, ciprofloxacin, sulfamethoxazole-trimethoprim, tetracycline, and chloramphenicol) was tested by the disk diffusion method (7). E. coli ATCC 25922 was used as a quality control strain. Broad-spectrum cephalosporin-resistant isolates were selected for further studies (one isolate per sample), and they were screened for ESBL production according to the CLSI criteria (7).
The presence of genes encoding TEM, SHV, OXA, CTX-M, and CMY type beta-lactamases was studied by specific PCRs (1, 13, 23). All obtained amplicons were sequenced on both strands, and sequences were compared with those included in the GenBank database and in the website http://www.lahey.org/Studies/ to identify the beta-lactamase genes. The genetic environment of blaCTX-M genes was also tested by PCR and by sequencing with previously reported primers (10, 15, 22).
The presence of other antibiotic resistance genes, associated with chloramphenicol (cmlA), tetracycline (tetA and tetB), streptomycin (aadA), and sulfonamide (sul1, sul2, and sul3) resistance, among our isolates was also analyzed by PCR and sequencing (21). The presence of the intI1 and intI2 genes, encoding class 1 and 2 integrases, respectively, and the composition of the variable regions of class 1 and 2 integrons were studied by PCR and sequencing (21). The identification of the major phylogenetic groups among our isolates was determined by PCR (6). Positive and negative controls from the bacterial collection of the University of La Rioja, Logroño, Spain, were used in all assays.
E. coli isolates were detected in Levine CTX plates from 11 of the 57 (19.3%) fecal samples studied. All 11 isolates obtained from these samples were intermediate or resistant to CTX and/or ceftazidime and had a positive screening test for ESBL production. Only 1 of the 11 E. coli isolates (GV-10) showed resistance to amoxicillin plus clavulanic acid. The beta-lactamase genes detected in these isolates were the following (numbers of isolates are in parentheses): blaTEM-52 (8), blaCTX-M-1 plus blaOXA-1 (1), blaCTX-M-14a (1), and blaCTX-M-32 (1) (Table 1). It is interesting that 73% of the ESBL-positive isolates of seagulls harbored the blaTEM-52 gene and that 27% of the isolates harbored the blaCTX-M gene. A high prevalence of TEM-52 has also been recently observed in E. coli isolates from healthy food-producing animals and chicken meat products from Portugal (16).
TABLE 1.
Characteristics of the ESBL-positive fecal E. coli isolates recovered from seagulls of Berlengas Island in Portugal
| E. coli isolate | Phylogenetic group | Non-beta-lactams to which isolates were resistanta | Type of ESBL | Class 1 integrons
|
Class 2 integrons
|
Other gene(s) detected | ||
|---|---|---|---|---|---|---|---|---|
| Presence of intI1 | Gene cassettes | Presence of intI2 | Gene cassettes | |||||
| GV-5 | D | NAL, CIP, TET, STR, SXT, CHL | TEM-52 | + | dfrA1 + aadA1 | − | tetA, sul1, sul3, cmlA | |
| GV-6 | B1 | NAL, CIP, TET, STR | TEM-52 | − | + | dfrA1 + sat + aadA1 | tetA | |
| GV-8 | B2 | NAL, CIP, TET, STR, SXT, CHL | TEM-52 | + | sat + psp + aadAb | + | dfrA1 + sat + aadA1 | tetA, sul2, sul3, cmlA |
| GV-9 | B1 | NAL, CIP, TET, STR, SXT, CHL | TEM-52 | + | dfrA1 + aadA1 | − | tetA, sul1, sul2, sul3, cmlA | |
| GV-33 | A | NAL, TET | TEM-52 | − | − | tetB | ||
| GV-51 | B1 | NAL, CIP, TET, SXT | TEM-52 | − | + | dfrA1 + sat + aadA1 | tetA, sul2 | |
| GV-52 | A | NAL, CIP, TET | TEM-52 | − | + | dfrA1 + sat + aadA1 | tetA | |
| GV-54 | D | NAL, CIP, TET, STR, SXT | TEM-52 | − | + | dfrA1 + sat + aadA1 | tetB, sul2 | |
| GV-23 | A | TET, CHL | CTX-M-1 | + | blaOXA-1 + aadA1 | − | tetB, su1l | |
| GV-10 | A | NAL, TET, STR | CTX-M-14a | − | − | tetB, aadA | ||
| GV-12 | B1 | NAL, CIP, TET, STR | CTX-M-32 | + | sat + aadA1b | − | tetB | |
TET, tetracycline; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole.
The qacEΔ-plus-sul1 3′ conserved region was absent in two intI1-positive E. coli isolates containing blaTEM-52 and blaCTX-M-32.
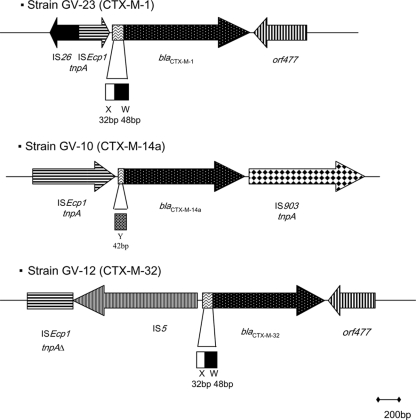
Different genetic environments surrounding the blaCTX-M genes were detected (Fig. 1). The sequence of the fragment obtained by PCR upstream of the blaCTX-M-1 gene in the E. coli GV-23 strain revealed the presence of a region of the IS26 transposase flanking a partially truncated ISEcp1 followed by an intergenic region; this whole structure has been previously found in E. coli (13, 22). The presence of ISEcp1 and IS903 surrounding the blaCTX-M-14a gene in E. coli GV-10 was identified, and the genetic environment of the blaCTX-M-32 gene detected in E. coli GV-12 included ISEcp1/IS5 upstream of the bla gene and orf477 downstream, as also detected by others (25).
FIG. 1.
Genetic environment of blaCTX-M genes in three E. coli isolates recovered from seagull fecal samples (the intergenic X, Y, and W regions have been previously reported [10]).
A variety of resistance genes (cmlA, tetA, tetB, aadA, sul1, sul2, and sul3) were observed among our ESBL-producing E. coli isolates (Table 1). Five isolates harbored class 1 integrons with the following gene cassettes in their variable regions: dfrA1 plus aadA1 (two isolates), sat plus psp plus aadA2 (one isolate), sat plus aadA1 (1 isolate), and blaOXA-1 plus aadA1 (one isolate). Five isolates harbored class 2 integrons, and the gene cassette arrangement dfrA1 plus sat plus aadA1 was identified in all of them. E. coli GV-8 contained simultaneously class 1 and 2 integrons. Eight of the ESBL-positive isolates corresponded to the A and B1 phylogenetic groups, two isolates corresponded to the D group, and only one blaTEM-52 isolate was assigned to the B2 phylogenetic group (Table 1). Previous studies have reported the association of E. coli isolates of the B2 group with extraintestinal infections (20), and the fecal origin of our isolates could explain the low prevalence of this phylogroup.
It is important to note the high prevalence and moderate diversity of ESBLs detected in fecal E. coli from seagulls that inhabit a natural reserve, as is the case for Berlengas Island. As previously indicated, the population of seagulls on an island that is not too far away from the Portuguese coast has significantly increased in recent years. The possibility that these animals eat the remains of human food cannot be excluded. This study gives new evidence for the wide dissemination of ESBLs in E. coli isolates from wild animals, as is the case for seagulls. More studies of this nature should be performed in the future to analyze the prevalence of this type of resistant bacteria in different ecosystems.
Acknowledgments
This work was partly financed by project SAF2006-14207-C02 of the Ministry of Science and Education of Spain. We also thank Pfizer for partial financial support. L. Vinué was supported by a fellowship from the Spanish Ministry of Education and Science (SAF2006-14207-C02-01), and S. Somalo was supported by a fellowship from the Government of La Rioja, Spain (Colabora 2007/15).
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Bertrand, S., F. X. Weill, A. Cloeckaert, M. Vrints, E. Mairiaux, K. Praud, K. Dierick, C. Wildemauve, C. Godard, P. Butaye, H. Imberechts, P. A. Grimont, and J. M. Collard. 2006. Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc, V., R. Mesa, M. Saco, S. Lavilla, G. Prats, E. Miró, F. Navarro, P. Cortés, and M. Llagostera. 2006. ESBL- and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118:299-304. [DOI] [PubMed] [Google Scholar]
- 3.Briñas, L., M. A. Moreno, T. Teshager, Y. Sáenz, M. C. Porrero, L. Domínguez, and C. Torres. 2005. Monitoring and characterization of extended-spectrum β-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob. Agents Chemother. 49:1262-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantón, R., A. Novais, A. Valverde, E. Machado, L. Peixe, F. Baquero, and T. M. Coque. 2008. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14:144-153. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli, A. 2008. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1):117-123. [DOI] [PubMed] [Google Scholar]
- 6.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI/NCCLS M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Costa, D., P. Poeta, L. Brinas, Y. Saenz, J. Rodrigues, and C. Torres. 2004. Detection of CTX-M-1 and TEM-52 beta-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 54:960-961. [DOI] [PubMed] [Google Scholar]
- 9.Costa, D., P. Poeta, Y. Sáenz, L. Vinué, B. Rojo-Bezares, A. Jouini, M. Zarazaga, J. Rodríguez, and C. Torres. 2006. Detection of Escherichia coli harbouring extended-spectrum β-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 59:1311-1312. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 11.Girlich, D., L. Poirel, A. Carattoli, I. Kempf, M. F. Lartigue, A. Bertini, and P. Nordmann. 2007. Extended-spectrum β-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 73:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Jouini, A., L. Vinué, K. B. Slama, Y. Sáenz, N. Klibi, S. Hammami, A. Boudabous, and C. Torres. 2007. Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J. Antimicrob. Chemother. 60:1137-1141. [DOI] [PubMed] [Google Scholar]
- 14.Kojima, A., Y. Ishii, K. Ishihara, H. Esaki, T. Asai, C. Oda, Y. Tamura, T. Takahashi, and K. Yamaguchi. 2005. Extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49:3533-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 16.Machado, E., T. M. Coque, R. Cantón, J. C. Sousa, and L. Peixe. 2008. Antibiotic resistance integrons and extended-spectrum beta-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 62:296-302. [DOI] [PubMed] [Google Scholar]
- 17.Meunier, D., E. Jouy, C. Lazizzera, M. Kobisch, and J. Y. Madec. 2006. CTX-M-1 and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28:402-407. [DOI] [PubMed] [Google Scholar]
- 18.Morais, L., R. Santos, T. Goettel, and L. Vicente. 1995. Preliminary evaluation of the first yellow-legged herring gull Larus cachinnans population control at Berlenga Island, Portugal, p. 32. In M. L. Tasker (ed.), Threats to seabirds. International Seabird Group, Sandy, United Kingdom.
- 19.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sáenz, Y., L. Briñas, E. Domínguez, J. Ruiz, M. Zarazaga, J. Vila, and C. Torres. 2004. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saladin, M., V. T. B. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 23.Stapleton, P. D., K. P. Shannon, and G. L. French. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 45:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teale, C. J., L. Barker, A. P. Foster, E. Liebana, M. Batchelor, D. M. Livermore, and E. J. Threlfall. 2005. Extended-spectrum β-lactamase detected in E. coli recovered from calves in Wales. Vet. Rec. 156:186-187. [PubMed] [Google Scholar]
- 25.Vinué, L., M. Lantero, Y. Sáenz, S. Somalo, I. de Diego, F. Pérez, F. Ruiz-Larrea, M. Zarazaga, and C. Torres. 2008. Characterization of extended-spectrum beta-lactamases and integrons in Escherichia coli isolates in a Spanish hospital. J. Med. Microbiol. 57:916-920. [DOI] [PubMed] [Google Scholar]