Abstract
Purpose
Axenfeld-Rieger syndrome (ARS) is an autosomal dominant disorder characterized by extraocular anomalies and developmental defects of the anterior segment. PITX2 (paired-like homeodomain transcription factor 2) is considered the major causative gene. In this study, we characterized the molecular defect in PITX2 in a Chinese family with ARS.
Methods
Two generations of the family with ARS were enrolled in the present study. In addition to ophthalmologic examinations, polymerase chain reaction (PCR) amplification and nucleotide sequencing of all coding exons of PITX2 were performed. Exon 5 (region 1) was also sequenced in 100 healthy controls unrelated to the family for comparison.
Results
A novel PITX2 mutation, c.840G>T, was identified in all affected members of the family with ARS that causes an amino acid substitution from tryptophan to cysteine at codon 86.
Conclusions
We found a novel p.W86C mutation in PITX2 in a Chinese family with ARS. The tryptophan residue at position 86 is strictly conserved in PITX2a proteins from several species and in homeodomain proteins. We suggest that this mutation in PITX2 is the cause of typical ARS in patients. Our results may be useful for better understanding of the spectrum of PITX2 mutations and the role of PITX2 in the development and progression of ARS.
Introduction
Axenfeld-Rieger syndrome (ARS; OMIM 180500) is a rare multisystem autosomal dominant disorder, which is characterized by complete penetrance but variable expressivity [1]. ARS encompasses several conditions with overlapping phenotypes including Rieger syndrome, Axenfeld anomaly, and Rieger anomaly [2,3]. The classic ocular defects of ARS include iris hypoplasia, iridocorneal adhesion, corectopia, polycoria, posterior embryotoxon, and other less frequent features such as cataracts, retinal detachment, and microcornea [4,5]. Systemic anomalies associated with ARS include facial malformation (telecanthus, maxillary hypoplasia with protruding lower lip), dental abnormalities, and redundant periumbilical skin. Approximately half of the individuals affected with ARS also develop glaucoma [2,3].
The incidence of ARS is estimated to be approximately 1:200,000 births [6]. Mutations in several chromosomal loci have been implicated in ARS including PITX2 (paired-like homeodomain transcription factor 2; 4q25; OMIM 601542), FOXC1 (forkhead box C1; 6p25; OMIM 601090), PAX6 (paired box gene 6; 11p13; OMIM 607108) and MAF (v-MAF avian musculoaponeurotic fibrosarcoma oncogene homolog; 16q24; OMIM 177075). A yet to be identified gene at 13q14 (OMIM 601499) may also be found involved in ARS [7-11].
Full-spectrum ARS is caused primarily by mutations in PITX2 [1]. PITX2 is a member of the bicoid-like homeobox transcription factor family [7]. The homeobox gene family members play fundamental roles in the genetic control of development, particularly in pattern formation and the determination of cell fate [12-14]. PITX2 contains a 60 amino acid homeodomain with a lysine at residue 50 that is characteristic of the bicoid-related proteins [15-17]. Its expression in neural crest cells is necessary for optic stalk and anterior segment structure development [18]. PITX2 is produced as at least four different transcriptional and splicing isoforms, PITX2a, b, c, and d, which have different biological properties [19]. PITX2a, PITX2b, and PITX2c isoforms contain the same homeodomain and COOH-terminus but differ at the NH2-terminus. The PITX2d isoform has a truncated, nonfunctional homeodomain [19]. Although the role of PITX2 in the pathogenesis of ARS has yet to be clearly defined, a deficiency in normal PITX2 protein (haploinsufficiency) is suggested to be one of the major molecular mechanisms for the development of ARS [20,21]. Except a few intronic mutations [22,23], most of the mutations detected in human PITX2 are point mutations in the homeodomain or COOH-terminal domains. A correlation between the severity of ARS phenotypes and the levels of normal PITX2 protein was also noted [24-26]. In the present study, we analyzed the PITX2a isoform for mutations in a family affected by ARS.
ARS is relatively rare in China. The purpose of this study was to describe the clinical and molecular characteristics of a northeastern Chinese family affected by Axenfeld-Rieger syndrome (ARS).
Methods
Study participants
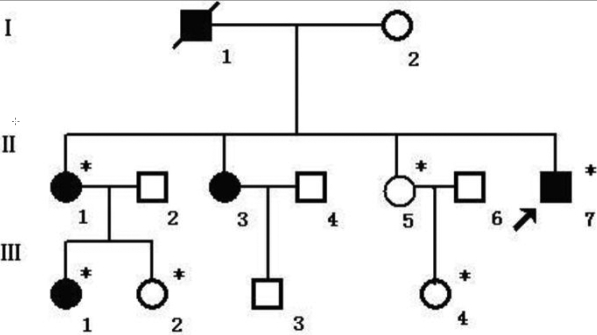
We studied two generations of a Chinese family with a history of Axenfeld-Rieger syndrome (ARS) from the Heilongjiang province in northeastern China (Figure 1). Three patients, three non-carrier relatives, and 100 healthy normal controls were recruited for this study. The study was approved by the Institutional Review Board of Harbin Medical University and informed consent was obtained from each of the participants. All subjects underwent full ophthalmologic examination including visual acuity, slit lamp, funduscopy, gonioscopy, and the measurement of intraocular pressure (IOP).
Figure 1.
Pedigree of a family with Axenfeld-Rieger syndrome. Autosomal dominant transmission of the disease is evident. The asterisks indicate subjects who underwent clinical and molecular analysis. Black symbols represent affected members. The arrow signals the proband.
Genetic analysis
Blood samples were drawn by venipuncture, and genomic DNA was extracted using the TIANamp Blood DNA Kit (Tiangen Biltech Co. Ltd, Beijing, China). All coding exons of PITX2 were amplified by polymerase chain reaction (PCR) using specific primers (Table 1). The primers for exons 3-5 were adapted from those described by Cella and coworkers [27]. The PCR fragments were purified with a TIANgel Midi Purification Kit (Tiangen Biltech Co. Ltd) and sequenced with an ABI BigDye Terminator Cycle Sequencing kit v3.1, (ABI Applied Biosystems, Foster City, CA).
Table 1. Primer sequences and the sizes of their corresponding PCR products.
| PITX2 exon | Sequences (5′-3′) | Amplicon (bp) |
|---|---|---|
|
PITX2-exon 1 |
F: CCGCTTCTTACAGCCTTCCT |
247 |
| |
R: CTGGCGATTTGGTTCTGATT |
|
|
PITX2-exon 2 |
F: TGGGTCTTTGCTCTTTGTCC |
400 |
| |
R: GCGGAGTGTCTAAGTTCAAGC |
|
|
PITX2-exon 3 |
F: GGGGCAGTAGCCAAGGACT |
289 |
| |
R: CAGCTAAGCGGGAATGTCTG |
|
|
PITX2-exon 4 |
F: GGCATGCTGACGGGAAAG |
300 |
| |
R: CTGTACCTCCACAACATCCTC |
|
|
PITX2-exon 5 (region 1) |
F: CACTGTGGCATCTGTTTGCT |
324 |
| |
R: GGACGACATGCTCATGGAC |
|
|
PITX2-exon 5 (region 2) |
F: TATGAACGTCAACCCCCTGT |
400 |
| R: CCATCCGGCAAGGTCCTA |
Results
PITX2 mutation analysis
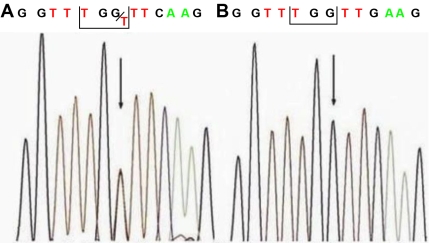
After direct sequencing of PITX2, a single heterozygous G to T mutation at nucleotide position 840 of PITX2 was detected (Figure 2) in all affected members. This resulted in a W86C mutation (tryptophan to cysteine substitution) at the protein level. None of the healthy family members or any of the 100 control subjects was positive for this mutation.
Figure 2.
Partial nucleotide sequence of PITX2. A: The sequence in an affected subject shows a heterozygous G>T transversion (indicated by the arrow). The nucleotide substitution at codon 86 results in a change from tryptophan to cysteine. B: Unaffected family members and the general population lack this nucleotide change.
Clinical findings
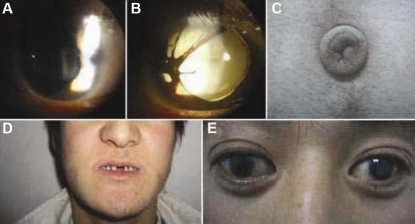
The proband (II:7) was 26 years old. He had experienced defective vision since childhood. His visual acuity was 20/100 in the right eye and counting fingers in the left eye. Although both eyes were affected, the clinical aspects of his eyes were different. In the right eye, ocular examination displayed corectopia associated with partial aniridia and iris hypoplasia (Figure 3A). Funduscopy showed glaucomatous atrophy of the optic nerve, and the IOP was 30 mmHg. External examination of the left eye showed displaced pupils along with iris atrophy, polycoria, and congenital cataract (Figure 3B). The complete cataract prevented ocular fundus examination. The IOP was 26 mmHg. In addition to the various ocular abnormalities, posterior embryotoxon was also found in both eyes following gonioscopic examination. Non-ocular abnormalities of the proband consisted of characteristic facial features with telecanthus, a thin upper lip, and a protruding lower lip (Figures 3D,E). In addition to redundant periumbilical skin, dental anomalies were also present (Figure 3C,D).
Figure 3.
Ocular characteristics and systemic anomalies of the proband with Axenfeld-Rieger syndrome. Slit lamp photographs are shown of the proband showing iris hypoplasia and corectopia associated with partial aniridia in the right eye (A), and iris atrophy and polycoria along with congenital cataract in the left eye (B). Systemic anomalies of the proband included redundant periumbilical skin (C), dental anomalies, a protruding lower lip (D), and telecanthus (E).
Individual II:1, 32 years old, is one of the proband’s sisters. Ocular examination of the right eye showed corectopia and iris hypoplasia. In the left eye, only mild iris hypoplasia was detected. She had posterior embryotoxon in each eye, but the fundus and IOP was normal. The non-ocular abnormalities included dental anomalies and characteristic redundant periumbilical skin. Her daughter, III:1, was nine years old. She had a bilateral posterior embryotoxon unaccompanied by any other ocular abnormality. She had normal teeth but had redundant periumbilical skin. Her IOP was also normal. Individual III:2 who was another daughter of individual II:1, individual II:5 who was another sister of the proband, and her daughter, III:4, did not show any clinical sign of ARS and had normal IOP.
Discussion
In this study, we described a novel mutation (c.840G>T) in PITX2a in a northeastern Chinese family with ARS. None of the proband’s unaffected relatives exhibited the mutation.
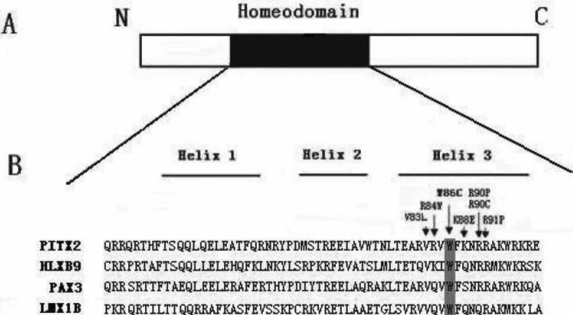
PITX2a is a 33 kDa homeodomain protein. The homeodomain is responsible for recognizing specific DNA sequences to bring the transcription factors to proper target genes. This 60 amino acid domain is composed of three helices and a flexible NH2-terminal arm [13,28]. Its integrity is essential for binding DNA and is critical for PITX2 to act as a transcription factor. The third helix is also called the recognition helix, which is the site of DNA recognition and binding, and is thought to fit into the major groove of DNA and to establish contact with specific residues of the bicoid binding sequence [13,28-30].
The function of the mutated PITX2 proteins can be analyzed in vitro through mutational and functional analyses. It showed that PITX2 mutations can alter PITX2 nuclear localization, DNA binding, and transactivation activity [31]. Thus far, several missense mutations resulting in single amino acid substitutions within helix 3 of the PITX2a homeodomain have been identified [7,24,32-34] (Figure 4), and these PITX2 mutations alter protein function to varying degrees. For example, the R84W mutant protein has a normal DNA binding specificity and could transactivate the Dlx2 promoter [25]. However, the K88E mutation is defective in its capacity to bind DNA and transactivation activity [26,35]. In 2002, Quentien et al. [36] demonstrated the R91P PITX2a mutation, which is dominant negative but can still bind DNA. The potential disruption of the helix 3 structure in the R91P mutant may affect the interaction of the factor with a limiting cofactor when bound to DNA. Interestingly, Priston et al. [24] reported the mutation, V83L, in PITX2a affects DNA binding and transactivation differently. Its mutation results in reduced DNA binding but also leads to a greater than 200% increase in transactivation activity compared to wt PITX2.
Figure 4.
Schematic diagram of PITX2a and amino acid sequence alignments of PITX2 homeodomain with other homeodomain proteins. A: The structure of the PITX2a isoform. The black region represents the homeodomain (HD). B: Alignments of the human PITX2 homeodomain and related homeodomain-containing proteins (the three α-helices are also indicated) are shown. The arrows indicate the previously characterized mutations within the helix 3 of PITX2a homeodomain. The tryptophan residue at position 86 is conserved among these homeodomain proteins.
The specific amino acid substitution, a change from tryptophan to cysteine, described in the present paper (p.W86C) occurs in the helix 3 of the homeodomain (Figure 4). The tryptophan residue at position 86 is strictly conserved in PITX2a proteins from different species such as Xenopus laevis, Mus musculus, Rattus norvegicus, Xenopus tropicalis, Canis lupus familiaris, Pan Troglodytes, Gallus gallus, and Bos Taurus (BLAST). Tryptophan residues are also conserved in other homeodomain proteins such as antennapedia, engrailed, and goosecoid. This high level of conservation indicates that this region may be important for the structure and/or function of the protein. Amino acid changes at this particular residue also have been shown to have a disease-causing effect in the case of the homeodomain transcription factors, HLXB9 (Currarino syndrome), LMX1B (Nail-patella syndrome), and PAX3 (Warrensburg syndrome). The mutational effects may be a disruption of protein stability [37].
In summary, we provide clinical and molecular evidence supporting the occurrence of a novel p.W86C mutation to PITX2a within a northeastern Chinese family with inherited ARS. Our results may improve the understanding of the role that PITX2 plays in ARS and helps expand the knowledge of the genetic causes of anterior segment disorders.
Acknowledgments
The authors are grateful to all patients, their families, and normal volunteers for their participation in this investigation.
References
- 1.Hjalt TA, Semina EV. Current molecular understanding of Axenfeld-Rieger syndrome. Expert Rev Mol Med. 2005;7:1–17. doi: 10.1017/S1462399405010082. [DOI] [PubMed] [Google Scholar]
- 2.Shields MB. Axenfeld-Rieger syndrome: a theory of mechanism and distinctions from the iridocorneal endothelial syndrome. Trans Am Ophthalmol Soc. 1983;81:736–84. [PMC free article] [PubMed] [Google Scholar]
- 3.Alward WL. Axenfeld-Rieger syndrome in the age of molecular genetics. Am J Ophthalmol. 2000;130:107–15. doi: 10.1016/s0002-9394(00)00525-0. [DOI] [PubMed] [Google Scholar]
- 4.Spallone A. Retinal detachment in Axenfeld-Rieger syndrome. Br J Ophthalmol. 1989;73:559–62. doi: 10.1136/bjo.73.7.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamińska A, Sokołowska-Oracz A, Pawluczyk-Dyjecińska M, Szaflik JP. Variability of clinical manifestations in the family with Axenfeld-Rieger syndrome. Klin Oczna. 2007;109:321–6. [PubMed] [Google Scholar]
- 6.Alkemade PPH. Dysgenesis mesodermalis of the iris and cornea: a study of Rieger’s Syndrome and Peters’ Anomaly. 1969; The Netherlands: Van Gorcum ad Comp NV Assen. [Google Scholar]
- 7.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–9. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 8.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerbäck S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–28. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GC. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- 10.Riise R, Storhaug K, Brøndum-Nielsen K. Rieger syndrome is associated with PAX6 deletion. Acta Ophthalmol Scand. 2001;79:201–3. doi: 10.1034/j.1600-0420.2001.079002201.x. [DOI] [PubMed] [Google Scholar]
- 11.Phillips JC, del Bono EA, Haines JL, Pralea AM, Cohen JS, Greff LJ, Wiggs JL. A second locus for Rieger syndrome maps to chromosome 13q14. Am J Hum Genet. 1996;59:613–9. [PMC free article] [PubMed] [Google Scholar]
- 12.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 13.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wüthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–23. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 14.Kumar J, Moses K. Transcription factors in eye development: a gorgeous mosaic? Genes Dev. 1997;11:2023–8. doi: 10.1101/gad.11.16.2023. [DOI] [PubMed] [Google Scholar]
- 15.Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–83. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–3. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- 17.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–47. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans AL, Gage PJ. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum Mol Genet. 2005;14:3347–59. doi: 10.1093/hmg/ddi365. [DOI] [PubMed] [Google Scholar]
- 19.Cox CJ, Espinoza HM, McWilliams B, Chappell K, Morton L, Hjalt TA, Semina EV, Amendt BA. Differential regulation of gene expression by PITX2 isoforms. J Biol Chem. 2002;277:25001–10. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- 20.Amendt BA, Semina EV, Alward WL. Rieger syndrome: a clinical, molecular, and biochemical analysis. Cell Mol Life Sci. 2000;57:1652–66. doi: 10.1007/PL00000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lines MA, Kozlowski K, Kulak SC, Allingham RR, Héon E, Ritch R, Levin AV, Shields MB, Damji KF, Newlin A, Walter MA. Characterization and prevalence of PITX2 microdeletions and mutations in Axenfeld-Rieger malformations. Invest Ophthalmol Vis Sci. 2004;45:828–33. doi: 10.1167/iovs.03-0309. [DOI] [PubMed] [Google Scholar]
- 22.Maciolek NL, Alward WL, Murray JC, Semina EV, McNally MT. Analysis of RNA splicing defects in PITX2 mutants supports a gene dosage model of Axenfeld-Rieger syndrome. BMC Med Genet. 2006;7:59. doi: 10.1186/1471-2350-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Houssaye G, Bieche I, Roche O, Vieira V, Laurendeau I, Arbogast L, Zeghidi H, Rapp P, Halimi P, Vidaud M, Dufier JL, Menasche M, Abitbol M. Identification of the first intragenic deletion of the PITX2 gene causing an Axenfeld-Rieger Syndrome: case report. BMC Med Genet. 2006;7:82. doi: 10.1186/1471-2350-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin AV, Walter MA, Héon E. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld-Rieger syndrome. Hum Mol Genet. 2001;10:1631–8. doi: 10.1093/hmg/10.16.1631. [DOI] [PubMed] [Google Scholar]
- 25.Espinoza HM, Cox CJ, Semina EV, Amendt BA. A molecular basis for differential developmental anomalies in Axenfeld–Rieger syndrome. Hum Mol Genet. 2002;11:743–53. doi: 10.1093/hmg/11.7.743. [DOI] [PubMed] [Google Scholar]
- 26.Saadi I, Semina EV, Amendt BA, Harris DJ, Murphy KP, Murray JC, Russo AF. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J Biol Chem. 2001;276:23034–41. doi: 10.1074/jbc.M008592200. [DOI] [PubMed] [Google Scholar]
- 27.Cella W, de Vasconcellos JP, de Melo MB, Kneipp B, Costa FF, Longui CA, Costa VP. Structural assessment of PITX2, FOXC1, CYP1B1, and GJA1 genes in patients with Axenfeld-Rieger syndrome with developmental glaucoma. Invest Ophthalmol Vis Sci. 2006;47:1803–9. doi: 10.1167/iovs.05-0979. [DOI] [PubMed] [Google Scholar]
- 28.Wolberger C. Transcription factor structure and DNA binding. Curr Opin Struct Biol. 1993;3:3–10. [Google Scholar]
- 29.Dave V, Zhao C, Yang F, Tung CS, Ma J. Reprogrammable recognition codes in bicoid homeodomain-DNA interaction. Mol Cell Biol. 2000;20:7673–84. doi: 10.1128/mcb.20.20.7673-7684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott MP, Tamkun JW, Hartzell GW. The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 31.Walter MA. PITs and FOXes in ocular genetics: the Cogan lecture. Invest Ophthalmol Vis Sci. 2003;44:1402–5. doi: 10.1167/iovs.02-0618. [DOI] [PubMed] [Google Scholar]
- 32.Alward WL, Semina EV, Kalenak JW, Héon E, Sheth BP, Stone EM, Murray JC. Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol. 1998;125:98–100. doi: 10.1016/s0002-9394(99)80242-6. [DOI] [PubMed] [Google Scholar]
- 33.Perveen R, Lloyd IC, Clayton-Smith J, Churchill A, van Heyningen V, Hanson I, Taylor D, McKeown C, Super M, Kerr B, Winter R, Black GC. Phenotypic variability and asymmetry of Rieger syndrome associated with PITX2 mutations. Invest Ophthalmol Vis Sci. 2000;41:2456–60. [PubMed] [Google Scholar]
- 34.Phillips JC. Four novel mutations in the PITX2 gene in patients with Axenfeld-Rieger syndrome. Ophthalmic Res. 2002;34:324–6. doi: 10.1159/000065602. [DOI] [PubMed] [Google Scholar]
- 35.Saadi I, Kuburas A, Engle JJ, Russo AF. Dominant negative dimerization of a mutant homeodomain protein in Axenfeld-Rieger syndrome. Mol Cell Biol. 2003;23:1968–82. doi: 10.1128/MCB.23.6.1968-1982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quentien MH, Pitoia F, Gunz G, Guillet MP, Enjalbert A, Pellegrini I. Regulation of prolactin, GH, and Pit-1 gene expression in anterior pituitary by Pitx2: An approach using Pitx2 mutants. Endocrinology. 2002;143:2839–51. doi: 10.1210/endo.143.8.8962. [DOI] [PubMed] [Google Scholar]
- 37.Chi YI. Homeodomain revisited: a lesson from disease-causing mutations. Hum Genet. 2005;116:433–44. doi: 10.1007/s00439-004-1252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]