Abstract
NuGO, the European Nutrigenomics Organization, utilizes 31 powerful computers for, e.g., data storage and analysis. These so-called black boxes (NBXses) are located at the sites of different partners. NuGO decided to use GenePattern as the preferred genomic analysis tool on each NBX. To handle the custom made Affymetrix NuGO arrays, new NuGO modules are added to GenePattern. These NuGO modules execute the latest Bioconductor version ensuring up-to-date annotations and access to the latest scientific developments. The following GenePattern modules are provided by NuGO: NuGOArrayQualityAnalysis for comprehensive quality control, NuGOExpressionFileCreator for import and normalization of data, LimmaAnalysis for identification of differentially expressed genes, TopGoAnalysis for calculation of GO enrichment, and GetResultForGo for retrieval of information on genes associated with specific GO terms. All together, these NuGO modules allow comprehensive, up-to-date, and user friendly analysis of Affymetrix data. A special feature of the NuGO modules is that for analysis they allow the use of either the standard Affymetrix or the MBNI custom CDF-files, which remap probes based on current knowledge. In both cases a .chip-file is created to enable GSEA analysis. The NuGO GenePattern installations are distributed as binary Ubuntu (.deb) packages via the NuGO repository.
Keywords: GenePattern, GetResultsForGo, LimmaAnalysis, NuGOArrayQualityAnalysis, NuGOExpressionFileCreator, TopGoAnalysis
Introduction
NuGO (http://www.nugo.org) provides, to each partner, a local NuGO Black Box (NBX: a powerful computer to, e.g., store (nutri-BASE: http://www.ebi.ac.uk/~oyeniran/) and analyze (GenePattern: http://www.broad.mit.edu/cancer/software/GenePattern/) [6] NuGO and Affymetrix microarrays. NuGO arrays are custom made Affymetrix arrays. NuGO defined the probes on these chips, but kept the same design and setup as seen on standard Affymetrix expression arrays. The NuGO technology work package has put much effort into helping NuGO members to analyze their “omics” data. In this paper, we focus on the GenePattern modules created by NuGO to help NuGO scientists with their microarray analyses.
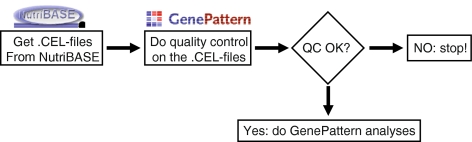
GenePattern, provided by the Broad institute, is a simple interface to a large number of analytic tools for genomics data. Modules are written in Java, MATLAB, Perl, or R/Bioconductor. The user friendly graphical interface allows biologist to easily enter their data and choose suitable settings in order to perform complex analyses (quality control, statistics, gene enrichment analysis, and so on), without detailed knowledge of the underlying programming language, algorithms and settings, allowing them to concentrate their efforts on interpretation of biologically meaningful results. A GenePattern user can run individual modules or create a pipeline, combining the use of various modules. A typical GenePattern pipeline could look as depicted in Fig. 1.
Fig. 1.
An example of a GenePattern workflow. Before performing statistics on a microarray experiment, it must be verified whether the array quality is acceptable
GenePattern managers can build their own modules. In the next paragraphs, we will present some modules, created by NuGO, which allow complex and easy to use analysis of affymetrix microarray data in GenePattern.
NuGOArrayQualityAnalysis
As depicted in Fig. 1, all microarray analysis should start with assessing the quality of your microarrays. Only if you are sure that the expression values derived from the arrays reflect your experimental setting (answers your scientific question), you can safely continue with the statistical analyses described below. The module takes the .CEL-files, the typical output format of GCOS affymetrix software where intensity calculations on the pixel values of the .dat files (the raw image data from affymetrix chip scanner) are stored (packed in a single zip-file), and submits them to a dedicated NuGO R-server via web services (written in Perl), executes the quality control pipeline utilizing a Bioconductor script, and returns the result as a single zip-file. The quality control procedure is identical to the implementation in MADMAX [3] (https://madmax.bioinformatics.nl) and the web services are just computer protocols to handle file transfers (via secure http) to execute the Bioconductor calculations, and to return the results (http://nugo-r.bioinformatics.nl/MADMAX_services/MADMAX_services.html) [5].
NuGOExpressionFileCreator
Utilizing GenePattern requires a function to import (and normalize) the data in such a way that all other modules can work with it. NuGOExpressionFileCreator is an enhanced version from the standard ExpressionFileCreator module that is present in GenePattern. It uses the most up-to-date Bioconductor version and supports the MBNI Custom CDF-files (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp) [2]. NuGO provides an up-to-date R and Bioconductor installation for the NBXses including annotation libraries for many Custom CDF-files (http://nugo-r.bioinformatics.nl/NuGO_R.html) and support libraries for the NuGO arrays. The NuGO GenePattern installation depends on these Ubuntu (.deb) R and Bioconductor packages. Consequently, NuGOExpressionFileCreator can create a so-called .chip-file that is specific for the utilized custom CDF-file. The .chip-file is required for properly executing the Gene Set Enrichment Analysis (GSEA: http://www.broad.mit.edu/gsea/) [7] later on in GenePattern (if the biologist wants to).
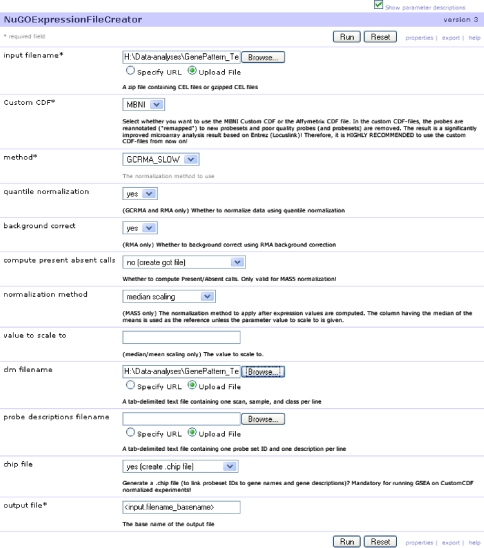
Figure 2 shows the input as a zip-file containing the .CEL-files and a so-called .clm file that defines the groups that the .CEL-files belong to (GenePattern help describes the proper file format). Some settings, e.g., normalization method, are required but the default values are usually convenient. Typically, the user loads the zip-file and the .clm-file and clicks on the “run” button. The output files are a .gct file and a .cls file: common GenePattern input files that can be used by many modules.
Fig. 2.
A screenshot of the NuGOExpressionFileCreator module at initialization of a run
LimmaAnalysis
The NuGO GenePattern module LimmaAnalysis implements the bioconductor package Limma (available at http://www.bioconductor.org/packages/release/bioc/html/limma.html). The Limma package uses moderated t and F statistics based on linear modelling in order to perform differential gene expression analysis for data arising from microarray experiments. The main advantage of Limma over traditional t or F tests is that for the estimation of variances and standard errors of a single gene information is borrowed from other genes, which stabilizes the analysis particularly for small sample sizes [8, 9]. The GenePattern module LimmaAnalysis provides an easy to use interface, allowing Limma analysis to be performed by researchers unfamiliar with the R and Bioconductor programming environment. At present, the LimmaAnalysis module can handle one-factorial experimental designs with a potential additional blocking factor. This covers many relevant study designs in nutritional research as for example the comparison of different dietary treatments on the same subject. The module takes the .gct file and .cls file obtained from NuGOExpressionFileCreator as input and returns the Limma results table including P values, Benjamini-Hochberg corrected P values and average log-ratios for all possible pairwise comparisons in separate files. The result tables give scientists an overview about which genes are significantly differentially expressed but they can also be used for further analysis with, e.g., the NuGO modules TopGoAnalysis and GetResultForGO.
TopGoAnalysis and GetResultForGO
The GenePattern module TopGoAnalysis implements the Bioconductor package topGO (available at http://bioconductor.org/packages/release/bioc/html/topGO.html). The Bioconductor module topGO is a gene enrichment analysis tool, which integrates the knowledge about the relationship between GO terms for the calculation of statistical significance. It allows to choose from three methods for Gene Ontology term scoring: Classic Method (classic), Eliminating Genes method (elim), or Weighting Genes method (weight) and the application of two test statistics: Fisher exact test (FIS) [4] or Kolmogorov–Smirnov test (KS) [7]. For further details on the topGO Bioconductor package see [1]. The GenePattern module TopGoAnalysis provides an intuitive user interface and implements five tests, combining the three methods for Gene Ontology term scoring with the two test statistics. For each test it returns the results table of the top 100 enriched GO identifiers plus a .pdf file containing the GO-graph for each test performed (Table 1).
Table 1.
An overview of the utilized GO test statistics combined with the GO terms scoring
| TopGoAnalysis test | topGO method for ontology scoring | topGO test statistic |
|---|---|---|
| ClassicFis | Classic | Fisher exact test |
| ElimFis | Elim | Fisher exact test |
| WeightFis | Weight | Fisher exact test |
| ClassicKS | Classic | KS statistics |
| ElimKS | Elim | KS statistics |
TopGoAnalysis is a quick way to gain information about the main GO molecular functions (MF), biological processes (BP), or cellular components (CC) affected by any treatments applied in nutritional research. This information is useful as a guide for the design of follow up experiments, because if, for example, a nutritional treatment affects many genes associated with a particular MF, BP, or CC this is likely to be much more relevant compared to any effects observed on single genes. To provide help with the design of follow-up experiments, any GO identifiers of interest can be further investigated with the GenePattern module GetResultForGO. This module utilizes an R script, which allows users to filter the LimmaAnalysis results table for genes annotated with a particular GO identifier of interest. This is useful for, e.g., examining whether genes associated with a particular GO identifier are predominantly up or down regulated in response to a nutritional or other intervention. It also allows the identification of those genes associated with a particular GO identifier, which responded most strongly to a particular treatment to help determine the most suitable target genes for validation of microarray data with real-time PCR.
GSEA and GSEALeadingEdgeViewer
These modules are already useful in the form provided in GenePattern. For GSEA, the biologist should provide, next to the .gct and .cls files, the proper .chip-file (provided by NuGOExpressionFileCreator). After finishing the GSEA analysis, the GSEALeadingEdgeViewer can be started to more thoroughly examine the GSEA results.
Conclusion
NuGO provides a number of user-friendly tools that implement the most relevant and up-to-date Bioconductor modules with respect to Affymetrix microarray analysis, which biologists can run in GenePattern, without knowledge of the R programming language. This approach allows biologists to focus on the underlying biology. Please note that GenePattern provides modules not only for microarray, but also for SNP, proteomics, and sequence analysis.
Acknowledgments
The authors thank the NuGO community for the financial support, user feedback, expert help, and notes of appreciation for our efforts. Furthermore, we thank the Broad institute for their help and their permission to maintain (and share) a NuGO binary GenePattern distribution.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22(13):1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 2.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acid Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavai AK, de Groot PJ, Lin K, Hooiveld G, Liu Y, Nijveen H, Neerincx P, Müller M, Leunissen JAM (2008) MADMAX—Management and analysis database for multiplatform microArray experiments. BMC Bioinformatics (submitted)
- 4.Lehman EL. Testing statistical hypothesis. Springer texts in statistics. 2. New York: Springer; 1986. [Google Scholar]
- 5.Neerincx PBT, de Groot PJ, Müller MR, Leunissen JAM (2008) MADMAX services—BioMoby web services for Affymetrix microarray normalization and quality control (in preparation)
- 6.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15555. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biolgy. 2004;1:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 9.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, Heidelberg