Abstract
Background
Rett syndrome (RTT) is an X linked neuro‐developmental disorder affecting mostly girls. Mutations in the coding region of MECP2 are found in 80% of classic RTT patients. Until recently, the region encoding MECP2 was believed to comprise exons 2, 3, and 4 with the ATG start site located at the end of exon 2 (MeCP2_e2).
Methods
Recent reports of another mRNA transcript transcribed from exon 1 (MeCP2_e1) prompted us to screen exon 1 among RNA samples from 20 females with classic or atypical RTT.
Results
A previously reported 11 base pair deletion in exon 1 was detected in one subject with a milder phenotype. Although RNA expression for both protein isoforms was detected from the mutant allele, evaluation of MeCP2 protein in uncultured patient lymphocytes by immunocytochemistry revealed that MeCP2 protein production was restricted to only 74–76% of lymphocytes. X chromosome inactivation studies of genomic DNA revealed similar XCI ratios at the HUMARA locus (73:27 with HpaII and 74:26 with McrBC). We have demonstrated that translation but not transcription of the MeCP2_e2 isoform is ablated by the 11 nucleotide deletion, 103 nucleotides upstream of the e2 translation start site.
Conclusions
These findings reveal that nucleotides within the deleted sequence in the 5′‐UTR of the MeCP2_e2 transcript, while not required for transcription, are essential for translation.
Keywords: 5′‐UTR, MeCP2, MECP2 , Rett syndrome, RNA secondary structure
RTT syndrome (RTT) is a rare neuro‐developmental disorder affecting 1 in 10 000 girls and is mostly caused by mutations in the X linked methyl CpG binding protein 2 gene (MECP2), which is subject to X inactivation.1,2,3 Until recently, its protein coding region was believed to be restricted to sequence within exons 2, 3, and 4 with the translational start site positioned near the 3′ end of exon 2. Reports in 2004 of an alternate mRNA transcript arising from exon 1, which is alternately spliced to exons 3 and 4, reveal a higher level of complexity than was previously appreciated. The protein isoform arising from exon 1 has been designated MeCP2_e1 (e1), while the originally defined protein isoform translated from exon 2 is now designated MeCP2_e2 (e2). Further, expression levels of the MeCP2_e1 mRNA transcript and protein isoform are much higher in brain than the expression levels of MeCP2_e2 mRNA transcript and its corresponding protein isoform.4,5
Mutations in exons 3 and 4 are detected in 80% of patients with clinically diagnosed classic RTT and in a lower proportion of atypical RTT cases.6,7 To date mutations have not been detected in exon 2, although a limited number of RTT causing mutations in exon 1 have now been reported.5,8,9 Using primers in exons 1 and 3, we screened RNA prepared from fresh lymphocytes from 20 RTT cases that had been classified as either classic or atypical RTT within the Australian Rett Syndrome Database10 and in whom earlier mutation screening had yielded wild type results. In one subject, we identified an 11 base pair deletion in exon 1 that has been previously reported.8,9 The deletion lies in the AGG repeat region of exon 1 and was confirmed by direct sequence analysis of genomic DNA. Subsequent assessment of MeCP2 protein production in uncultured fresh lymphocytes by immunocytochemical analysis revealed absence of detectable immunostaining in 24–26% of cells. Since the deletion lies in the 5′‐UTR region of e2, we hypothesised that expression of the e2 RNA transcript may be disrupted. However, fragment length analysis of amplified cDNA revealed all four transcripts, two from the wild type allele and two from the mutant allele. We validated our immunocytochemistry results by quantitative assessment of the X inactivation ratio at the HUMARA locus in DNA prepared from the same uncultured patient cells and observed similar ratios of 73:27 with HpaII, and 74:26 with McrBC. Our observations suggest that the nucleotides within the deleted sequence in the 5′‐UTR of the e2 RNA transcript are essential for MeCP2_e2 protein translation.
Methods
Sample collection
Molecular and immunocytochemical studies were conducted on blood samples collected in tubes with ACD anti‐coagulant from 20 female patients with classic or atypical RTT identified from the Australian Rett Syndrome Database and in whom earlier sequencing and MLPA screening of exons 2, 3, and 4 had not revealed a mutation. Prior approval for the study had been provided by the ethics committee of Princess Margaret Hospital for Children, Perth. Clinical details of the second case were provided from InterRett.11
DNA analysis
DNA was extracted from 1200 μl of blood using the Puregene DNA extraction kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's instructions. PCR analysis was performed on 50 ng of DNA as described previously.12
mRNA analysis
Total RNA was extracted from 1200 μl of blood using the Purescript RNA extraction kit (Gentra Systems) as per the manufacturer's instructions. RT‐PCR was performed on RNA using the Titan one step RT‐PCR kit (Roche Applied Science, Castle Hill, NSW, Australia). In brief, 50 ng of RNA was added to the RT‐PCR mix containing dNTPs (10 mM), DTT (125 mM), 5× RT‐PCR buffer, 50 ng each of forward (Ex1F2) and reverse (Ex4R2) primers in exons 1 and 4, respectively, and 1 U of enzyme (refer to table 1 for primer sequences). After reverse transcription at 48°C (30 min), the samples were PCR amplified at an annealing temperature of 61°C for 25 cycles. A 1 μl aliquot of the product from the primary PCR was further amplified in a semi‐nested PCR reaction using primers in exon 1 (Ex1F2) and exon 3 (Ex3R2) and Tth polymerase enzyme at an annealing temperature of 61°C for 25 cycles. PCR products were assessed visually by electrophoresis on a 2% agarose gel.
Table 1 Sequences of primers used in reverse transcription and PCR analysis.
| Name | Sequence | Location |
|---|---|---|
| 1F2 | 5′ ‐TAAAAGCCGTCCGGAAAAT | Exon 1 |
| 3R2 | 5′ ‐CTCAGCAGAGTGGTGGGCTGA | Exon 3 |
| 4R2 | 5′ ‐AGTTTGAAAGGCATCTTGAC | Exon 4 |
Purification and sequencing of PCR products
PCR products were purified using the Qiaquick PCR purification kit or gel extracted using the Qiaex II Gel purification kit (Qiagen, Doncaster, Vic, Australia). The purified products were sequenced using BigDye terminator chemistry version 3.1 on an ABI 3730 (Applied Biosystems, Foster City, CA, USA). Sequence chromatograms were analysed using Chromas software (Technelysium, Tewantin, Qld, Australia).
Immunocytochemical analysis
Buffy coat was separated from blood samples within 24 h of venesection and frozen in DMSO with liquid nitrogen. Samples from healthy individuals were used as normal (positive staining) controls for immunostaining in each assay. For every analysis, autofluorescence controls (containing no primary or secondary antibody) and mock controls (containing no primary antibody but stained with secondary antibody) were set up as negative controls from each sample. When required for immunochemical studies, frozen cells from a patient and at least one healthy individual were thawed simultaneously and resuspended in RBC lysis buffer for 10 min. The residual white cell pellet was collected by centrifugation and fixed with 5% formaldehyde in PBS. Cells were permeabilised for 10 min in PBS containing 0.1% Triton X‐100 and blocked in PBS containing 1% BSA and 2% FCS before incubation with primary antibody (N‐terminal antibody generated using a peptide of amino acids 9–27 of human MeCP2 and C‐terminal antibody to amino acids 329–348 of human MeCP2, amino acid positions with reference to the e2 protein isoform) in a dilution of 1:100 in PBS containing 1% BSA, 2% FCS, and 0.1% Triton X for 45 min at room temperature. After three washes in PBS containing 1% BSA and 2% FCS, cells were incubated with a secondary goat anti‐rabbit antibody conjugated with FITC (Jackson Immunochemicals, West Grove, PA, USA) in a dilution of 1:100 in permeabilisation buffer containing 1% BSA and 2% FCS for 30 min at room temperature. The cells were then washed three times and cytospun onto slides. Slides were mounted in DAPI and cells were counted and analysed under a fluorescent microscope (Olympus, Melville, NY, USA).
Fragment length analysis
Fragment length analysis was performed on the products of RT‐PCR with FAM labelled forward primer 1F2 and reverse primer 3R2 (table 1). Products were analysed using GeneScan software (Applied Biosystems).
X chromosome inactivation studies
X chromosome inactivation analysis was conducted on 2 μg of DNA sample with two different methylation sensitive restriction enzymes. In one tube DNA was digested with HpaII, which digests unmethylated DNA, and in the other with McrBC, which digests methylated DNA. The digested DNA was amplified using previously published labelled primers for the androgen receptor gene and analysed using GeneScan software according to previously published protocols.13,14 For quantification, the peak area underneath the smaller allele was divided by the sum of the peak areas for both alleles after correction.
Analysis of the MeCP2_e2 5′‐UTR
We analysed the secondary structure of the wild type and mutant sequence of MeCP2_e2 5′‐UTR using the Mfold program available from the Burnet Institute, Melbourne at http://mfold.burnet.edu.au/. This program predicts optimal and suboptimal secondary structures of RNA based on free energy minimisation.15 The 170 nucleotide 5′‐UTR sequence (GenBank accession number AY523575) up to the AUG start codon and a sequence of 159 nucleotides (170 del 11) depicting the deletion of 11 nucleotides in the 5′‐UTR were folded using the default parameters. Planar tree graphs were constructed for comparison of secondary structures.16
Results
Proband
The proband was a female born by spontaneous vertex delivery after a pregnancy complicated only by hypertension requiring induction at term. The proband's mother had reported reduced fetal movements earlier in the pregnancy. Birth weight was 3317 g (10th–50th percentiles) and head circumference was 35 cm (50th percentile). The proband was initially dusky with an Apgar score of 7 at 1 min. She was breast fed but later experienced difficulty sucking a bottle.
She sat at age 5.5 months. Concerns about her development emerged between 6 and 10 months. At 10 months she was placid and bottom shuffling. Crawling began at 12 months. By 13 months she was pulling herself up on furniture and eventually walked at 21 months.
The family were initially worried, both because of her motor delay as well as her overall slowing down of development. Around 18 months she appeared to be socially disengaged and had autistic traits. She spoke her first words at 18 months but at about 30 months (at which time she had been speaking single words) her speech began to deteriorate. Gradual loss of hand use and development of hand stereotypies then emerged at 36 months. At the age of 13 years, she remains able to pick up a grape and finger feed.
Seizures, which started at around 3 years, have responded partially to carbamazepine. Currently, she does not have scoliosis but suffers from constipation, grinds her teeth, and has periodic rapid breathing. At the age of 13 years she remains independently mobile and is well nourished with no growth or other major health problems. Her clinical features are summarised in table 2 alongside the clinical features of the first reported case5 notified to InterRett.17
Table 2 Comparative table of phenotypical features from two reported cases with exon 1 mutation.
| Patient 212 (Australian case) | Patient 052 (InterRett case published in Mnatzakanian et al5) | |
|---|---|---|
| Pregnancy | ||
| Maternal health | Maternal hypertension requiring | No problems |
| induction at term | ||
| Pregnancy duration | 40 weeks | 40 weeks |
| Birth and first week of life | ||
| Birth weight | 3317 g | 3430 g |
| Birth head circumference | 35 cm | Not available |
| APGAR at 1 min | 7 | 9 |
| First week of life | No problems | No problems |
| Early development | ||
| Parental comments about period up to 6 months | Breast fed satisfactorily. Would not suck | Vomited frequently |
| on a bottle | ||
| Parental comments about period from 6 | Placid and rather slow. Wouldn't crawl | Unable to sit up unaided until 11 months. |
| to 10 months | Continued to vomit often in the morning. | |
| Age at independent walking | 21 months | 18 months |
| Age at use of words with meaning | 18 months | 18 months |
| Parental concerns | ||
| First concerns | Reduced social engagement at 18 months | Speech delay |
| First diagnosis | Autism | Autism |
| Age and year of clinical diagnosis of Rett syndrome | 3 years (1994) | 9 years (1985) |
| Features of regression | ||
| Level of speech prior to deterioration | Single words | 3 word sentences |
| Age at loss of speech | 30 months | 36 months |
| Nature of deterioration | Gradual | Gradual |
| Loss of hand use | Yes | Yes |
| Age at deterioration of hand use | 36 months | 48 months |
| Age at development of hand stereotypies | 36 months | 48 months |
| Current status | ||
| Age in 2005 | 13 years | 28 years |
| Speech at present | No | No |
| Ability to walk | Walks independently | Walks independently |
| Ability to finger feed | Yes | Able to fill and use a spoon with help |
| Best level of hand use today | Can pick up a grape and finger feed | Can finger feed, picks up a grape, holds a glass |
| to drink, and points to sign/objects with palm | ||
| Other features | ||
| Presence of epilepsy | Yes | Yes mild, fully suppressed with Tegretol |
| Age at diagnosis of seizures | 3 years | 11 years |
| Presence of scoliosis | No | Yes |
Detection of an 11 nucleotide deletion
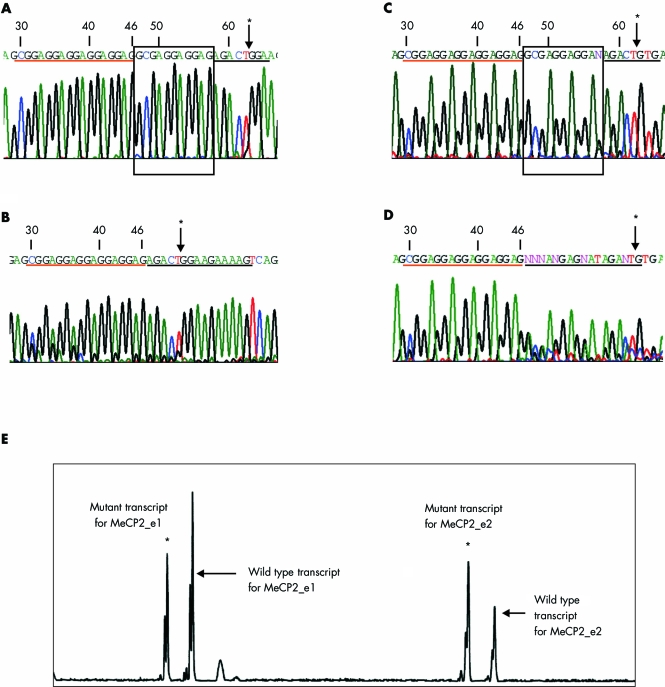
We determined the sequence of mRNA extracted from 20 RTT patients (10 atypical, 10 classic) in whom mutation screening of exons 2, 3, and 4 by sequencing and MLPA had yielded wild type results. In all patients, amplification with primers in exon 1 and exon 3 yielded two bands, which were confirmed by sequence analysis using forward and reverse primers to be wild type e1 and e2 mRNA transcripts. In a sample from one atypical RTT patient (212), a smaller band was also observed, which upon sequencing revealed a previously reported 11 nucleotide deletion in exon 1 (c.47_57del GCGAGGAGGAG) (fig 1A,B). Genomic DNA obtained from the patient was assessed by sequencing and the deletion was again detected in exon 1 (fig 1C,D).
Figure 1 Sequence chromatograms and fragment length analysis. Numbers in the chromatograms correspond to c.DNA sequence for MeCP2_e1 with the A of ATG start site numbered as 1. Arrows marked with an asterisk indicate the exon/exon junction in RNA sequences shown in panels A and B and exon/intron junction in DNA sequences shown in panels C and D. Nucleotides underlined in red are upstream of the deletion, while those in black are downstream. The 11 nucleotide deletion is shown in the box in sequences from normal individuals. (A, B) Sequence chromatograms from the RNA of a normal individual (A) and patient 212 (B). The nucleotides in the box in panel A were found to be deleted in the chromatogram obtained from the RNA of patient 212 shown in panel B. (C, D) Sequence chromatograms from the DNA of a normal individual (C) and patient 212 (D). The deletion is seen as a mixed chromatogram beneath the underlined sequence in the patient DNA in panel D. (E) Fragment length analysis of the mRNA transcripts with GeneScan software. All four RNA transcripts, two arising from the normal allele (indicated by arrows) and two from the mutant allele (indicated by asterisks), are clearly visible. The mutant RNA transcripts differ from the wild type RNA transcripts by 11 nucleotides.
Absence of MeCP2 in the peripheral lymphocytes
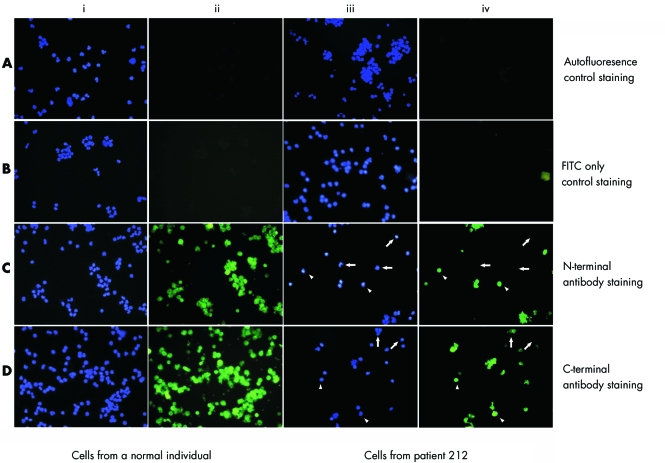
To investigate the effects of the 11 nucleotide deletion on protein translation, peripheral lymphocytes from the patient were analysed immunocytochemically with anti‐MeCP2 antibodies directed against the N‐ and C‐terminals which detect both MeCP2 protein isoforms. Peripheral lymphocytes from a normal subject were used as a positive control for the assay (fig 2A–D, panels i and ii). FITC staining was not detectable in cells from autofluorescence and mock controls from the normal individual (fig 2A, i and ii and fig 2B, i and ii) and patient 212 (fig 2A, iii and iv and fig 2B, iii and iv). We noticed that cells positively stained with MeCP2 antibodies show variable intensity of staining (fig 2C,D, i–iv). We counted 250 cells from positive control slides stained with the N‐terminal antibody and found 99.2% of cells (lower end of 95% CI 97 to 99) staining positively (fig 2C, i and ii). Similarly, from the positive control slides stained with the C‐terminal antibody (fig 2D, i and ii), we counted 289 cells and found positive staining in 98.6% of cells (lower end of 95% CI 96 to 99). In cells from patient 212, a mixture of fluorescing (positive, indicated by arrowheads) and non‐fluorescing (negative, indicated by arrows) cells were observed (fig 2C,D, iii and iv), indicating, in our interpretation, loss of both the MeCP2_e1 and MeCP2_e2 protein isoforms. We counted 321 cells from slides stained with the antibody against the N‐terminal epitope (fig 2C, iii and iv) and found positive staining in 76% of cells (95% CI 70 to 80). Similarly, on slides stained with the C‐terminal antibody (fig 2D, iii and iv), 74% (95% CI 68 to 78) of 285 cells counted, stained positively, with the remainder showing absence of staining.
Figure 2 Immunofluorescence analysis of peripheral blood lymphocytes. Panels i and ii show cells from a normal individual immunostained with DAPI (blue) and FITC (green), respectively. Panels iii and iv show cells from patient 212 stained with DAPI in blue and FITC in green, respectively. Panel A shows autofluorescent controls where no primary or secondary antibody was used during staining. Panel B shows FITC only staining controls, where cells were stained with the secondary antibody (mouse anti‐rabbit FITC) alone. Note green staining of cells is not detectable in the negative controls from a normal subject (i and ii) and patient 212 (iii and iv) shown in panels A and B. Cells stained with the antibody against the N‐terminal epitope are shown in panel C and cells stained with the antibody against the C‐terminal epitope are shown in panel D. All cells from a normal individual show positive green staining with antibodies against the N‐terminal and C‐terminal shown in panels i and ii. Note the mixture of green staining positives (indicated by arrowheads) and non‐staining negatives (indicated by arrows) in cells from the patient shown in panels iii and iv. In patient cells, 76% of cells stained positive with the antibody against the N‐terminal of MeCP2 and 74% stained positive with the antibody against the C‐terminal of MeCP2.
cDNA fragment length analysis
Fragment length was analysed by GeneScan on the RT‐PCR products to confirm the presence of deletions in both the e1 and e2 transcripts (fig 1E). Four transcripts, two wild type fragments (indicated by arrows in fig 1E) and two corresponding mutant fragments that were 11 base pairs shorter than the wild type lengths (indicated by asterisks in fig 1E), were revealed and this confirmed the presence of two RNA transcripts from the mutant allele.
X chromosome inactivation studies
Quantitative XCI results from the HpaII digest revealed a ratio of 73:27. An independent parallel assay with McrBC yielded a confirmatory ratio of 74:26.
Analysis of 5′‐UTR sequence
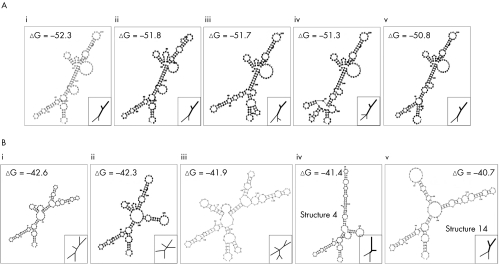
Since the 11 nucleotide deletion is upstream of the AUG start codon for the e2 protein isoform, we examined its 5′‐UTR sequence for features that could explain the influence of the deletion on the initiation of translation. The Mfold program predicted six secondary structures for the wild type 5′‐UTR RNA sequence and 16 for the deletion mutant RNA. The five most stable secondary structures for wild type sequence and the mutant sequence are shown in fig 3A,B. The Gibbs free energy of formation, which is approximated as a sum of negative contributions from helices plus positive contributions from the loss of entropy when loops and bulges form, ranged from −52.3 kcal/mol to −49.8 kcal/mol for the wild type sequence and from −42.6 kcal/mol to −40.5 kcal/mol for the deletion mutant. Structural analysis of the two transcripts revealed that the mutant sequence formed a more complicated secondary structure than the wild type sequence. Tree graphs of the secondary structures revealed a consistent Y shaped structure in the wild type sequence, whereas only two out of the 16 predicted structures of the mutant sequence (structure numbers 4 and 14, fig 3B, iv and v) had a Y shaped tree graph similar to the wild type sequence. Inspection revealed that the main stem of the Y shaped structure is made of 10 uninterrupted stacked pairs with three GU wobble bases in five out of six predicted structures for the wild type sequences. In contrast, the main stem of the Y shaped tree graph in structures 4 and 14 of mutant sequence is made of four uninterrupted stacked pairs with no GU wobble bases. In the wild type sequence, the double stranded stem is interrupted by a multi‐loop at one end consisting of 11–15 single stranded nucleotides and a bulge of five to nine single stranded bases at the other end, but in the mutant structures 4 and 14, the stem is interrupted by multi‐loops at both ends consisting of a maximum number of eight single stranded nucleotides at one end and two at the other. The multi‐loop and bulge interrupting the stem in the wild type structures have a GU wobble pair as the external closing pair whereas no such closing pair was observed in the multi‐loops interrupting the stem in the mutant sequence.
Figure 3 Predicted secondary structure analysis of wild type and mutant 5′‐UTR of MeCP2_e2 using Mfold. (A) The five most stable secondary structure predictions of the wild type MeCP2_e2 5′‐UTRs are shown in panels i–v. (B) Panels i–iv show the four most stable secondary structures predicted for the mutant MeCP2_e2 5′‐UTR, including structure 4. Panel v shows predicted structure 14. Simplified tree graphs drawn for each structure are depicted as insets. The Y shaped structure is indicated in bold within the insets. Only structures 4 and 14 of the mutant sequence show similarity to the tree graphs of the wild type structures with subtle differences that are discussed in the text.
Discussion
We have identified a case with an exon 1 mutation, which has been previously reported, by screening samples from 20 mutation negative classic and atypical RTT patients identified from the Australian Rett Syndrome Database.10 The mutation is an 11 base pair deletion 46 bases downstream from the start codon in exon 1 (c.47_57del GCGAGGAGGAG). The deletion results in a frame shift and generates a missense protein sequence after amino acid position 15, which stops after amino acid position 36 in the MeCP2_e1 protein isoform. In contrast, the reading frame of the MeCP2_e2 protein isoform is unaffected, as the 11 nucleotide deletion is positioned within the 5′‐UTR starting 103 nucleotides upstream of the e2 protein translation initiation site. Although the e1 protein isoform would be absent from a proportion of cells in this patient, we expected positive immunostaining in all lymphocytes due to the presence of the intact e2 protein isoform in all cells, which in lymphocytes is presumably much more abundantly produced than the e1 protein isoform, in line with the observed RNA levels.5 However, evaluation of MeCP2 protein in uncultured peripheral lymphocytes from the patient with anti‐MeCP2 antibodies directed against both the N‐ and C‐terminal epitopes, demonstrated no discernible protein in 24% (95% CI 19 to 29) and 26% (95% CI 21 to 31) of cells, respectively, which is significantly elevated compared with 0.8–1.4% (95% CI 0.2 to 3) of cells in the positive control slides that failed to stain.
To investigate further the reason for the absence of protein in cells, we looked for evidence of a shorter MeCP2_e2 mRNA species, which, if present, would indicate transcription from the mutant allele. Demonstration of the mutant RNA transcripts for both isoforms (indicated by asterisks in fig 1E) in the patient sample, from which RNA was extracted 24 h after venesection, suggests that RNA instability is very unlikely to be responsible for the absence of protein.
To assess the validity of our immunostaining results, we sought additional indirect evidence from quantitative XCI. Two independent assays at the HUMARA locus yielded XCI ratios that closely matched our observation of 74–76% of cells staining with antibodies against MeCP2 protein. Our immunostaining and X inactivation findings reveal that the wild type allele is active in the majority of patient lymphocytes, although it is appreciated that the X inactivation ratio in peripheral blood lymphocytes may not reflect the ratio in brain tissue.
It is known that the 5′‐UTR plays an important role in tissue specific protein translation and in some instances has been shown to regulate the level of protein translation without affecting transcription.18,19,20,21 From observations in a mouse model, which revealed that even mild overproduction of MeCP2 protein may cause neurological deficits,22 it appears that MeCP2_e2 protein levels in vivo are tightly regulated. It is known that the upstream AUG in exon 1 interferes with the translation of MeCP2_e2 protein isoform.4 Since the translation efficiency of a transcript is also affected by the length and secondary structure of its 5′‐UTR,23,24 we considered the possibility that the upstream AUG combined with altered RNA secondary structure may adversely affect translation24,25 and examined the predicted secondary structures arising from mutant 5′‐UTR compared with those predicted for the wild type. The Gibbs free energy of formation for the predicted secondary structures of the wild type and mutant 5′‐UTRs indicates that both are stable, which strongly suggests that additional factors are required for initiation of translation.24,26,27 It is interesting to note that part of the deleted sequence (GCGAGGAGGAG) bears similarity to the Shine‐Dalgarno sequence (AGGAGG), which is important for initiation of translation in prokaryotes28,29 and archaea,30 although its significance in mammalian cells is unknown. The deleted sequence also participates in the formation of a nine nucleotide repeat GAGGAGGAG around GC (GAGGAGGAGGCGAGGAGGAG). Such repeats in the 5′‐UTR could provide binding sites for proteins involved in translation.24 The tree diagrams clarify the general shape of the predicted folds and reveal that five out of the six predicted structures for the wild type sequence retain a similar overall structure, consisting of a long double stranded stem interrupted by single stranded loops. The deleted sequence contributes to the double stranded stem in all the predicted structures for the wild type sequence. This double stranded stem is shortened upon deletion of the 11 bases causing a change in secondary structure. Whether such subtle differences in secondary structure are sufficient to cause ablation of MeCP2_e2 protein translation remains to be determined experimentally.
Alternatively, it is possible that the e1 protein isoform is abundantly produced in lymphocytes, while the e2 protein isoform remains poorly translated despite higher levels of e2 mRNA transcript, because of interference from the upstream AUG in exon 1. It has been shown in vitro that the start codon in exon 1 interferes with the initiation of translation at the start site in exon 2, resulting in low levels of e2 protein isoform.4 Furthermore, the levels of native e2 protein isoform were noted to be 10 times lower than the levels of native e1 protein isoform in the brain tissue. As the deletion does not disrupt the upstream AUG in our subject, it seems likely that the phenomenon of translational interference has remained intact and, in combination with the RNA secondary structural changes induced by the mutation, results in a major reduction in the levels of the e2 protein isoform. Additional in vivo studies designed to determine endogenous levels of e1 and e2 protein isoforms in lymphocytes and other tissues would indicate whether the upstream AUG interferes with translation from the MeCP2_e2 start site in these tissues.
Thus, at this stage, the exact molecular mechanism leading to the unexpected absence of MeCP2_e2 protein isoform in a proportion of cells in this patient remains undefined. It cannot be explained on the basis of protein instability, as the mutation is upstream of the coding region of the e2 RNA transcript. Nonsense mediated decay is ruled out by our observation of the retention of the mutant e1 RNA transcript containing the premature termination codon, as well as the mutant e2 RNA transcript with the normal coding sequence. Combining the results of our analysis of the secondary structures of mutant and wild type 5′‐UTR and a previous study on upstream AUG in exon 14 suggests that protein translation from the e2 mRNA transcript is regulated in part by the upstream AUG and factors binding within or to sequences overlapping the 11 deleted nucleotides in its 5′‐UTR, or to secondary structures induced by the deleted sequence. Further experiments designed to elucidate these binding factors should yield some insights into the exact mechanism of regulation. Nevertheless, our protein and RNA level analysis of this non‐coding mutation extends knowledge of the biological consequences of this particular mutation. The splice site mutation reported by Amir et al9 also has the potential to disrupt both protein isoforms, although an aberrantly spliced RNA transcript was not observed in that study. Importantly, our findings do not support a gathering view that the mutation we have studied ablates only the MeCP2_e1 protein isoform and that an RTT‐like phenotype may occur in the presence of a normal MeCP2_e2 protein isoform.8
Comparison of the clinical features of the Australian and the Canadian case, which was originally reported by Mnatzakanian et al5 and subsequently notified to the international InterRett database,17 reveals some interesting similarities. Although the two subjects have different exon 1 mutations at the genomic DNA level, because of tri‐nucleotide repeats in the region of deletions, the 11 nucleotide deletion reported in Mnatzakanian et al results in the generation of the same mutant DNA and RNA sequence as in our subject. Consequently, both the mutations generate identical missense proteins that then terminate after amino acid 36. Both subjects have a phenotype that is slightly milder than usual. Regression in both subjects occurred later than normally expected and motor function (including hand function) was better than average for subjects with classic Rett syndrome. The Australian patient is 15 years younger than the Canadian subject, who, at 28 years, has a slightly lower level of mobility. Quantitative X inactivation studies on the samples from the Canadian case revealed a ratio of 64:46 (personal communication, Dr Patrick MacLeod). We note that the atypical case reported by Amir et al,9 who has the same 11 base pair deletion as our case, had random X inactivation although no additional clinical details were provided. Until more clinical data are published on these exon 1 cases, it is not possible to comment further on the most likely biological basis of the observed phenotypes, in particular whether they reflect patterns of X inactivation, bearing in mind the immunostaining finding in the Australian case that reveals the wild type allele is active in the majority of cells, a finding that is consistent with the observed milder clinical features in this child. Alternatively, this particular mutation may be characteristically milder in its clinical consequences, as has been found for mutations such as R133C and R306C.31,32
In conclusion, as well as revealing the biological importance of the deleted non‐coding region of RNA for protein translation, our findings confirm the emerging consensus that mutations in exon 1 are responsible for only a minority of RTT cases, which is not surprising given the small proportion of nucleotides contributed by exon 1 to the gene's total coding sequence. Pooling the findings among our 20 cases with the earlier published findings from exon 1 screening reveals that a total of seven exon 1 mutations have been detected among 209 mutation negative RTT patients (2/19 patients in Mnatzakanian et al,3 2/63 in Amir et al,9 0/97 in Evans et al, 2/10 in Ravn et al,8 and 1/20 in this study). At a practical level, this permits an estimate to be made of the prevalence of exon 1 mutations among mutation negative RTT cases of 3.3% (95% CI 1.6 to 6.7). As mutation negative RTT patients represent approximately one in five of all cases, the overall contribution of exon 1 mutations to the occurrence of RTT is less than 1%.
Acknowledgements
We would like to thank Dr Andrew Barker from the Laboratory for Cancer Medicine, WAIMR, UWA for guidance and critical reading of this manuscript, Mrs Carol Philippe for her work in facilitating sample collection, and Dr Wendy Robinson from the Department of Medical Genetics, University of British Columbia, Canada, for X inactivation analysis on the Canadian subject. We would also like to express our gratitude to all the families who have participated in the study, the Australian Paediatric Surveillance Unit, and the Rett Syndrome Association of Australia who continue to facilitate case ascertainment of Rett syndrome in Australia.
Electronic‐database information
The web site of the Burnet Institute, Melbourne can be found at http://mfold.burnet.edu.au/
Footnotes
The authors would like to acknowledge the Raine Foundation and Rett Syndrome Australian Research Fund for funding of this project as well as the National Institute of Child Health and Human Development for its current funding of the Australian Rett Syndrome Database under NIH grant number 1 R01 HD43100‐01A1 PI. InterRett is funded by the International Rett Syndrome Association. DR is supported by a grant from the University of Western Australia, HL is funded by NHMRC program grant 353514, and JC is funded by NHMRC project grants 185202 and 346603.
Competing interests: none declared
Patient details are published with consent
The web site of the Burnet Institute, Melbourne can be found at http://mfold.burnet.edu.au/
References
- 1.Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F. Rett syndrome: the complex nature of a monogenic disease. J Mol Med 200381(6)346–354. [DOI] [PubMed] [Google Scholar]
- 2.Colvin L, Leonard H, de Klerk N, Davis M, Weaving L, Williamson S, Christodoulou J. Refining the phenotype of common mutations in Rett syndrome. J Med Genet 200441(1)25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quaderi N A, Meehan R R, Tate P H, Cross S H, Bird A P, Chatterjee A, Herman G E, Brown S D. Genetic and physical mapping of a gene encoding a methyl CpG binding protein, Mecp2, to the mouse X chromosome. Genomics 199422(3)648–651. [DOI] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Bird A. The major form of MeCP2 has a novel N‐terminus generated by alternative splicing. Nucleic Acids Res 200432(5)1818–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mnatzakanian G N, Lohi H, Munteanu I, Alfred S E, Yamada T, MacLeod P J, Jones J R, Scherer S W, Schanen N C, Friez M J, Vincent J B, Minassian B A. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet 200436(4)339–341. [DOI] [PubMed] [Google Scholar]
- 6.Hagberg B. Clinical manifestations and stages of Rett syndrome. Ment Retard Dev Disabil Res Rev 20028(2)61–65. [DOI] [PubMed] [Google Scholar]
- 7.Amir R E, Van den Veyver I B, Wan M, Tran C Q, Francke U, Zoghbi H Y. Rett syndrome is caused by mutations in X‐linked MECP2, encoding methyl‐CpG‐binding protein 2. Nat Genet 199923(2)185–188. [DOI] [PubMed] [Google Scholar]
- 8.Ravn K, Nielson J, Schwartz M. Mutations found within exon 1 of MECP2 in Danish patients with Rett syndrome. Clin Genet 200567(6)532–533. [DOI] [PubMed] [Google Scholar]
- 9.Amir R E, Fang P, Yu Z, Glaze D G, Percy A K, Zoghbi H Y, Roa B B, Van den Veyver I B. Mutations in exon 1 of MECP2 are a rare cause of Rett syndrome. J Med Genet 200542(2)e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colvin L, Fyfe S, Leonard S, Schiavello T, Ellaway C, De Klerk N, Christodoulou J, Msall M, Leonard H. Describing the phenotype in Rett syndrome using a population database. Arch Dis Child 200388(1)38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fyfe S, Cream A, de Klerk N, Christodoulou J, Leonard H. InterRett and RettBASE: International Rett Syndrome Association databases for Rett syndrome. J Child Neurol 200318(10)709–713. [DOI] [PubMed] [Google Scholar]
- 12.Evans J C, Archer H L, Whatley S D, Kerr A, Clarke A, Butler R. Variation in exon 1 coding region and promoter of MECP2 in Rett syndrome and controls. Eur J Hum Genet 200513(1)124–126. [DOI] [PubMed] [Google Scholar]
- 13.Pegoraro E, Schimke R N, Arahata K, Hayashi Y, Stern H, Marks H, Glasberg M R, Carroll J E, Taber J W, Wessel H B.et al Detection of new paternal dystrophin gene mutations in isolated cases of dystrophinopathy in females. Am J Hum Genet 199454(6)989–1003. [PMC free article] [PubMed] [Google Scholar]
- 14.Karasawa M, Tsukamoto N, Yamane A, Okamoto K, Maehara T, Yokohama A, Nojima Y, Omine M. Analysis of the distribution of CAG repeats and X‐chromosome inactivation status of HUMARA gene in healthy female subjects using improved fluorescence‐based assay. Int J Hematol 200174(3)281–286. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol 199425267–294. [DOI] [PubMed] [Google Scholar]
- 16.Gan H H, Pasquali S, Schlick T. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design. Nucleic Acids Res 200331(11)2926–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore H, Leonard H, Fyfe S, de Klerk N N L. InterRett ‐ the application of bioinformatics to international Rett syndrome research. Ann Hum Biol 200532(2)228–236. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol 1991115(4)887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer H A, Thomas A A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′‐untranslated region of an mRNA. Biochem J 2002367(Pt 1)1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkie G S, Dickson K S, Gray N K. Regulation of mRNA translation by 5′‐ and 3′‐UTR‐binding factors. Trends Biochem Sci 200328(4)182–188. [DOI] [PubMed] [Google Scholar]
- 21.Lammich S, Schobel S, Zimmer A K, Lichtenthaler S F, Haass C. Expression of the Alzheimer protease BACE1 is suppressed via its 5′‐untranslated region. EMBO Rep 20045(6)620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins A L, Levenson J M, Vilaythong A P, Richman R, Armstrong D L, Noebels J L, David Sweatt J, Zoghbi H Y. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet 200413(21)2679–2689. [DOI] [PubMed] [Google Scholar]
- 23.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol 199814399–458. [DOI] [PubMed] [Google Scholar]
- 24.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 20023(3)reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray N K, Hentze M W. Regulation of protein synthesis by mRNA structure. Mol Biol Rep 199419(3)195–200. [DOI] [PubMed] [Google Scholar]
- 26.Koromilas A E, Lazaris‐Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non‐coding region translate efficiently in cells overexpressing initiation factor eIF‐4E. EMBO J 199211(11)4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Velden A W, Thomas A A. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol 199931(1)87–106. [DOI] [PubMed] [Google Scholar]
- 28.Shine J, Dalgarno L. Terminal‐sequence analysis of bacterial ribosomal RNA. Correlation between the 3′‐terminal‐polypyrimidine sequence of 16‐S RNA and translational specificity of the ribosome. Eur J Biochem 197557(1)221–230. [DOI] [PubMed] [Google Scholar]
- 29.Shinedling S, Gayle M, Pribnow D, Gold L. Mutations affecting translation of the bacteriophage T4 rIIB gene cloned in Escherichia coli. Mol Gen Genet 1987207(2–3)224–232. [DOI] [PubMed] [Google Scholar]
- 30.Karlin S, Brocchieri L, Campbell A, Cyert M, Mrazek J. Genomic and proteomic comparisons between bacterial and archaeal genomes and related comparisons with the yeast and fly genomes. Proc Natl Acad Sci U S A 2005102(20)7309–7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard H, Colvin L, Christodoulou J, Schiavello T, Williamson S, Davis M, Ravine D, Fyfe S, de Klerk N, Matsuishi T, Kondo I, Clarke A, Hackwell S, Yamashita Y. Patients with the R133C mutation: is their phenotype different from patients with Rett syndrome with other mutations? J Med Genet 200340(5)e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schanen C, Houwink E J, Dorrani N, Lane J, Everett R, Feng A, Cantor R M, Percy A. Phenotypic manifestations of MECP2 mutations in classical and atypical Rett syndrome. Am J Med Genet 2004126A(2)129–140. [DOI] [PubMed] [Google Scholar]