Abstract
Background
Parkinson's disease is a genetically complex disease with mixed mode of inheritance. Recently, a haplotype across the sepiapterin reductase (SPR) gene, which is located in the PARK3 linkage region, was shown to modulate age of onset of Parkinson's disease in sibships from North America.
Objective
To make a thorough assessment of the SPR gene region in sporadic Parkinson's disease.
Methods
A linkage study in 122 European sibship families with five microsatellite and 17 single nucleotide polymorphism (SNP) markers in and around the SPR gene region, and an association analysis in 340 sporadic cases of Parkinson's disease and 680 control subjects from Germany with 40 SNPs. Linkage was evaluated by non‐parametric linkage scores and genotypic or haplotype association was tested by regression analysis, assuming different risk effect models.
Results
Significant LOD scores between 2 and 3 were obtained at the two SPR‐flanking markers D2S2110 and D2S1394 and seven SNP markers around the SPR gene. We found the previously reported promoter SNP rs1876487 also significantly associated with age of onset in our sib pair families (p‐value 0.02). One strong linkage disequilibrium (LD) block of 45 kb including the entire SPR gene was observed. Within this LD block all 14 inter‐correlated SNPs were significantly associated with Parkinson's disease affection status (p‐value 0.004).
Conclusions
DNA polymorphisms in a highly intercorrelated LD block, which includes the SPR gene, appear to be associated with both sporadic and familial Parkinson's disease. This confirms a previous study showing that SPR potentially modulates the onset of or risk for Parkinson's disease.
Keywords: linkage disequilibrium, PARK3, Parkinson's disease, sepiapterin reductase
Parkinson's disease is one of the most common neurodegenerative disorders, affecting around 2% of the population above 65 years of age.1 Pathological features include degeneration of dopaminergic neurones of the substantia nigra pars compacta, and the presence of eosinophilic inclusions known as Lewy bodies in affected brain areas.2
Six genes for monogenically inherited forms of Parkinson's disease, which account for only a small fraction of all Parkinson's disease cases, have been identified. It has been shown that mutations in the parkin gene (PARK2), in the PINK1 gene (PARK6), and in the DJ‐1 gene (PARK7) cause autosomal recessive early onset parkinsonism.3,4,5 The α‐synuclein gene (SNCA) has been implicated in rare forms of dominantly inherited Parkinson's disease (PARK1), either by missense mutations or gene multiplications.6,7,8 Leucine‐rich repeat kinase 2 (LRRK2) has been identified as the disease causing gene for late onset autosomal dominant parkinsonism (PARK8).9,10 In addition, various genetic studies have detected linkage to several other chromosomal regions, for which the disease causing genes have yet to be detected: PARK3 (2p13), PARK9 (1p36), PARK10 (1p32), and PARK11(2q13).11 Eight genome scans evaluating linkage with affection status as well as with age of onset have been carried out using sibling pair cohorts as well as multiplex families from North American and European populations.12,13,14,15,16,17,18
We have mapped a locus for autosomal dominant Parkinson's disease on chromosome 2p13 (PARK3)19 which we further refined to 2.5 megabases.20 Three other genome scans confirmed the PARK3 locus.14,16,17 In a refinement of the locus position, DeStefano et al found evidence for association between later Parkinson's disease age at onset and allele 174 of marker D2S1394, which is close to the SPR gene.18 Karamohamed et al extended the work by genotyping single nucleotide polymorphisms (SNPs) in the vicinity of marker D2S1394.21 They reported association of one SNP and a haplotype across the SPR gene with age of onset in sibships of North American origin.
Here, we thoroughly assess the SPR gene region in sporadic Parkinson's disease cases ascertained from Germany, and in familial Parkinson's disease samples from five different European countries. After characterising the linkage disequilibrium (LD) structure in the SPR gene region, we further refined association signals for Parkinson's disease susceptibility to haplotypes and SNPs within an LD block comprising the SPR gene.
Methods
We conducted our study in two parts. First, we genotyped five short tandem repeat (STR) markers and 17 SNPs from the PARK3 core region centred around the SPR gene (table 1, fig 1) in a cohort of European affected siblings (122 sibships). In the second part of our study, an additional 23 SNPs, chosen to cover the SPR gene and flanking regions (fig 1), were genotyped in 340 German sporadic Parkinson's disease patients and 680 controls.
Table 1 Non‐parametric logarithm of odds (LOD) scores of single nucleotide polymorphisms with affection status (sibship data).
| SNP | Germany | UK | France | Italy | Netherlands | All |
|---|---|---|---|---|---|---|
| rs1876488 | 0.51 | 0.25 | 0.61 | 0.23 | 0.45 | 2.91* |
| rs1396107 | 0.37 | 0.27 | −0.25 | −0.17 | 0.53 | 0.35 |
| rs1508060 | 0.42 | 0.00 | 1.89 | 0.00 | 0.00 | 2.18* |
| rs1567230 | −0.27 | 0.25 | −0.07 | −0.10 | 0.34 | 0.22 |
| rs2135985 | −0.01 | 0.73 | 0.76 | −0.71 | 0.41 | 0.98 |
| rs1876491 | −0.02 | −0.13 | −0.06 | −0.16 | 0.59 | −0.14 |
| rs2421095 | −0.13 | 0.48 | 0.60 | −0.08 | 0.45 | 2.70* |
| rs1876487 | 0.36 | 0.30 | −0.06 | −0.15 | 0.43 | 0.40 |
| rs1150500 | −0.01 | −0.01 | −0.85 | −0.41 | 0.11 | 1.41 |
| rs1561244 | 0.00 | 0.02 | −0.17 | −0.27 | 0.05 | −0.16 |
| rs1561245 | −0.11 | 0.39 | 0.60 | 0.23 | 0.45 | 1.56 |
| rs4852903 | 0.02 | 0.49 | 0.60 | 0.23 | 0.45 | 2.19* |
| rs989040 | 1.62 | 0.48 | 0.61 | 0.23 | 0.45 | 2.63* |
| rs1561247 | 0.12 | 1.38 | 0.00 | 0.00 | 0.00 | 2.26* |
| rs999494 | −0.33 | 0.60 | 0.64 | −0.41 | 0.72 | 0.78 |
| rs1465805 | 1.58 | 0.00 | 0.00 | 0.00 | 0.00 | 2.48* |
| rs3980960 | 1.49 | 0.83 | 0.53 | −0.51 | 0.33 | 0.74 |
*p value <0.05.
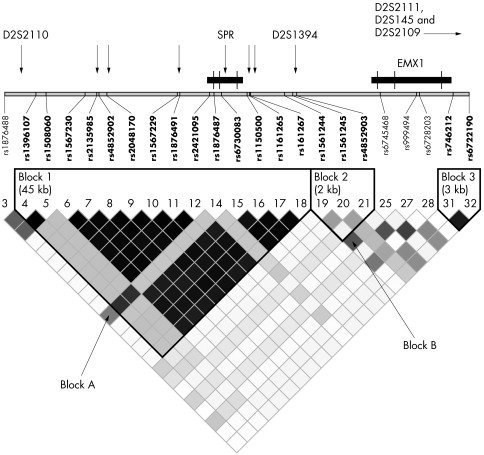
Figure 1 Linkage disequilibrium (LD) structure around the sepiapterin reductase (SPR) gene region. All marker positions and genes are indicated on the physical map. Black cells = high pairwise r2 values; grey cells = intermediate pairwise r2 values; white cells = low pairwise r2 values. Additional SNPs genotyped are indicated by arrows. Exons are indicated by vertical bars.
Recruitment of Parkinson's disease families
In all, 122 families from five European countries were ascertained through the European Consortium on Genetic Susceptibility in Parkinson's disease (GSPD). We included families having at least two affected siblings. A total of 35 families from Germany, 26 from the UK, 12 from the Netherlands, 33 from France, and 16 from Italy were ascertained. Characteristics of the families are given in table 2.
Table 2 Characteristics of the recruited families.
| Country | Number of families | Number of siblings | % Male | % Female | Age at onset (years)* | |
|---|---|---|---|---|---|---|
| Affected genotyped | Unaffected genotyped | |||||
| Germany | 35 | 100 | 44 | 48.8 | 51.2 | 57.4 (10.4) |
| UK | 26 | 65 | 24 | 50.6 | 49.4 | 61.2 (7.3) |
| France | 33 | 84 | 36 | 49.4 | 50.6 | 56.4 (12.8) |
| Italy | 16 | 54 | 12 | 42.3 | 57.7 | 52.8 (16.6) |
| Netherlands | 12 | 23 | 22 | 52.1 | 47.9 | 58.2 (11.1) |
| Total | 122 | 326 | 138 | 48.6 | 51.3 | 57.6 (11.4) |
*Mean (SD).
After appropriate informed consent was obtained, blood samples were drawn from the individuals for DNA extraction. Families suggestive for dominant inheritance were screened for known LRRK2 mutations. In families suggestive for recessive inheritance with early onset (<45 years) in at least one affected member, the parkin, PINK1 and DJ‐1 genes were completely sequenced and mutations were excluded.
Recruitment of sporadic Parkinson's disease patients
Parkinson's disease patients were recruited mainly from the departments of neurology at the Universities of Munich and Tübingen. Specialists in movement disorders examined the patients. Diagnosis was established according to UK Brain Bank criteria.22 The median age at onset was 55.4 years (range ±19.1). As controls we used 680 healthy, age and sex matched subjects from the KORA (Cooperative Research in the Region of Augsburg) Survey 2000, which involved a large, population based sample.
Genotyping of STRs
We genotyped the five STR markers D2S2110, D2S1394, D2S2111, D2S145, and D2S2109 with an average spacing of 0.2 cM from the PARK3 core region, as defined by West et al.20 The new marker order was obtained from the Marshfield Genetic Laboratories Map. Mendelian inconsistencies in the genotypic data were checked by using the program PedCheck.23
Genotyping of SNPs
Within the PARK3 core region we focused on the SPR gene region (4 kb SPR gene plus 94 kb flanking regions; in all 98 kb; see fig 1). Forty SNPs with an average spacing of 2.5 kb were identified using public databases. All SNPs showed high genotyping quality and Hardy–Weinberg equilibrium in the control subjects. Genotyping was undertaken using the matrix assisted laser desorption/ionisation time of flight (MALDI‐TOF) mass spectrometry method (Sequenom, San Diego, California, USA).
Statistical analysis
We used the non‐parametric linkage scores (NPL) for the analysis of the sibship data.24 NPL compares the estimated proportion of alleles shared by identical by descent siblings with the null hypothesis of no linkage. The Sall function in Genehunter was used for calculation. The age of onset effect in siblings was tested by the quantitative transmission disequilibrium test (QTDT) using an orthogonal and monks model.25 This allows one to incorporate pedigrees without parental genotypes. Whenever there are more than two siblings in the families, allelic transmission scores were used to assess the association.
The method of Gabriel et al, as implemented in Haploview, was used to construct LD blocks from SNPs with minor allele frequencies (MAF) of more than 0.05.26 In the sporadic patients, logistic regression was used to test for association between SNP alleles or haplotypes and Parkinson's disease affection status as implemented in UNPHASED.27 A stepwise approach was employed to determine which variants independently influence the risk of the disease. This approach allows the effect at a locus to be assessed for conditioning on alleles at other loci. A linear regression model was used to assess the age of onset effect.
Results
Analysis of sibship families
We obtained overall non‐parametric LOD scores with affection status of 2.12 and 1.96 at the SPR‐neighbouring markers D2S2110 and D2S1394, respectively. After the exclusion of six families with PARK8 mutations the LOD scores increased to 2.76 and 2.08 at these two markers, as shown in table 3.
Table 3 Non‐parametric logarithm of odds (LOD) scores of short tandem repeat (STR) markers with affection status (sibship data).
| Marker | Distance (cM) | Germany | UK | France | Italy | Netherlands | All |
|---|---|---|---|---|---|---|---|
| D2S2110 | 89.76 | 1.73 | 2.17* | 0.91 | 1.14 | −0.41 | 2.76* |
| D2S1394 | 90.29 | 1.29 | 0.97 | 1.41 | 0.41 | 0.20 | 2.08* |
| D2S2111 | 90.29 | 0.95 | 0.97 | 1.04 | −0.25 | 0.17 | 1.43 |
| D2S145 | 90.82 | 0.99 | 0.08 | 1.56 | 0.41 | −1.15 | 1.21 |
| D2S2109 | 90.82 | −0.46 | −0.18 | 1.78 | 0.82 | 0.38 | 1.12 |
*p values <0.05.
Family sets from single countries showed the same trend but were underpowered to obtain significant LOD scores except for the UK sample. Seven of the 17 SNPs located between the STR markers D2S2110 and D2S1394 also showed significant LOD scores (table 1).
We further analysed the age of onset effect in our sibship samples. The STR marker D2S2110 (p = 0.04) and the SNP marker rs1876487 (p = 0.04) showed significant association with age of onset. D2S2110 is the only genotyped STR marker which is located within the SPR LD block described below. The SPR promoter SNP rs1876487 was also associated with age of onset in the study by Karamohamed et al.21
Analysis of sporadic patients
Seventeen of the 40 genotyped SNPs had MAF <0.05 and were excluded from further analysis. In all, 23 SNPs were used in the final analysis. One highly intercorrelated LD block of 45 kb (block A) can be defined around the SPR gene between SNP rs1396107 and rs1161267 (fig 1).
All 14 SNPs within block A were significantly associated (p<0.05) with Parkinson's disease, as shown in table 4.
Table 4 Genotypic association tests with each allele as the recessive risk allele (sporadic Parkinson's disease data).
| SNP ID | Allele major/minor | Distance | Gene position | Frequency of major allele | Recessive risk model of major allele | Recessive risk model of minor allele | |||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Control s | p Value | Odds ratio | p Value | Odds ratio | ||||
| (95% CI) | (95% CI) | ||||||||
| rs1876488 | G/T | 72981628 | 0.809 | 0.825 | 0.410 | 0.73 (0.34 to 1.54) | 0.507 | 1.09 (0.83 to 1.44) | |
| rs1396107 | C/T | 72988240 | 0.708 | 0.751 | 0.464 | 0.82 (0.50 to 1.75) | 0.024 | 1.35 (1.03 to 1.75) | |
| rs1508060 | T/C | 72990328 | 0.716 | 0.756 | 0.486 | 0.83 (0.49 to 1.40) | 0.040 | 1.31 (1.01 to 1.71) | |
| rs1567230 | T/C | 72998602 | 0.897 | 0.931 | 0.817 | 1.21 (0.23 to 6.29) | 0.004 | 1.67 (1.16 to 2.37) | |
| rs2135985 | C/A | 73001132 | 0.899 | 0.928 | 0.625 | 1.48 (0.29 to 7.39) | 0.012 | 1.56 (1.09 to 2.20) | |
| rs4852902 | G/A | 73001656 | 0.898 | 0.927 | 0.624 | 1.48 (0.29 to 7.41) | 0.011 | 1.56 (1.10 to 2.20) | |
| rs2048170 | A/G | 73003323 | 0.900 | 0.926 | 0.590 | 1.53 (0.30 to 7.63) | 0.025 | 1.49 (1.04 to 2.12) | |
| rs1567229 | T/C | 73018249 | SPR promoter | 0.900 | 0.927 | 0.620 | 1.49 (0.30 to 7.45) | 0.017 | 1.52 (1.07 to 2.15) |
| rs1876491 | T/G | 73018800 | SPR promoter | 0.906 | 0.926 | 0.290 | 2.97 (0.35 to 24.7) | 0.049 | 1.42 (0.09 to 2.03) |
| rs2421095 | G/A | 73025179 | SPR promoter | 0.899 | 0.929 | 0.812 | 1.22 (0.23 to 6.32) | 0.011 | 1.57 (1.10 to 2.23) |
| rs1876487 | G/T | 73026007 | SPR promoter | 0.677 | 0.731 | 0.146 | 0.71 (0.45 to 1.15) | 0.015 | 1.38 (1.06 to 1.79) |
| rs6730083 | A/G | 73027652 | SPR intron 2 | 0.905 | 0.935 | 0.787 | 1.25 (0.24 to 6.49) | 0.008 | 1.16 (1.12 to 2.32) |
| rs1150500 | C/T | 73033098 | SPR 3′UTR | 0.901 | 0.930 | 0.987 | 0.98 (0.18 to 5.41) | 0.014 | 1.54 (1.08 to 2.19) |
| rs1161265 | A/G | 73033597 | SPR 3′UTR | 0.902 | 0.929 | 1.000 | 1.00 (0.18 to 5.48) | 0.025 | 1.49 (1.04 to 2.12) |
| rs1161267 | G/A | 73033836 | SPR 3′UTR | 0.904 | 0.930 | 0.970 | 0.97 (0.17 to 5.36) | 0.031 | 1.47 (1.03 to 2.10) |
| rs1561244 | G/A | 73041041 | 0.789 | 0.826 | 0.174 | 0.58 (0.27 to 1.27) | 0.078 | 1.28 (0.98 to 1.70) | |
| rs1561245 | C/T | 73042822 | 0.597 | 0.636 | 0.473 | 0.87 (0.60 to 1.26) | 0.060 | 1.29 (0.98 to 1.70) | |
| rs4852903 | G/C | 73043640 | 0.804 | 0.810 | 0.909 | 1.01 (0.77 to 1.34) | 0.468 | 0.78 (0.40 to 1.51) | |
| rs6745468 | G/C | 73061450 | EMX1 intron2 | 0.801 | 0.836 | 0.225 | 0.59 (0.25 to 1.39) | 0.068 | 1.29 (0.98 to 1.70) |
| rs999494 | G/A | 73069050 | EMX1 intron2 | 0.818 | 0.829 | 0.491 | 1.13 (0.83 to 1.46) | 0.993 | 1.00 (0.48 to 2.09) |
| rs6728203 | A/G | 73069674 | EMX1 | 0.765 | 0.800 | 0.532 | 0.86 (0.40 to 1.58) | 0.061 | 1.29 (0.98 to 1.68) |
| rs746212 | C/T | 7376889 | EMX1 | 0.687 | 0.705 | 0.140 | 0.72 (0.47 to 1.14) | 0.840 | 1.02 (0.78 to 1.33) |
| rs6722190 | C/G | 73080248 | EMX1 | 0.672 | 0.689 | 0.090 | 0.69 (0.46 to 1.06) | 1.000 | 1.00 (0.76 to 1.30) |
*SNP positions are based on human genome assembly (hg17) and NCBI build 35.
SNP, single nucleotide polymorphism; SPR, sepiapterin reductase gene.
After testing different allelic and genotypic risk effect models, we show only the recessive risk effect model, which best fitted to the observed genotype data. The associated SNPs comprised variants with risk allele frequencies of either ∼7% and ∼27% in the control population, and were highly intercorrelated, with r2 values of 0.81 to 0.92. The 11 SNPs with MAF of 7% seem to be more strongly associated than the more common SNPs, which include the SNP rs1876487 identified by Karamohamed et al. The estimated relative risks ranged from 1.21 to 1.57. In a stepwise regression procedure which tested the allelic association of all significant SNPs conditioned on alleles at the other significant loci, we were unable to find an independent signal. It appears that all significant SNPs within block A (fig 1) potentially referred to a single causal variant.
A haplotype within block A with a population frequency of 7.6% was also significantly associated with Parkinson's disease (p = 0.010), as shown in table 5.
Table 5 Association analysis of common (>1%) block A haplotypes with affection status; sporadic Parkinson's disease data.
| Haplotype | Case frequency | Control frequency | p Value |
|---|---|---|---|
| CTTCGATTAGACAG | 0.678 | 0.731 | 0.012 |
| CTTCGATTATACAG | 0.032. | 0.027 | 0.571 |
| TCTCGATTATACAG | 0.189 | 0.165 | 0.230 |
| TCCAAGCGGTGTGA | 0.097 | 0.065 | 0.010 |
Interestingly, this haplotype is differentiated by 11 mutational steps compared with the next related common haplotype (frequency 17.5%) in the same block. This “Yin–Yang” pattern of haplotype relations may indicate recent positive or balancing selection operating on the block A variants.28
We further evaluated the three‐locus haplotype (rs2421095–rs1876487–rs1561244) described by Karamohamed et al,21 which extends between block A and block B (fig 1). The haplotype AGG, which was most significantly associated with age of onset in Karamohamed's study, was also most significantly associated in our study (p = 0.002; table 6), albeit with affection status.
Table 6 Association analysis of common (>1%) 3‐locus haplotype with affection status; sporadic Parkinson's disease data.
| rs2421095 | rs1876487 | rs1561244 | Haplotype frequency, cases | Haplotype frequency, controls | p Value |
|---|---|---|---|---|---|
| A | G | A | 0.067 | 0.062 | 0.54 |
| A | G | G | 0.603 | 0.670 | 0.002 |
| A | T | A | 0.139 | 0.111 | 0.06 |
| A | T | G | 0.087 | 0.083 | 0.76 |
| G | T | G | 0.098 | 0.072 | 0.03 |
We found neither single marker nor haplotype association with age of onset in our sporadic Parkinson's disease sample.
Discussion
There is accumulating evidence that the SPR gene is one of the likely candidates for the PARK3 locus. First, in a large sample of multiplex families (the GenePD study), one microsatellite marker D2S1394, which is only 9 kb away from the SPR gene, showed association with Parkinson's disease age at onset.18 The associated allele “174” of this marker is also common to the segregating core haplotype observed in two PARK3 families.19,20 Second, a haplotype harbouring the SPR gene and the promoter SNP rs1876487 was reported to be associated with onset age in the GenePD study.21 Third, we obtained significant LOD scores with Parkinson's disease susceptibility at the two microsatellite markers D2S2110 and D2S1394, which encompass the SPR gene, in an independent large sample of multiplex families (European GSPD study). Interestingly, the English families with the highest mean age of onset contributed most to this linkage signal. This result fits well to the finding that the European genome scan reports a prominent linkage peak at the SPR region only for late age of onset families.17 We also found association with age of onset at the markers D2S2110 and rs1876487 in the GSPD family sample. Fourth, we found association with Parkinson's disease susceptibility at several intercorrelated SNP markers and haplotypes in an LD block spanning the SPR gene in a German sample of patients with sporadic Parkinson's disease.
Given all converging evidence, DNA variant(s) in or around the SPR gene appear to influence Parkinson's disease onset. Coding regions of potential PARK3 candidate genes including SPR have been screened for pathogenic mutations in Parkinson's disease families.20 The failure to detect such mutations could indicate that the functional variant affects expression or splicing regulation rather than the protein structure itself.
A recently published study has revealed a mutation in the 5′ UTR of the SPR gene responsible for causing dopa responsive dystonia.29 SPR is an interesting candidate gene because it catalyses the conversion of 6‐pyrovyl‐tetrahydropterin (PTP) to tetrahydrobiopterin (BH4). Previous studies have shown that BH4 acts not only as a cofactor for TH4 and is therefore important for dopamine biosynthesis, but also stimulates NOS isoforms (inducible (iNOS), neural (NOS), and endothelial (eNOS)).30 It has been suggested that iNOS confers protection to Parkinson's disease.
Across studies, there is some variability of the Parkinson's disease phenotype mapped to the SPR region. Whereas the GenePD studies report associations with Parkinson's disease onset age,18 the study based on the European GSPD samples reveals associations with Parkinson's disease susceptibility. Of note, however, we could also directly replicate the age of onset effect at the promoter marker rs1876487 found by Karamohamed et al.21
It is obvious that affection status and age of onset are related phenotypes in late onset disorders that show preclinical and postclinical progression. A factor that influences the preclinical history of Parkinson's disease (for example, the start and rate of the progression of neuronal loss) will affect the onset age of Parkinson's disease if a progression threshold is assumed for the transition from healthy to disease status. As general mortality (independent of Parkinson's disease) increases with age, an age of onset factor may also be seen as a susceptibility factor, because early onset factors are upweighted in an association study dealing with affection status. Different age structures in the analysed samples may shift the emphasis (detectability) between the extremes of pure age of onset and pure susceptibility effects.
In summary, we show that the SPR gene is a likely PARK3 candidate. The association signal appears to be confined to a haplotype block of 45 kb surrounding the SPR gene. Our data suggest a single risk variant or age of onset factor of about 7% frequency with a relative risk of about 1.4 within haplotype block A. Further evidence of this association signal comes from the haplotype pattern, which is indicative for recent natural selection in this genomic region.31
Electronic database information
The URLs for the data presented in this paper are as follows:
Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for the chromosome 2p13 genetic map)
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Parkinson's disease, PARK1, PARK2, PARK3, PARK5, PARK6, PARK7, PARK8, PARK9, PARK10, PARK11).
Haploview: http://www.broad.mit.edu/personal/jcbarret/haploview
Abbreviations
LD - linkage disequilibrium
MAF - minor allele frequency
NPL - non‐parametric linkage scores
SNP - single nucleotide polymorphism
SPR - sepiapterin reductase
STR - short tandem repeat
Footnotes
We would like to thank the patients and their family members for their support. The KORA group consists of H E Wichmann (speaker), H Löwel, C Meisinger, T Illig, R Holle, J John and their coworkers, who are responsible for the design and conduct of the KORA studies. This study was supported by the German Ministery for Education and Research (BMBF – Competence Network Parkinson (01G10201), the German National Genome Research Network NGFN (01GS0116), and the Hertie‐Institute for Clinical Brain Research, Tübingen, Germany.
Conflicts of interest: none declared
References
- 1.de Rijk M C, Tzourio C, Breteler M M, Dartigues J F, Amaducci L, Lopez‐Pousa S, Manubens‐Bertran J M, Alperovitch A, Rocca W A. Prevalence of parkinsonism and Parkinson's disease in Europe: the EUROPARKINSON Collaborative Study. European Community Concerted Action on the Epidemiology of Parkinson's disease. J Neurol Neurosurg Psychiatry 19976210–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calne D. A definition of Parkinson's disease. Parkinsonism Relat Disord 200511(suppl 1)S39–S40. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998392605–608. [DOI] [PubMed] [Google Scholar]
- 4.Bonifati V, Rizzu P, Van Baren M J, Schaap O, Breedveld G J, Krieger E, Dekker M C, Squitieri F, Ibanez P, Joosse M, van Dongen J W, Vanacore N, van Swieten J C, Brice A, Meco G, van Duijn C M, Oostra B A, Heutink P. Mutations in the DJ‐1 gene associated with autosomal recessive early‐onset parkinsonism. Science 2002299256–259. [DOI] [PubMed] [Google Scholar]
- 5.Valente E M, Abou‐Sleiman P M, Caputo V, Muqit M M, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio A R, Healy D G, Albanese A, Nussbaum R, Gonzalez‐Maldonado R, Deller T, Salvi S, Cortelli P, Gilks W P, Latchman D S, Harvey R J, Dallapiccola B, Auburger G, Wood N W. Hereditary early‐onset Parkinson's disease caused by mutations in PINK1. Science 20043041158–1160. [DOI] [PubMed] [Google Scholar]
- 6.Polymeropoulos M H, Lavedan C, Leroy E, Ide S E, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos E S, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson W G, Lazzarini A M, Duvoisin R C, Di Iorio G, Golbe L I, Nussbaum R L. Mutation in the α‐synuclein gene identified in families with Parkinson's disease. Science 19972762045–2047. [DOI] [PubMed] [Google Scholar]
- 7.Singleton A B, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson M R, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn‐Hardy K. alpha‐Synuclein locus triplication causes Parkinson's disease. Science 2003302841. [DOI] [PubMed] [Google Scholar]
- 8.Ibanez P, Bonnet A M, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha‐synuclein gene duplication and familial Parkinson's disease. Lancet 20043641169–1171. [DOI] [PubMed] [Google Scholar]
- 9.Paisan‐Ruiz C, Jain S, Evans E W, Gilks W P, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil A M, Khan N, Johnson J, Martinez J R, Nicholl D, Carrera I M, Pena A S, de Silva R, Lees A, Marti‐Masso J F, Perez‐Tur J, Wood N W, Singleton A B. Cloning of the gene containing mutations that cause PARK8‐linked Parkinson's disease. Neuron 200444595–600. [DOI] [PubMed] [Google Scholar]
- 10.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti R J, Calne D B, Stoessl A J, Pfeiffer R F, Patenge N, Carbajal I C, Vieregge P, Asmus F, Muller‐Myhsok B, Dickson D W, Meitinger T, Strom T M, Wszolek Z K, Gasser T. Mutations in LRRK2 Cause autosomal‐dominant parkinsonism with pleomorphic pathology. Neuron 200444601–607. [DOI] [PubMed] [Google Scholar]
- 11.Bertram L, Tanzi R E. The genetic epidemiology of neurodegenerative disease. J Clin Invest 20051151449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankratz N, Nichols W C, Uniacke S K, Halter C, Rudolph A, Shults C, Conneally P M, Foroud T, Parkinson Study Group Genome screen to identify susceptibility genes for Parkinson disease in a sample without parkin mutations. Am J Hum Genet 200271124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankratz N, Nichols W C, Uniacke S K, Halter C, Murrell J, Rudolph A, Shults C W, Conneally P M, Foroud T, Parkinson Study Group Genome‐wide linkage analysis and evidence of gene‐by‐gene interactions in a sample of 362 multiplex Parkinson disease families. Hum Mol Genet 2003122599–2608. [DOI] [PubMed] [Google Scholar]
- 14.Pankratz N, Uniacke S K, Halter C A, Rudolph A, Shults C W, Conneally P M, Foroud T, Nichols W C, Parkinson Study Group Genes influencing Parkinson disease onset: replication of PARK3 and identification of novel loci. Neurology 2004621616–1618. [DOI] [PubMed] [Google Scholar]
- 15.Li Y J, Scott W K, Hedges D J, Zhang F, Gaskell P C, Nance M A, Watts R L, Hubble J P, Koller W C, Pahwa R, Stern M B, Hiner B C, Jankovic J, Allen F A, Goetz C G, Mastaglia F, Stajich J M, Gibson R A, Middleton L T, Saunders A M, Scott B L, Small G W, Nicodemus K K, Reed A D, Schmechel D E, Welsh‐Bohmer K A, Conneally P M, Roses A D, Gilbert J R, Vance J M, Haines J L, Pericak‐Vance M A. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet 200270985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeStefano A L, Golbe L I, Mark M H, Lazzarini A M, Maher N E, Saint‐Hilaire M, Feldman R G, Guttman M, Watts R L, Suchowersky O, Lafontaine A L, Labelle N, Lew M F, Waters C H, Growdon J H, Singer C, Currie L J, Wooten G F, Vieregge P, Pramstaller P P, Klein C, Hubble J P, Stacy M, Montgomery E, MacDonald M E, Gusella J F, Myers R H. Genome‐wide scan for Parkinson's disease: the GenePD Study. Neurology 2001571124–1126. [DOI] [PubMed] [Google Scholar]
- 17.Martinez M, Brice A, Vaughan J R, Zimprich A, Breteler M M, Meco G, Filla A, Farrer M J, Betard C, Hardy J, De Michele G, Bonifati V, Oostra B, Gasser T, Wood N W, Durr A, French Parkinson's Disease Genetics Study Group; European Consortium on Genetic Susceptibility in Parkinson's Disease Genome‐wide scan linkage analysis for Parkinson's disease: the European genetic study of Parkinson's disease. J Med Genet 200441900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeStefano A L, Lew M F, Golbe L I, Mark M H, Lazzarini A M, Guttman M, Montgomery E, Waters C H, Singer C, Watts R L, Currie L J, Wooten G F, Maher N E, Wilk J B, Sullivan K M, Slater K M, Saint‐Hilaire M H, Feldman R G, Suchowersky O, Lafontaine A L, Labelle N, Growdon J H, Vieregge P, Pramstaller P P, Klein C, Hubble J P, Reider C R, Stacy M, MacDonald M E, Gusella J F, Myers R H. PARK3 influences age at onset in Parkinson disease: a genome scan in the GenePD study. Am J Hum Genet 2002701089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasser T, Müller‐Myhsok B, Wszolek Z K, Oehlmann R, Calne D B, Bonifati V, Bereznai B, Fabrizio E, Vieregge P, Horstmann R D. A susceptibility locus for Parkinson's disease maps to chromosome 2p13. Nat Genet 199818262–265. [DOI] [PubMed] [Google Scholar]
- 20.West A B, Zimprich A, Lockhart P J, Farrer M, Singleton A, Holtom B, Lincoln S, Hofer A, Hill L, Muller‐Myhsok B, Wszolek Z K, Hardy J, Gasser T. Refinement of the PARK3 locus on chromosome 2p13 and the analysis of 14 candidate genes. Eur J Hum Genet 20019659–666. [DOI] [PubMed] [Google Scholar]
- 21.Karamohamed S, DeStefano A L, Wilk J B.et al A haplotype at the PARK3 locus influences onset age for Parkinson's disease: the GenePD study. Neurology 200361(11)1557–1561. [DOI] [PubMed] [Google Scholar]
- 22.Hughes A J, Daniel S E, Kilford L, Lees A J. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico‐pathological study of 100 cases [see comments]. J Neurol Neurosurg Psychiatry 199255181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connell J R, Weeks D E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 199863259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruglyak L, Daly M J, Reeve Daly M P, Lander E S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996581347–1363. [PMC free article] [PubMed] [Google Scholar]
- 25.Abecasis G R, Cardon L R, Cookson W O. A general test of association for quantitative traits in nuclear families. Am J Hum Genet 200066279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett J C, Fry B, Maller J, Daly M J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 200521263–265. [DOI] [PubMed] [Google Scholar]
- 27.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 200325115–121. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Rowe W L, Clark A G, Buetow K H. Genomewide distribution of high‐frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet 2003731073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberger D, Blau N, Goriuonov D, Bitsch J, Zuker M, Hummel S, Muller U. Heterozygous mutation in 5′‐untranslated region of sepiapterin reductase gene (SPR) in a patient with dopa‐responsive dystonia. Neurogenetics 20045187–190. [DOI] [PubMed] [Google Scholar]
- 30.Klatt P, Schmid M, Leopold E, Schmidt K, Werner E R, Mayer B. The pteridine binding site of brain nitric oxide synthase. Tetrahydrobiopterin binding kinetics, specificity, and allosteric interaction with the substrate domain. J Biol Chem 199426913861–13866. [PubMed] [Google Scholar]
- 31.Bamshad M, Wooding S P. Signatures of natural selection in the human genome. Nat Rev Genet 2003499–111. [DOI] [PubMed] [Google Scholar]