Abstract
Introduction
The majority of hearing loss in children can be accounted for by genetic causes. Non‐syndromic hearing loss accounts for 80% of genetic hearing loss in children, with mutations in DFNB1/GJB2 being by far the most common cause. Among the second tier genetic causes of hearing loss in children are mutations in the DFNB9/OTOF gene.
Methods
In total, 65 recessive non‐syndromic hearing loss families were screened by genotyping for association with the DFNB9/OTOF gene. Families with genotypes consistent with linkage or uninformative for linkage to this gene region were further screened for mutations in the 48 known coding exons of otoferlin.
Results
Eight OTOF pathological variants were discovered in six families. Of these, Q829X was found in two families. We also noted 23 other coding variant, believed to have no pathology. A previously published missense allele I515T was found in the heterozygous state in an individual who was observed to be temperature sensitive for the auditory neuropathy phenotype.
Conclusions
Mutations in OTOF cause both profound hearing loss and a type of hearing loss where otoacoustic emissions are spared called auditory neuropathy.
Keywords: OTOF, DFNB9, otoferlin, auditory neuropathy, hearing loss, temperature sensitive
The majority of hearing loss in children can be accounted for by genetic causes. Non‐syndromic hearing loss accounts for 80% of genetic hearing loss in children, with mutations in DFNB1/GJB2 being by far the most common cause.1 Among the second tier genetic causes of hearing loss in children are mutations in DFNB9/OTOF (OMIM: 603681). This type of hearing loss is further complicated in that at an early stage it may present itself as an auditory neuropathy and thus evade detection by newborn hearing screening based on otoacoustic emissions testing.2,3,4
Auditory neuropathy/auditory dys‐synchrony (AN/AD) is a unique type of hearing loss diagnosed when tympanographs are normal, and acoustic reflexes (AR) and auditory brainstem response (ABR) are absent or severely abnormal, but outer hair cell (OHC) function is normal as indicated by the presence of otoacoustic emissions (OAE) and/or cochlear microphonics (CM). These test results indicate that the auditory pathway up to and including the OHC is functioning but the auditory signal is not transmitted to the brainstem, suggesting that the lesion lies at the level of the inner hair cells (IHC), the IHC synapse to the afferent nerve fibres, or the auditory nerve itself. Individuals with AN/AD can have various degrees of hearing loss as measured by pure tone audiometry. However, they generally have disproportionately poor speech understanding. In contrast to persons with non‐AN/AD hearing loss, hearing aids may provide little help in speech understanding in most individuals with AN/AD. Cochlear implantation has been shown to help with speech understanding in some cases of AN/AD,5,6,7,8,9,10 but others have not had favourable results.11,12
The term “auditory neuropathy” was first coined in 1996.13 However, many early reports describe cases now thought to have been AN/AD.14,15,16 At the time, such cases were considered paradoxical because test findings were inconclusive or contradictory. However, advances in the measurement of OHC function allowed further characterisation of this condition.
Approximately 50% of AN/AD patients have no defined aetiology.17 These patients are generally non‐syndromic, and some pedigrees suggest a recessive pattern of inheritance based on the presence of at least two affected siblings and unaffected parents. Recently mutations in OTOF, the gene encoding otoferlin were found to cause a non‐syndromic recessive AN/AD.2,3,4 There have been several reports of mutations in OTOF associated with non‐syndromic recessive hearing loss (NSRHL).18,19,20,21,22,23 None of the NSRHL reports defined the status of the OHC in affected individuals, so it could not be determined whether these individuals had AN/AD.
In adult mouse cochlea, otoferlin mRNA is detectable in the IHCs but is not evident in OHCs.23 Several isoforms of otoferlin exist, owing to variable start sites and alternative splicing. At least two other members of this gene family have been found in mammals: DYSF, encoding dysferlin, and MYOF, encoding myoferlin.24 Dysferlin was recently found to be involved in membrane repair.25 Otoferlin, dysferlin, and myoferlin are predicted to be signal anchor membrane proteins with the greater part of the protein, including the N‐terminus, facing the cytoplasm anchored by the C‐terminal transmembrane domain.26,27 They contain multiple C2 domains, and all bear homology to the synaptotagmins, proteins involved in synaptic vesicle fusion.28
We report here the genetic screen of 65 hearing loss families, the discovery of novel otoferlin mutations, and corresponding clinical data on hearing loss in families with pathological mutations. One of these mutant alleles is of particular interest because the phenotype associated with this allele is temperature sensitive.
MATERIALS AND METHODS
Patient sample and medical information
DNA samples and medical information were collected under institutional review board protocols that have been approved by the respective institutions. Sampling included the proband and, if possible, all full biological siblings, both biological parents, and all biological grandparents. In some cases, research participants were asked to undergo complete physical examinations and other appropriate testing, including audiological, vestibular, neurological, and ophthalmological testing. Audiological testing included air and bone conduction puretone audiometry, ABR, tympanometry, AR thresholds, OAE, OAE suppression, speech awareness threshold (SAT) and/or speech reception threshold (SRT) testing. The following scale was used to classify the degree of hearing loss (HL) using pure tone audiometry: 0–20 dB HL as normal, 21–40 dB HL as mild, 41–60 dB HL as moderate, 61–80 dB HL as severe, and >80 dB HL as profound.
Genotyping and mutation screening
All families in this study were previously screened for DFNB1/GJB2 mutations. In the Nebraska group of 47 NSRHL families (recruited at BTNRH or LSU), genotyping of the chromosomal region around the OTOF locus was performed as previously described.2
To determine if a difference exists between patient DNA and the control OTOF BAC DNA (RP11‐638P8; Research Genetics, Inc.), mutation detection enhancement (MDE) heteroduplex analysis was performed as previously described, with slight modifications.29 PCR products were amplified from both genomic sample DNA and OTOF BAC control DNA, mixed, heated at 95°C for 3 minutes, and then cooled to 25°C over a 45 minute period. The re‐annealed reaction products were then run on an electrophoresis gel at 850 V for 18–24 hours, depending on the size of the fragment. Following electrophoresis, the bands were visually assessed using ethidium bromide staining. The PCR primers used to amplify OTOF exons and flanking sequence have been described previously.2
Denaturing high performance liquid chromatography (DHPLC, using the Transgenomic WAVE DNA Fragment Analysis System; Transgenomic, Inc.) was also used to identify mutations according to the manufacturer's directions.
In the Iowa group (families recruited at the University of Iowa), 18 DFNB1 negative NSRHL families were analysed for allele status with STRP markers (D2S405, D2S158, and D2S1360). Three families with markers segregating consistent with linkage or uninformative for linkage were further screened using single strand conformational polymorphism (SSCP) for all 48 known OTOF exons. Mutations recognised by SSCP were then identified by sequencing. Families where only one mutation was detected were screened by sequencing all 48 known OTOF exons.
DNA samples showing a positive heteroduplex pattern by either MDE gel, DHPLC, or SSCP were sequenced using one of two kits (ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) or CEQ Dye Terminator Cycle Sequencing Quick Start kit (Beckman Coulter)). Analysis of the sequence data was performed using Sequence Analysis software (version 3.4), the Lasergene suite (DNASTAR, Inc), and the Wisconsin package (Pharmacopeia, Inc). Mutations were checked in unrelated normal hearing control individuals using restriction enzyme digestion or heteroduplex gel/WAVE DHPLC and DNA sequencing if no restriction enzyme was available. The control population consisted of Americans of white European descent with no report of hearing problems. In this study all families with individuals affected with hearing loss were also American of white European descent except for one family from the UK. Mutations corresponding to the coding region are numbered as in the human brain otoferlin long form AF183185.1, beginning with the starting methionine, AUG (A as number 1). Splice site mutations are designated by their adjacent exon.23
RESULTS
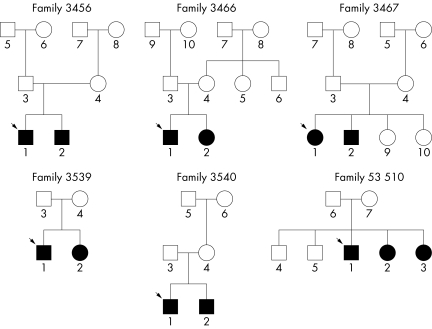
Mutations were revealed during two independent screens of NSRHL families. In the Nebraska group, STRP genotyping of the OTOF locus was performed on 47 families (38 NSRHL families and nine families with non‐syndromic recessive auditory neuropathy (NSRAN) families), 17 (14 NSRHL and three NSRAN) of whom were consistent with linkage to the OTOF locus by haplotyping and were therefore included in the genetic screening. Sixteen families were uninformative for linkage (12 NSRHL and four NSRAN). Fourteen families (12 NSRHL and two NSRAN) were informative and not consistent with linkage. The 17 families consistent for linkage and the 16 uninformative for linkage were included in the mutation screen. Seven mutations believed to be pathogenic were found in five families in this group (table 1, fig 1).
Table 1 OTOF mutations including mutations from genetic screening of 65 NSRAN and NSRHL families (in bold) .
| Exon or IVS | Mutation | Codon | Controls, no. of chromosomes | Family | Diagnosis | Origin | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 709C→T | R237X | 0/36 | NSRHL | UAE | 19 | ||||||||
| 8 IVS | IVS8−2A→G | 0/218 | NSRHL | India | 23 | |||||||||
| 15 | 1469C→A | P490Q | 0/220 | NSRHL | Turkey | 21 | ||||||||
| 15 | 1544T→C | I515T | 0/178, 0/220 | 3467 | TS‐NSRAN, NSRHL | USA, Turkey | 21 | |||||||
| 16 | 1651delG | 0/188 | NSRAN | USA | 2 | |||||||||
| 17 | 1886_1887insA | K629fs | 0/188 | 3466 | NSRAN | USA | ||||||||
| 18 IVS | IVS18 +1G→T | 0/188 | 3539 | NSRAN | USA | |||||||||
| 19 | 2122C→T | R708X | 0/100 | NSRHL | Spain | 3 | ||||||||
| 21 | 2348delG | G783fs | 0/184 | 3466 | NSRAN | USA | ||||||||
| 21 | 2381G→A | R794H | 0/160 | 53510 | NSRHL | USA | ||||||||
| 22 | 2485C→T | Q829X | 0/172, 0/400 | 3456, 3540 | NSRAN, NSRHL | UK, USA, Spain, Cuba | 3,20 | |||||||
| 24 IVS | IVS24 +1G→A | 0/240 | NSRHL | Druze/Israel | 18 | |||||||||
| 26 | 3032T→C | L1011P | 0/100 | NSRAN | Turkey | 4 | ||||||||
| 28 IVS | IVS28 −2A→C | 0/184 | 3456 | NSRAN | UK | |||||||||
| 36 | 4275G→A | W1425X | 0/100 | NSRAN | Spain | 3 | ||||||||
| 36 IVS | IVS36 +2T→G | 0/100 | NSRAN | Spain | 3 | |||||||||
| 37 | 4491T→A | Y1497X | 0/212 | NSRHL | Lebanon | 22 | ||||||||
| 39 IVS | IVS39 +1G→C | 0/188 | NSRAN | USA | 2 | |||||||||
| 44 | 5473C→G | P1825A | 0/200 | NSRHL | Spain | 20 | ||||||||
| 48 | 5860_5862delATC | I1954del | 0/100 | NSRAN | Spain | 3 | ||||||||
| 48 | 6014G→A | R1939Q | 0/188 | NSRAN | USA | 2 | ||||||||
| 48 | 6158C→G | P1987R | 0/188 | NSRAN | USA | 2 |
IVS, intervening sequences or intron.
Figure 1 Pedigrees of families with OTOF mutations. Proband is identified by arrow.
In the Iowa group, 18 NSRHL families were screened with markers linked to OTOF. None of the Iowa families had been previously diagnosed with auditory neuropathy. Of these 18 families, all were informative for linkage but only three were consistent for linkage to OTOF. These families were screened by SSCP. A single missense mutation deemed pathogenic was identified in family 53510 in this group (table 1, fig 1). An audiological summary is given for each family (table 2).
Table 2 Summary of audiological data.
| NSRAN individual | Age tested | Audiometric loss | Audiogram shape | ABR | CM | OAE | Acoustic reflexes | Diagnosis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3456‐1 | 3 years | Profound | Corner | –* | +† | + | NT‡ | NSRAN | ||||||||
| 3456‐2 | 15 months | Profound | Corner | – | + | + | NT | NSRAN | ||||||||
| 3466‐1 | 4 years | Profound | Flat | – | NI§ | + | NI | NSRAN | ||||||||
| 3466‐2 | 16 months | Profound | Flat | – | NI | + | NI | NSRAN | ||||||||
| 3467‐1 37°C | 6 years | Mild to normal | Rising | Abnormal | + | + | – | TS‐NSRAN | ||||||||
| 3467‐1 37.8°C | Mild to moderate | Cookie bite | NT | NT | NT | NT | TS‐NSRAN | |||||||||
| 3467‐1 38.1°C | R: profound, L: severe to profound | R: flat, L: rising | – | NT | + | – | TS‐NSRAN | |||||||||
| 3467‐2 37°C | 2 years | mild to normal | Rising | Abnormal | + | + | – | TS‐NSRAN | ||||||||
| 3539‐1 | 6 years | Moderate to severe | Rising | – | + | + | – | NSRAN | ||||||||
| 3539‐2 | 3 years | Moderate to severe | Rising | – | + | + | – | NSRAN | ||||||||
| 3540‐1 | 3 years | NT | NT | – | + | + | – | NSRAN | ||||||||
| 53510‐1 | 26 years | Profound | Corner | NT | NT | – | NT | NSRHL | ||||||||
| 53510‐2 | 33 years | Severe to profound | Corner | NT | NT | NT | NT | NSRHL | ||||||||
| 53510‐3 | 35 years | Profound | Corner | NT | NT | NT | NT | NSRHL |
*Absent bilaterally; †present bilaterally; ‡not tested. ABR, auditory brainstem response; CM, cochlear microphonics;OAE, otoacoustic emissions.
We found a high degree of neutral variation within the subject population during the mutation screen; 23 coding polymorphic changes were noted (*denotes alleles found in the Spanish population).20 Eleven of these were missense alleles (158C→T, A53V; 244C→T*, R82C*; 1723G→A, V575M; 2317C→T*, R773S*; 2464C→T, R822W; 3247G→C, A1083P; 3470G→A, R1157Q; 3966C→G, D1322E; 4874G→A, V1625M; 4936C→T, P1646S; and 5663G→A, G1888D), and 12 were silent alleles (372A→G*, T124T*; 945G→A*, K315K*; 1926C→T, N642N; 2022CvT, D674D; 2025G→A, E675E; 2580C→G*, V860V*; 2736G→C*, L919L*; 3189G→A, A1063A; 4677G→A, V1559V; 4767C→T, R1589R; 5391C→T*, F1797F*; and 5655C→T*, R1885R*).
Probably the most unusual family of the group is 3467, a family with temperature sensitive AN/AD, with two affected siblings. The variant 1544T→C, I515T was found heterozygously in the father and the two affected children of this temperature sensitive NSRAN family. The maternal mutation is still unknown.
The audiogram for individual 3467‐1, when she was afebrile, showed a mild low frequency hearing loss and speech comprehension was below the 10th percentile for both quiet and noise. Tympanometry was normal and AR were absent. ABR was abnormal, but CM were present. On two occasions, testing was performed during febrile illness. Her core temperature was defined approximately 2 hours before testing. At a temperature of 38.1°C, her pure tone thresholds decreased to profound deafness in the low frequencies, rising to severe hearing loss in the high frequencies. SAT was 80 dB HL, but she was unable to repeat any of the test spondee words. Tympanometry and OAE were normal, but AR and ABR were absent. With a temperature of 37.8°C, she was tested again and showed a mild to moderate hearing loss and zero speech comprehension. ABR and OAE were not tested. The following day her auditory functions returned to normal after her fever abated. She has reported to her parents that her hearing becomes affected “suddenly” when she is febrile.
Individual 3467‐2 was examined twice when afebrile. He has a mild low frequency hearing loss, normal tympanograms, absent AR, and abnormal ABR. CM and OAE were present bilaterally. We were unable to test 3467‐2 when febrile, but his parents report that under those conditions he experienced hearing loss similar to his sister. OAE suppression was tested to determine medial olivocochlear (MOC) neurone integrity. Activation of these neurones by presenting sound to the contralateral cochlea will induce a suppression of ipsilateral OAEs.30 Patients with AN/AD cannot suppress their OAE during this test.31 As expected, the OAE of 3467‐2 were not suppressed, which indicates that the sound signal reached the OHC, but not the efferent MOC neurones that feed back to suppress the OHC. We have tested the parent carrying the I515T mutation and no abnormality of pure tone threshold, speech comprehension in quiet and noise, ABR, AR, or OAE was identified. The clinical details of this family have been published previously.32
Both OTOF pathological alleles have been found in family 3456. The maternally inherited mutation is a novel splice site mutation, IVS2 –2A→C. The mutation inherited through their father is a previously published nonsense mutation 2485C→T, Q829X, found in a group of Spanish families and one Cuban family.3,20 Family 3456 is a white family from England with no known Hispanic ancestry, with two sons both having AN/AD. Pregnancy and birth history were unremarkable for the two affected boys except for slight jaundice in the older boy (individual 3467‐1). Motor milestones were met at an appropriate age. As is consistent with AN/AD, ABR was absent and OAE were present. Both boys have profound hearing loss and corner audiograms. OAE suppression was tested in individual 3467‐1 and, as expected, his OAE were not suppressed, but were instead increased in amplitude. Vestibular function testing in the older boy indicated that there may be a slight hypofunction on the left side. The younger was not tested for vestibular function. Computed tomography scans were normal in both boys, and magnetic resonance imaging in the older boy. Both boys have had positive experiences with their cochlear implants and consider them to be beneficial.
Both mutations in family 3466 result in a frameshift. The maternally inherited mutation in exon 17 contains an insertion 1886_1887insA (K629fs). The paternally inherited mutation is a deletion in exon 21, 2348delG (G783fs). This family has two affected children, a boy (3466‐1) and a girl (3466‐2) with normal OAE and absent ABR. Birth histories were uneventful and developmental milestones, except speech, were attained at a normal age. The children underwent neurological examinations at the age of 5 years (3466‐1) and 7 years (3466‐2), which were normal. However, 3466‐1 may also have a vestibular neuropathy or hypoactive vestibular function, as no nystagmus developed after spinning in a chair and his father reported that the child can spin without falling down.
Family 3539 has two affected children with normal tympanometry, OAE and CM, but absent AR and ABR. The daughter was born prematurely at 32 weeks gestation; otherwise, the birth histories were unremarkable. A splice site mutation, IVS18 +1G→T, was found in the maternal allele.
The Hispanic nonsense mutation 2485C→T, Q829X has been found heterozygously in family 3450. This family has Mexican ancestry. Both affected children are “typical” of NSRAN, in that OAE and CM are present, but ABR and AR are absent.
In the Iowa group of families, an OTOF mutation was found in one family. There were three affected children in family 53510, all heterozygous for 2381G→A, R794H. Two of these individuals had a profound hearing loss while one had a severe to profound hearing loss. All three individuals had a corner audiogram.
DISCUSSION
Seven OTOF mutations were discovered and believed to be pathogenic in five NSRAN families in the Nebraska group. One missense mutation was found in the Iowa group of NSRHL families. It is possible that the previously published OTOF NSRHL families were actually NSRAN families; however, proving this could be difficult owing to the fact that, in many cases, OHC function has been observed to decline with age. The mutation found in the Iowa group of families reported in this study may also have been NSRAN for the same reason. Recent studies, including this work, indicate that all individuals with OTOF pathological variants in both the maternal and paternal alleles will present with auditory neuropathy as young children if comprehensively tested (including OHC function).2,3,4 In this report, two families were heterozygous for the nonsense allele 2485C→T, Q829X, found recurrently in Spanish families.3,20 Two frameshift, two splice site, and two missense mutations have also been found (table 1). One of the missense alleles, 1544T→C, I515T is associated with a temperature sensitive NSRAN phenotype. It was previously described in cis with another missense mutation 1469C→A, P490Q not detected in the families reported here.21
The nonsense mutation 2485C→T, Q829X appears to be the most common otoferlin mutant allele discovered to date (Rodriguez‐Ballesteros et al,3 Migliosi et al,20 and this report). In the Spanish population, it is the third most common cause of genetic hearing loss in children.3 Two splice site mutations, IVS28 −2A→C and IVS18 +1G→T, were discovered, but their consequences have not been investigated. A single base insertion 1886_1887 insA, K629fs and single base deletion 2348delG, G783fs have been detected. Both of these mutations are predicted to cause a frameshift with premature termination, which should result in mRNA destabilisation via nonsense mediated mRNA decay.33
The two missense mutations occur at amino acids that are conserved by identity in human, mouse, chicken, and zebrafish otoferlin. Missense mutations were considered pathological if: (a) their segregation was consistent with affected status, (b) if no such variant was found in the control population, and (c) the reference amino acid was conserved across vertebrate lines. The I515T mutation is discussed in detail below. The arginine to histidine change, R794H, seen in family 53510 occurs as part of a coiled coil region identified by the SMART/COILS algorithm,34,35 which extends across otoferlin residues 792–820. This is a conservative change as both arginine and histidine are polar basic amino acids. However, arginine is more hydrophilic than histidine and differs significantly structurally. Coiled coil domains are characterised by alpha helixes that interact with one another, potentially forming a “peptide Velcro” interaction with other proteins.36 Weakening the hydrophilic interaction in such a structure could affect an important protein protein interaction disrupting otoferlin function. Marker analysis for the 53510 family was consistent and informative for linkage at DFNB9. None of the mutations considered pathological was detected in a hearing control population of at least 80 individuals.
This is the second study to discover a high degree of polymorphic variation at the OTOF locus.3 All the polymorphisms found in the Spanish population were also found in the population described here, and 15 additional neutral variants were discovered. The primary criterion for describing a variant as neutral was the detection of that mutation in a set of normal hearing controls.
The proband in family 3467 has abnormal ABR, present OAE, and relatively normal hearing until she becomes febrile, when OAE remain normal but hearing degrades, and ABR worsens from abnormal with unidentified waves I–III and delayed latency of the wave IV–V complex to being totally absent. The amount of decline in hearing in the proband is dependent on the degree of fever. A mild to moderate hearing loss was present when she had a temperature of 37.8°C and a profound hearing loss was present at 38.1°C.
Only one mutation has been found in family 3467 thus far, 1544T→C, I515T. This amino acid is conserved in human, mouse, chicken, and zebrafish otoferlin. It is a missense mutation occurring in the otoferlin C2C domain, which was previously found in a consanguineous family from Eastern Turkey with profound prelingual hearing loss.21 No mention was made of a temperature associated phenotype in this family. The affected members of the family from Eastern Turkey are homozygous for the I515T missense mutation and another missense mutation, 1469C→A, P490Q inherited in cis.
Both the P490Q and I515T mutations in the Turkish kindred were found in the C2C domain (third C2 domain), which is predicted to bind calcium.21 The alignment of otoferlin and otoferlin related proteins revealed remarkable conservation of amino acids within the human and mouse C2C domains.21 Mirghomizadeh et al predicted that either of these two mutations would severely disrupt the structure of the C2C domain, with the I515T mutation resulting in the creation of a new myristylation site.21 Temperature sensitive mutations have been previously recognised in several human genetic diseases.37,38,39
Eight OTOF mutations were found in six families in this study. In previous studies, four mutations were found in three families2 and 10 mutations were found in six families.3 All other OTOF mutations described are in consanguineous families or in Spanish families homozygous for Q829X.3,20OTOF is a large gene with multiple isoforms coded by at least 48 known exons. The screening of this gene focused on the transcribed part of the gene that is predicted to code for protein and the intron sequence flanking the splice sites of individual exons. The screens employed in this study depended on PCR amplification of genomic DNA and assumes that both alleles are represented equally in the amplified product. Heterogeneity at this locus may interfere with specific PCR primers annealing to specific alleles, preventing the amplification of one allele but not the other.
This report summarises the clinical findings associated with eight OTOF mutations, five of them novel, in 65 NSRHL and NSRAN families. One of these families has an allele associated with a temperature sensitive AN/AD phenotype. A temperature sensitive allele for otoferlin should provide a valuable tool to understand the function of otoferlin and its role in the auditory process.
ACKNOWLEDGEMENTS
This grant was supported by the NIH Center for Hearing Loss in Children 5P60DC00982, the NIH NIDCD Program Project (grant no. 2P01DC0813‐07 to W J Kimberling) and the Deafness Research Foundation (to P M Kelley). This study was also supported in part by a grant from the NIDCD (grant nos. R01‐DC02842 to R J H Smith and R01‐DC02618 to A Starr), and The Oberkotter Foundation to (to C I Berlin and L J Hood). Computing services were provided by the Genetic Sequence Analysis Facility at the University of Nebraska Medical Center and are supported by the Nebraska Research Initiative.
Abbreviations
ABR - auditory brainstem responses
AD - auditory dys‐synchrony
AN - auditory neuropathy
AR - acoustic reflexes
CM - cochlear microphonics
DHPLC - denaturing high performance liquid chromatography
HL - hearing loss
IHC - inner hair cells
MDE - mutation detection enhancement
MOC - medial olivocochlear
NSRAN - non‐syndromic recessive auditory neuropathy
NSRHL - non‐syndromic recessive hearing loss
OAE - otoacoustic emissions
OHC - outer hair cell
SAT - speech awareness threshold
SSCP - single strand conformational polymorphism
SRT - speech reception threshold
Footnotes
Competing interests: there are no competing interests
References
- 1.Van L L, Cryns K, Smith R J, Van C G. Nonsyndromic hearing loss. Ear Hear 200324275–288. [DOI] [PubMed] [Google Scholar]
- 2.Varga R, Kelley P M, Keats B J, Starr A, Leal S M, Cohn E, Kimberling W J. Non‐syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J Med Genet 20034045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez‐Ballesteros M, del Castillo F J, Martin Y, Moreno‐Pelayo M A, Morera C, Prieto F, Marco J, Morant A, Gallo‐Teran J, Morales‐Angulo C, Navas C, Trinidad G, Tapia M C, Moreno F, del C, I Auditory neuropathy in patients carrying mutations in the otoferlin gene (OTOF). Hum Mutat 200322451–456. [DOI] [PubMed] [Google Scholar]
- 4.Tekin M, Akcayoz D, Incesulu A. A novel missense mutation in a C2 domain of OTOF results in autosomal recessive auditory neuropathy. Am J Med Genet A 20051386–10. [DOI] [PubMed] [Google Scholar]
- 5.Madden C, Hilbert L, Rutter M, Greinwald J, Choo D. Pediatric cochlear implantation in auditory neuropathy. Otol Neurotol 200223163–168. [DOI] [PubMed] [Google Scholar]
- 6.Katada A, Nonaka S, Harabuchi Y. Cochlear implantation in an adult patient with auditory neuropathy. Eur Arch Otorhinolaryngol 2005262449–452. [DOI] [PubMed] [Google Scholar]
- 7.Shallop J K, Jin S H, Driscoll C L, Tibesar R J. Characteristics of electrically evoked potentials in patients with auditory neuropathy/auditory dys‐synchrony. Int J Audiol 200443(suppl)S22–S27. [PubMed] [Google Scholar]
- 8.Lin C Y, Chen Y J, Wu J L. Cochlear implantation in a Mandarin Chinese‐speaking child with auditory neuropathy. Eur Arch Otorhinolaryngol 2005262139–141. [DOI] [PubMed] [Google Scholar]
- 9.Peterson A, Shallop J, Driscoll C, Breneman A, Babb J, Stoeckel R, Fabry L. Outcomes of cochlear implantation in children with auditory neuropathy. J Am Acad Audiol 200314188–201. [PubMed] [Google Scholar]
- 10.Mason J C, De M A, Stevens C, Ruth R A, Hashisaki G T. Cochlear implantation in patients with auditory neuropathy of varied etiologies. Laryngoscope 200311345–49. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto R T, Kirk K I, Renshaw J, Hussain D. Cochlear implantation in auditory neuropathy. Laryngoscope 1999109181–185. [DOI] [PubMed] [Google Scholar]
- 12.Rance G, Beer D E, Cone‐Wesson B, Shepherd R K, Dowell R C, King A M, Rickards F W, Clark G M. Clinical findings for a group of infants and young children with auditory neuropathy. Ear Hear 199920238–252. [DOI] [PubMed] [Google Scholar]
- 13.Starr A, Picton T W, Sininger Y, Hood L J, Berlin C I. Auditory neuropathy. Brain 1996119741–753. [DOI] [PubMed] [Google Scholar]
- 14.Davis H, Hirsh S K. A slow brain stem response for low‐frequency audiometry. Audiology 197918445–461. [DOI] [PubMed] [Google Scholar]
- 15.Prieve B A, Gorga M P, Neely S T. Otoacoustic emissions in an adult with severe hearing loss (published erratum appears in J Speech Hear Res 1991;34:703). J Speech Hear Res 199134379–385. [DOI] [PubMed] [Google Scholar]
- 16.Worthington D W, Peters J F. Quantifiable hearing and no ABR: paradox or error? Ear Hear 19801281–285. [DOI] [PubMed] [Google Scholar]
- 17.Starr A, Sininger Y S, Pratt H. The varieties of auditory neuropathy. J Basic Clin Physiol Pharmacol 200011215–230. [DOI] [PubMed] [Google Scholar]
- 18.Adato A, Raskin L, Petit C, Bonne‐Tamir B. Deafness heterogeneity in a Druze isolate from the Middle East: novel OTOF and PDS mutations, low prevalence of GJB2 35delG mutation and indication for a new DFNB locus. Eur J Hum Genet 20008437–442. [DOI] [PubMed] [Google Scholar]
- 19.Houseman M J, Jackson A P, Al Gazali L I, Badin R A, Roberts E, Mueller R F. A novel mutation in a family with non‐syndromic sensorineural hearing loss that disrupts the newly characterised OTOF long isoforms. J Med Genet 200138e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migliosi V, Modamio‐Hoybjor S, Moreno‐Pelayo M A, Rodriguez‐Ballesteros M, Villamar M, Telleria D, Menendez I, Moreno F, del C, I Q829X, a novel mutation in the gene encoding otoferlin (OTOF), is frequently found in Spanish patients with prelingual non‐syndromic hearing loss. J Med Genet 200239502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirghomizadeh F, Pfister M, Apaydin F, Petit C, Kupka S, Pusch C M, Zenner H P, Blin N. Substitutions in the conserved C2C domain of otoferlin cause DFNB9, a form of nonsyndromic autosomal recessive deafness. Neurobiol Dis 200210157–164. [DOI] [PubMed] [Google Scholar]
- 22.Yasunaga S, Grati M, Cohen‐Salmon M, El‐Amraoui A, Mustapha M, Salem N, El‐Zir E, Loiselet J, Petit C. A mutation in OTOF, encoding otoferlin, a FER‐1‐like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 199921363–369. [DOI] [PubMed] [Google Scholar]
- 23.Yasunaga S, Grati M, Chardenoux S, Smith T N, Friedman T B, Lalwani A K, Wilcox E R, Petit C. OTOF encodes multiple long and short isoforms: genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet 200067591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil P L, Kirchhausen T. An emergency response team for membrane repair. Nat Rev Mol Cell Biol 20056499–505. [DOI] [PubMed] [Google Scholar]
- 25.Bansal D, Miyake K, Vogel S S, Groh S, Chen C C, Williamson R, McNeil P L, Campbell K P. Defective membrane repair in dysferlin‐deficient muscular dystrophy. Nature 2003423168–172. [DOI] [PubMed] [Google Scholar]
- 26.Wahlberg J M, Spiess M. Multiple determinants direct the orientation of signal‐anchor proteins: the topogenic role of the hydrophobic signal domain. J Cell Biol 1997137555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott C M, Lingappa V R. Integral membrane protein biosynthesis: why topology is hard to predict. J Cell Sci 20021152003–2009. [DOI] [PubMed] [Google Scholar]
- 28.Britton S, Freeman T, Vafiadaki E, Keers S, Harrison R, Bushby K, Bashir R. The third human FER‐1‐like protein is highly similar to dysferlin. Genomics 200068313–321. [DOI] [PubMed] [Google Scholar]
- 29.Keen T J, Inglehearn C F, Green E D, Cunningham A F, Patel R J, Peacock R E, Gerken S, White R, Weissenbach J, Bhattacharya S S. A YAC contig spanning the dominant retinitis pigmentosa locus (RP9) on chromosome 7p. Genomics 199528383–388. [DOI] [PubMed] [Google Scholar]
- 30.Berlin C I, Hood L J, Hurley A, Wen H. The First Jerger Lecture. Contralateral suppression of otoacoustic emissions: an index of the function of the medial olivocochlear system, Otolaryngol Head Neck Surg 19941103–21. [DOI] [PubMed] [Google Scholar]
- 31.Hood L J, Berlin C I, Bordelon J, Rose K. Patients with auditory neuropathy/dys‐synchrony lack efferent suppression of transient evoked otoacoustic emissions. J Am Acad Audiol 200314302–313. [PubMed] [Google Scholar]
- 32.Starr A, Sininger Y, Winter M, Derebery M J, Oba S, Michalewski H J. Transient deafness due to temperature‐sensitive auditory neuropathy. Ear Hear 199819169–179. [DOI] [PubMed] [Google Scholar]
- 33.Singh G, Lykke‐Andersen J. New insights into the formation of active nonsense‐mediated decay complexes. Trends Biochem Sci 200328464–466. [DOI] [PubMed] [Google Scholar]
- 34.Letunic I, Goodstadt L, Dickens N J, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley R R, Ponting C P, Bork P. Recent improvements to the SMART domain‐based sequence annotation resource. Nucleic Acids Res 200230242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci 199621375–382. [PubMed] [Google Scholar]
- 36.Mason J M, Arndt K M. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 20045170–176. [DOI] [PubMed] [Google Scholar]
- 37.Payne A S, Kelly E J, Gitlin J D. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper‐dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci USA 19989510854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berson J F, Frank D W, Calvo P A, Bieler B M, Marks M S. A common temperature‐sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J Biol Chem 200027512281–12289. [DOI] [PubMed] [Google Scholar]
- 39.Santana A, Salido E, Torres A, Shapiro L J. Primary hyperoxaluria type 1 in the Canary Islands: a conformational disease due to I244T mutation in the P11L‐containing alanine:glyoxylate aminotransferase. Proc Natl Acad Sci USA 20031007277–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]