Abstract
Based on binding studies undertaken by electrospray ionization-mass spectrometry, a synthetic pyrrole-inosine nucleoside, 1, capable of forming an extended three-point Hoogsteen-type hydrogen-bonding interaction with guanine, is shown to form specific complexes with two different quadruplex DNA structures [dTG4T]4 and d(T2G4)4 as well as guanine rich duplex DNA. The binding interactions of two other analogs were evaluated in order to unravel the structural features that contribute to specific DNA recognition. The importance of the Hoogsteen interactions was confirmed through the absence of specific binding when the pyrrole NH hydrogen-bonding site was blocked or removed. While 2, with a large blocking group, was not found to interact with virtually any form of DNA, 3, with the pyrrole functionality missing, was found to interact non-specifically with several types of DNA. The specific binding of 1 to guanine rich DNA emphasizes the necessity of careful ligand design for specific sequence recognition.
Keywords: electrospray ionization, oligonucleotides, non-covalent interactions, Hoogsteen binding
Introduction
A variety of DNA interactive ligands have been developed with a wide range of binding modes and targeted applications [1-4] such as ones designed to stall replication or transcription[4-6] or to stimulate apoptosis [7]. Common binding modes of these ligands to duplex DNA include minor groove binding and intercalation [1,3-4,8]. The former is common in the deep well of adenine/thymine rich regions, whereas the latter most often occurs in guanine/cytosine rich regions and involves insertion between base pairs in duplex DNA. Other ligands have been developed to bind to quadruplex DNA to limit binding and activity of telomerase [9-18]. There continues to be considerable effort to design and construct new DNA interactive agents, ones with greater selectivity for certain DNA structures or ones with higher affinities. The development of these DNA interactive ligands is facilitated by rapid, sensitive analytical screening methods that provide diagnostic feedback about key issues like binding selectivity and specificity of DNA recognition. Electrospray ionization mass spectrometry (ESI-MS) is one of the newer tools used to evaluate DNA interactive ligands and offers the advantages of high sensitivity, low sample consumption, and high throughput screening [19-21]. Mass spectrometric studies of DNA interactive ligands have been effective in characterizing binding selectivity and specificity [9-10,15-16,22], establishing the mode of binding [9-10,15,22], and correlating specific binding patterns with in vitro antitumor and antibacterial cytotoxicity [22].
Recently, work presented by our group [15,23] and others [9-10,14,17,24-27] has established the utility of using ESI-MS to evaluate the binding properties of potential quadruplex-interactive ligands. Although many of these prior studies have focused on DNA interactive ligands that have been developed as drugs, ESI-MS can also play a role in evaluating new DNA binding modalities in the early stages of ligand design, whether for new drug platforms, DNA sensors, or targeted DNA cleavage agents. Because ESI-MS offers fast analysis with minimal sample usage, it can be used conveniently to provide step-by-step feedback at each state of ligand design, thus allowing systematic development of structure/binding activity relationships and optimization of DNA recognition.
In this study, we use ESI-MS to examine a pyrrole-inosine based synthetic nucleoside, 1 (Chart 1) that is an offspring from a previous water insoluble analog that demonstrated the ability to bind DNA via an extended Hoogsteen-type interaction [28]. 1 was designed with appropriate functional elements to engage in a series of hydrogen-bonding interactions with a complementary nucleobase, in this case guanine (Scheme 1). The present work was undertaken to determine whether 1 binds quadruplexes specifically due to their guanine-rich composition and Hoogsteen-stabilized conformations or has a more general affinity for guanine bases. The interactions of 1 with two different quadruplex-forming oligonucleotides, dTG4T, which forms a four-stranded intermolecular quadruplex [dTG4T]4, and d(T2G4)4, which forms an intramolecular quadruplex, as well as several single-stranded and double-stranded oligonucleotide sequences were investigated to probe the selectivity of 1 towards different DNA secondary structures. Single strands and duplexes were chosen with both high and low GC content to determine guanine selectivity. Furthermore, comparative experiments were carried out to evaluate the binding of structural analogs of 1 in which the pyrrole NH was either removed (3) or the NH was blocked (2) (Chart 1).
Chart 1.
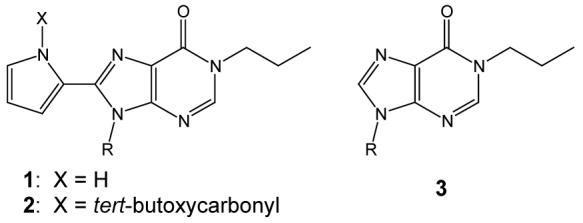
Structures of synthetic nucleosides, 1-3.
Scheme 1.
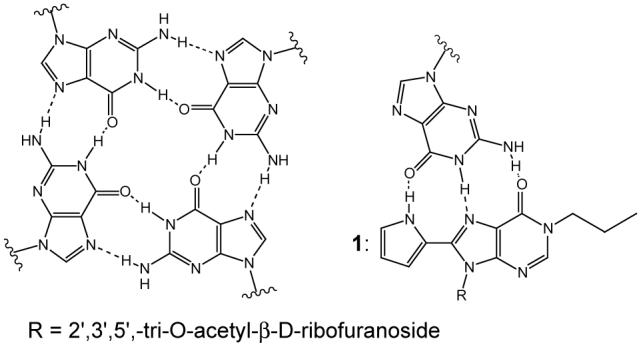
Hoogsteen binding in guanine tetrads and between 1 and guanine bases.
Experimental
Reagents
The general preparation strategy for the synthetic nucleosides 1, 2, and 3, albeit bound to different sugars, was described previously [28]. Details of the syntheses are provided in the supplemental information. Oligonucleotides were prepared and purified by reversed-phase HPLC by Integrated DNA Technologies (Coralville, IA). Ammonium acetate (98% purity) and A.C.S. Grade Spectranalyzed methanol was obtained from ThermoFisher Scientific (Waltham, MA). All purchased chemicals were used without further purification. Water was purified in house with an EASYpure UV deionizer (Barnstead International, Dubuque, IA).
Preparation of Samples for Analysis
Annealing of d(T2G4)4 (Q1), [dTG4T]4 (Q2),dGGCGTCGGCGTCGC/dGCGACGCCGACGCC (DS1), and dATAAAAACGAAAATA/dTATTTTCGTTTTTAT (DS2) was performed by heating a 1 mM solution of the oligonucleotide in 150 mM ammonium acetate to 90°C followed by cooling to room temperature over 2 hours. [14,16,28] Two single strand oligonucleotide sequences were also analyzed: dGGCGTCGGCGTCGC (SS1) and dATAAAAACGAAAATA (SS2). The annealed or single strand oligonucleotides were analyzed in a final ammonium acetate concentration of 50 mM and a solvent composition of 20% methanol. A ligand (20-50 μM) was incubated with DNA (10 μM) for 10 minutes prior to ESI-MS analysis.
Methods and Instrumentation
Mass spectrometric analyses were performed on a ThermoFinnigan LCQ Duo quadrupole ion trap mass spectrometer equipped with the stock ESI source (San Jose, CA). Oligonucleotide samples were analyzed in the negative ion mode with the heated capillary set at 80°C. Source conditions were tuned to minimize in-source fragmentation of complexes with a needle voltage of 3.5 kV and a nitrogen sheath gas of 40 arbitrary units. Solutions were infused directly at 3 μL min−1, and the spectra presented represent the average of 100 to 200 scans. Relative binding affinities for each ligand to each type of DNA are given as the fraction of bound DNA, expressed as a percentage, which relates the total peak areas of the various ligand:DNA complexes to all DNA species (both bound and free). The equation, which has been used in previous studies [29,30], to calculate the fraction bound DNA is:
where PAn:m represents the peak areas of the various ligand:DNA complexes and PADNA denotes the total peak area of free DNA ions. Peak areas were determined in Origin 7.0 (OriginLab, Northampton, MA). A high fraction bound value indicates a higher ligand:DNA binding efficiency. Measurement of the peak areas, rather than relative abundances, of the complexes permits the inclusion of salt adducts that may not be evident upon visual inspection of the mass spectra.
Results and Discussion
Interactions with Quadruplex DNA
Previous experiments have demonstrated the utility of ESI-MS for analyzing quadruplex DNA structures formed from short oligonucleotides [9-10,14-15,17,29,31-32]. For the present study, two of these oligonucleotides were selected to evaluate the interactions between quadruplex DNA and the three nucleosides. The first oligonucleotide, d(T2G4)4, forms an intramolecular quadruplex in which one strand is folded into a quadruplex with four G-tetrads (Q1). The second oligonucleotide, dTG4T, forms a parallel intermolecular quadruplex involving four strands of DNA (Q2), [dTG4T]4.
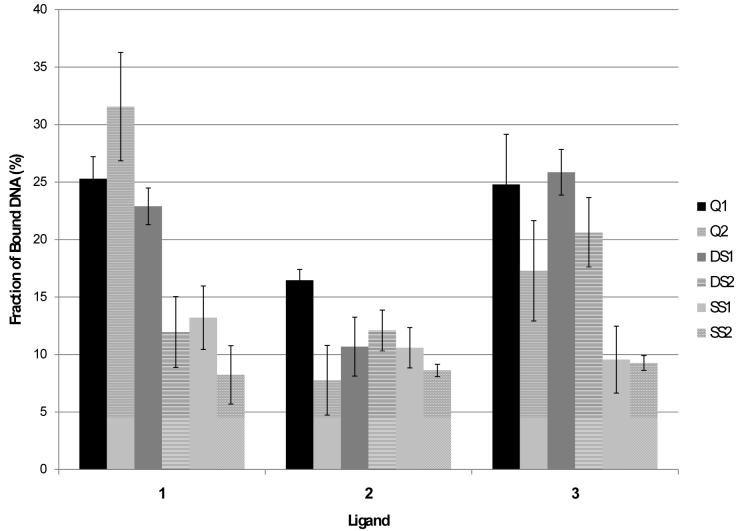
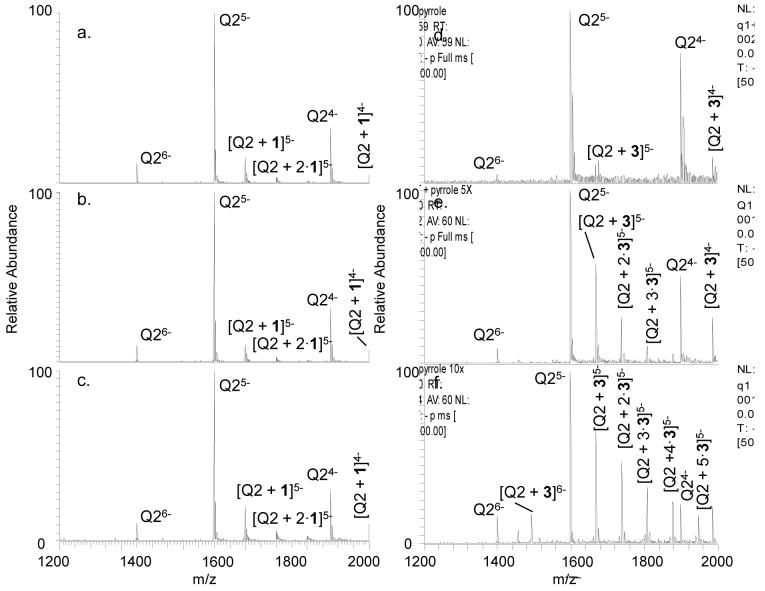
Examples of the ESI-mass spectra obtained for the quadruplex and quadruplex/ligand solutions are shown in Figure 1. Quadruplex ions in the -4, -5, and -6 charge states are observed for Q2 (Figure 1a), and the quadruplex/ligand complexes are easily discerned and assigned in Figures 1b-d based on the characteristic mass shifts caused by the bound ligand(s). A table listing m/z values for all mass spectra is given in the supplemental information (Table S-1). Ligand 1 exhibits the greatest relative extent of binding, whereas ligand 2 engages in very little quadruplex binding. The bulky tert-butoxycarbonate (BOC) group of ligand 2 appears to significantly suppress DNA binding. Ligand 3 exhibits lower binding affinity than 1, and this difference is ascribed to the lack of a third possible hydrogen bonding interaction which is possible for 1. For the three nucleosides, the relative quadruplex binding affinity follows the trend 1 > 3 > 2 indicating that the improved hydrogen bonding motif and lack of steric interference allows 1 to bind to Q2 more effectively than the other two compounds. A similar trend was observed for binding to Q1 albeit with greater binding between Q1 and 3 than was seen with Q2 (mass spectral results not shown), and the complete set of comparative results is shown in bar graph form in Figure 2 for easy visual comparison. The differences in ligand binding between Q1 and Q2 are most likely due to the different conformations at either end of each quadruplex since the quadruplexes themselves are of the same length. Since Q1 is an intermolecular quadruplex while Q2 is an intramolecular one, the thymidine nucleotides at either end of the quadruplexes will be in different conformations: unstructured for Q1 and in thymidine loops for Q2. This could create different binding pockets for ligand interaction.
Figure 1.
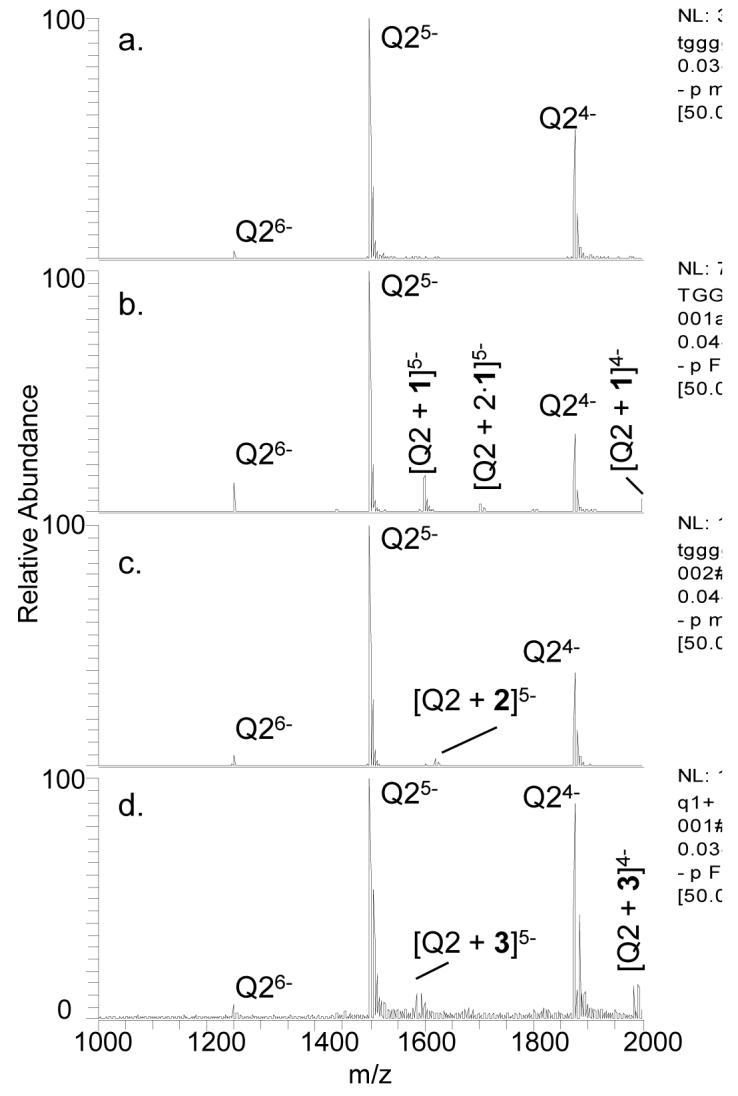
ESI-mass spectra of Q2 (10 μM) alone (a) and with 1 (b), 2 (c), or 3 (d) with each ligand at 20 μM. The label Q indicates the quadruplex form.
Figure 2.
Fraction of bound DNA for ligands 1, 2, and 3 to quadruplexes, duplexes and single strands. Error bars represent standard deviations after three replicates. The ratio of ligand to DNA was 2:1 in each case.
Concentration Dependence of Ligand Binding
To evaluate whether the interaction between 1 and quadruplex DNA is a specific association or a non-specific aggregation, solutions containing Q2 with varying concentrations of 1 were analyzed (Figure 3). The concentration of DNA was fixed at 10 μM, and the ligand concentration was varied from 20 to 50 to 100 μM. Over this range, the extent of complexation between 1 and Q2 remained unchanged. The lack of dependence of the extent of complexation on the concentration of 1 offers evidence against non-specific aggregation between 1 and the quadruplex DNA. Analogous concentration-dependent experiments were carried out with 3 and indicated a significant degree of non-specific binding, based on the substantial increase in the stoichiometries of the DNA/ligand complexes as well as the notable increase in the fraction of bound DNA as the relative concentration of ligand 3 increased (Figure 3). For example, up to five molecules of 3 are bound to one quadruplex in Figure 3f. The high number of bound ligands suggests that 3 is aggregating with the quadruplex in a non-specific manner, a situation that does not occur for 1. When analyzed alone, both 1 and 3 show the same extent of dimer and trimer formation indicating that the differences seen in binding are not related to simple ligand aggregation (data not shown). Results of the same experiment performed with 2 showed minimal binding even at ligand:DNA concentrations as high as 10:1, highlighting the impact of steric hindrance on DNA binding (data not shown).
Figure 3.
ESI-mass spectra of Q2 (10 μM) with varying concentrations of 1 or 3. Concentrations of 1 were 20 μM (a), 50 μM (b) and 100 μM (c). Concentrations of 3 were 20 μM (d), 50 μM (e), and 100 μM (f).
Interactions with Single-Stranded or Duplex DNA
To evaluate the selectivity of these ligands towards quadruplex DNA versus other alternative DNA structures, the ligands were incubated with single strand and duplex DNA. The single strands or duplexes were chosen due to their widely different GC versus AT base content. For instance, duplex DS1 contains over 80% GC base composition, whereas DS2 contains only about 25% GC base content. A decrease in binding of 1 to DS2 is expected if the binding is guanine specific. Examples of the DNA binding results are shown in Figure 4 for DS1 with all three ligands. It is evident from comparison of the ESI-mass spectra for solutions containing DS1 versus DS2 (data not shown) that 1 exhibited more substantial binding to the GC rich duplex than the AT rich duplex. In fact, the extent of binding to DS2 was similar to that observed for the individual single strands for ligand 1 (see bar graph comparison in Figure 2). This latter result suggests that the binding motif of 1 is G-selective but not necessarily uniquely specific for quadruplex or duplex structures. In the case of 3, the ligand bound well to both duplexes and with a similar level of binding to both Q1 and Q2, thus indicating a greater degree of non-specificity with this ligand. The bulky BOC group of ligand 2 suppressed binding to both duplexes.
Figure 4.
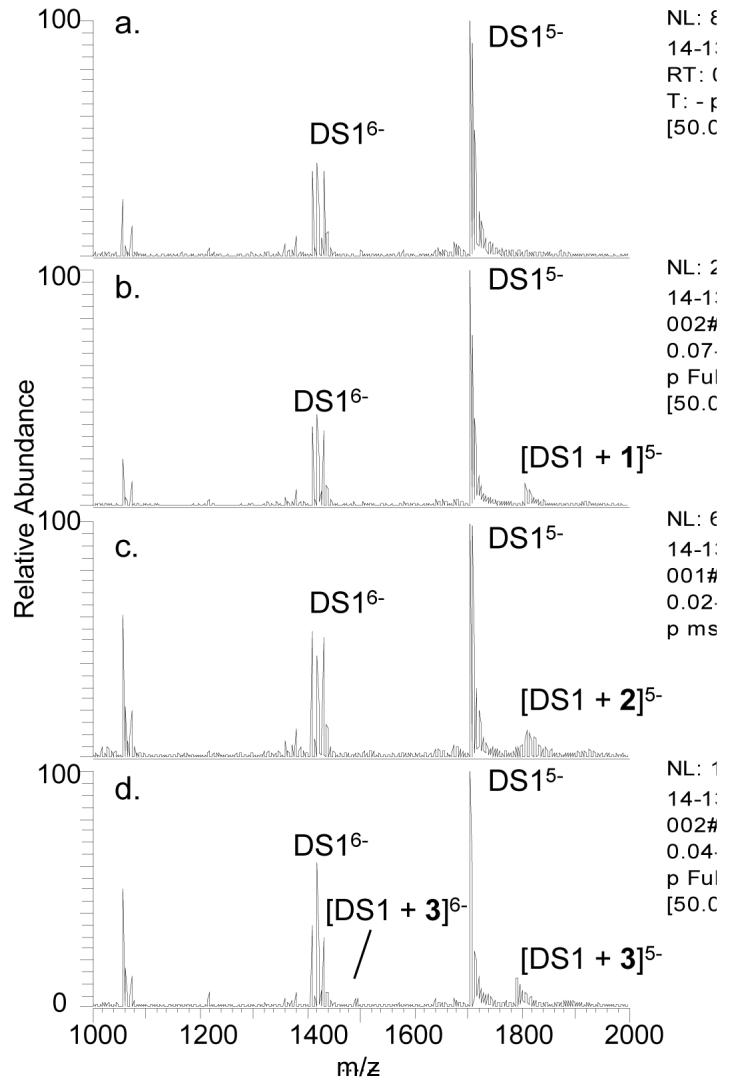
ESI-mass spectra of DS1 alone (a) and with 1 (b), 2 (c), or 3 (d). DS indicates the double strand. The duplex concentration was 10 μM and the ligand concentrations were 20 μM each.
Examples of the ESI-MS binding results involving the single strands are shown in Figure 5. Binding was lower for the single strands than for the duplexes or quadruplexes, even for the GC rich single strand. The low level of binding to the single strands as well as the consistency across the three nucleoside ligands suggests that the single strand binding may be due to simple aggregation with no base specificity.
Figure 5.
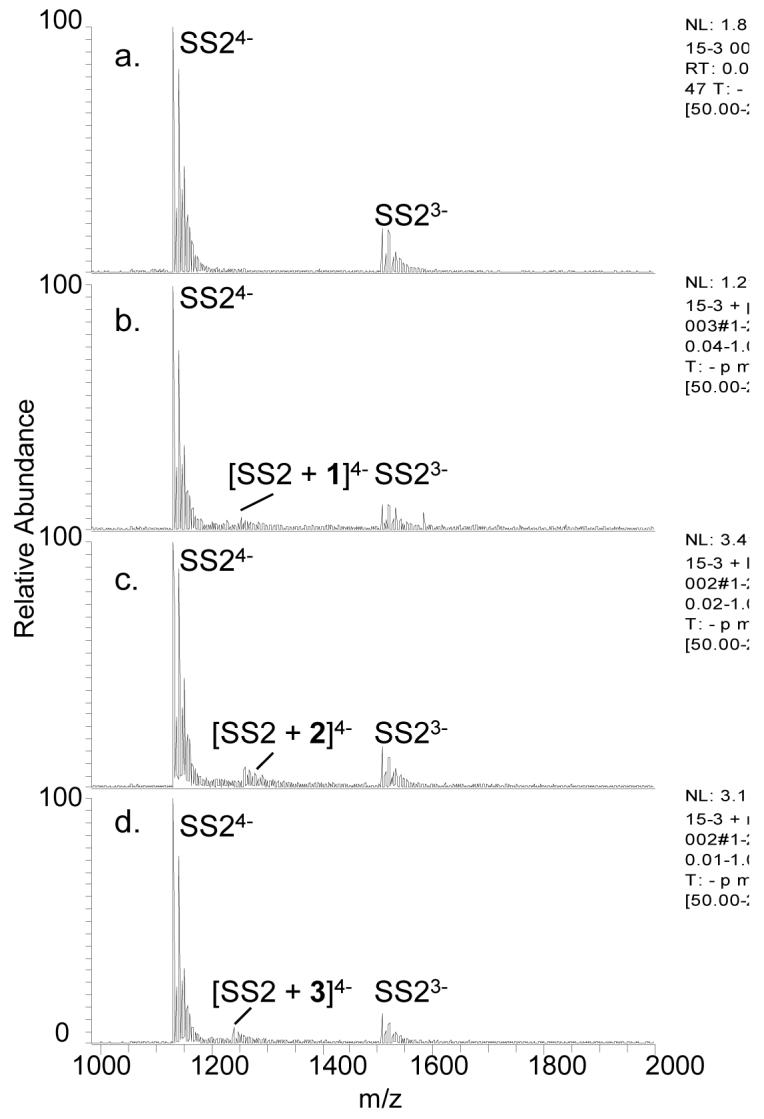
ESI-mass spectra of SS2 alone (a) and with 1 (b), 2 (c), and 3 (d). The single strand concentration was 10 μM and the ligand concentrations were 20 μM each.
The bar graph shown in Figure 2 summarizes the DNA binding trends for the three ligands and the various DNA structures. The binding to guanine-rich oligonucleotides was most significant for 1 which is attributed to its three appropriately-configured hydrogen-bonding sites. As this ligand is designed with a donor-acceptor-acceptor hydrogen-bonding pattern, it is complementary to the acceptor-donor-donor motif of Hoogsteen binding to guanine. However, lack of any evidence of destabilization of the quadruplexes and duplexes upon binding of 1 would indicate that base stacking is a likely binding motif. It is possible the pyrrole functionality enhances the binding to quadruplex and guanine rich duplex structures without disrupting the Hoogsteen or Watson-Crick interactions. For 2, the single donor hydrogen-bonding site is blocked which, as reflected in Figure 2, greatly reduces the ability of 2 to bind to guanine in addition to causing substantial steric hindrance. The steric hindrance factor is alleviated in the third ligand, 3; however, the lack of a donor hydrogen bonding site reduces its guanine specific binding affinity and makes it a less base-selective structural motif.
Collision Induced Dissociation of DNA/1 Complexes
The nature of the binding interactions between ligand 1 and DNA was further explored by collision induced dissociation (CID). Previous studies have shown that duplex/ligand complexes which incorporate groove binding interactions dissociate mainly by strand separation and base loss upon collisional activation, with some dependence on charge state [33]. In contrast, elimination of the ligand upon CID conveys that the binding mode is intercalation [33-34]. Our group and others have also reported different dissociation pathways for quadruplex/ligand complexes based on the binding mode [14-15,29]. For quadruplex groove binders, strand separation either with or without base retention has been observed, while end-stacking ligands are typically ejected from the complexes [14,29]. Dissociation of the Q2/1 complex resulted in loss of 1 rather than in strand separation (see Figure 6a), suggesting end-stacking as a possible binding mode. Dissociation of the -6 charge state of the DS1/1 complex resulted in loss of the ligand and no strand separation (see Figure 6b). While these CAD spectra cannot definitively confirm the binding mode, these dissociation patterns are consistent with intercalation or end stacking for the duplex and an end-stacking mode for the quadruplex.
Figure 6.
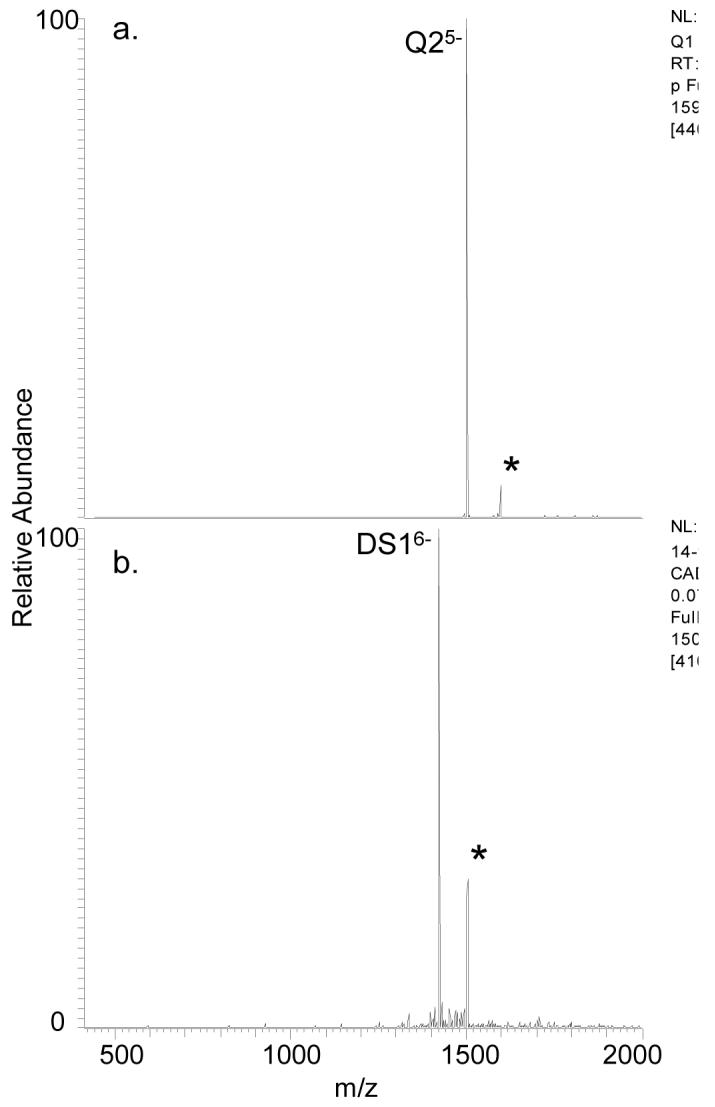
ESI-CAD mass spectra for Q2 and DS1. The complexes analyzed were [Q2 + 1]5- (a) and [DS1 + 1]6- (b) where the selected precursor ions are labeled with asterisks.
Conclusions
Ligand 1 bound two quadruplex structures, [dTG4T]4 and d(T2G4)4, as well as a guanine rich double strand. Ligand 1, with the capability of forming three-point hydrogen bonds with guanine, was found to have high affinity for guanine-rich sequences, and binding was consistent with specific interactions with guanine bases. The lack of aggregation of 1 with DNA at high concentration ratios supports that the binding is specific. Only low abundance complexes were observed with 2, thus indicating the impact of steric hindrance caused by the large BOC blocking group. Interactions between the various DNA structures and 3, especially at high ligand concentrations, indicate that this small ligand binds non-specifically. In previous work undertaken in a predominantly organic solvent, evidence of dissociation of guanine nucleoside clusters upon addition of a water insoluble analog of 1 was interpreted as indicating the ability of this ligand to engage in Hoogsteen interactions that disrupted guanine-guanine binding [28]. Similar results were not explicitly seen in this work suggesting that the binding mode may be more complex. For instance, CID spectra are consistent with end-stacking for the quadruplex/1 complex and intercalation as a possible binding mode for the DS1/1 complex, which could explain the apparent guanine selectivity possessed by 1. The differences in duplex binding between the three nucleosides may be due to the greater number of hydrogen-bonding sites of 1 or simply that the shape is sterically more conducive to intercalation than the others. This work showcases ESI-mass spectrometry as a tool for rapid screening of new synthetic ligands and its role in providing early stage feedback towards the design of base-selective DNA interactive ligands.
Supplementary Material
Acknowledgments
Support from the Welch Foundation (F-1155 to JSB and F-1018 to JLS) and NIH (RO1 GM65956) is gratefully acknowledged. SEP and CLS acknowledge fellowship support from an NSF/IGERT grant (D6E-03333080).
Supporting Information Available. Details of the synthesis of nucleosides 1, 2, and 3 including a reaction scheme for the formation of the three nucleosides as well as a table of m/z ratios for ions of interest from all mass spectra.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Wemmer DE. Annu. Rev. Biomol. Struct. 2000;29:439. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- [2].Gniazdowski M, Denny WA, Nelson SM, Czyz M. Curr. Med. Chem. 2003;10:909. doi: 10.2174/0929867033457683. [DOI] [PubMed] [Google Scholar]
- [3].Snyder RD, Hendry LB. Environ. Mol. Mutagen. 2005;45:100. doi: 10.1002/em.20096. [DOI] [PubMed] [Google Scholar]
- [4].Nelson SM, Ferguson LR, Denny WA. Mutat. Res.-Fund. Mol. M. 2007;623:24. doi: 10.1016/j.mrfmmm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- [5].Kumar CV, Barton JK, Turro NJ. J.Am. Chem. Soc. 1985;107:5518. [Google Scholar]
- [6].Geierstanger BH, Wemmer DE. Annu. Rev. Bioph. Biom. 1995;24:463. doi: 10.1146/annurev.bb.24.060195.002335. [DOI] [PubMed] [Google Scholar]
- [7].Lambert B, Jones BK, Roques BP, LePecq JB, Yeung AT. Proc. Natl. Acad. Sci. USA. 1989;86:6557. doi: 10.1073/pnas.86.17.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Snyder RD. Mutat. Res.-Fund. Mol. M. 2007;623:72. doi: 10.1016/j.mrfmmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- [9].Guittat L, Alberti P, Rosu F, Van Miert S, Thetiot E, Pieters L, Gabelica V, De Pauw E, Ottaviani A, Riou J-F, Mergny J-L. Biochimie. 2003;85:535. doi: 10.1016/s0300-9084(03)00035-x. [DOI] [PubMed] [Google Scholar]
- [10].Rosu F, De Pauw E, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou J-F, Mergny J-L. Biochemistry. 2003;42:10361. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- [11].Perry PJ, Reszka AP, Wood AA, Read MA, Gowan SM, Dosanjh HS, Trent JO, Jenkins TC, Kelland LR, Neidle S. J. Med. Chem. 1998;41:4873. doi: 10.1021/jm981067o. [DOI] [PubMed] [Google Scholar]
- [12].Mergny J-L, Lacroix L, Teulade-Fichou M-P, Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron J-P, Lehn J-M, Riou J-F, Garestier T, Hélène C. Proc. Nat. Acad. Sci. USA. 2001;98:3062. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Read MA, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. Proc. Nat. Acad. Sci. USA. 2001;98:4844. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Rapid Commun. Mass Spectrom. 2002;16:1729. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- [15].David WM, Brodbelt J, Kerwin SM, Thomas PW. Anal. Chem. 2002;74:2029. doi: 10.1021/ac011283w. [DOI] [PubMed] [Google Scholar]
- [16].Carrasco C, Rosu F, Gabelica V, Houssier C, De Pauw E, Garbay-Jaureguiberry C, Roques B, Wilson WD, Chaires JB, Waring MJ, Bailly C. ChemBioChem. 2002;3:1235. doi: 10.1002/1439-7633(20021202)3:12<1235::AID-CBIC1235>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- [17].Rosu F, Gabelica V, Shin-ya K, De Pauw E. Chem. Commun. 2003:2702. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- [18].Redon S, Bombard S, Elizondo-Riojas M-A, Chottard J-C. Nucleic Acids Res. 2003;31:1605. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Greig MJ, Robinson JM. J. Biomol. Screen. 2000;5:441. doi: 10.1177/108705710000500607. [DOI] [PubMed] [Google Scholar]
- [20].Wan KX, Shibue T, Gross ML. J.Am. Chem. Soc. 2000;122:300. [Google Scholar]
- [21].Hofstadler SA, Sannes-Lowery KA. Nat. Rev. Drug Discov. 2006;5:585. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- [22].Oehlers L, Mazzitelli CL, Brodbelt JS, Rodriguez M, Kerwin S. J. Am. Soc. Mass Spectrom. 2004;15:1593. doi: 10.1016/j.jasms.2004.07.015. [DOI] [PubMed] [Google Scholar]
- [23].Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. J. Am. Soc. Mass Spectrom. 2006;17:593. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [24].Baker ES, Lee JT, Sessler JL, Bowers MT. J. Am. Chem. Soc. 2006;128:2641. doi: 10.1021/ja0564968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li W, Zhang M, Zhang J, Li H, Zhang X, Sun Q, Qiu C. FEBS Letters. 2006;580:4905. doi: 10.1016/j.febslet.2006.08.007. [DOI] [PubMed] [Google Scholar]
- [26].Gabelica V, Baker ES, Teulade-Fichou M, De Pauw E, Bowers MT. J. Am. Chem. Soc. 2007;129:895. doi: 10.1021/ja065989p. [DOI] [PubMed] [Google Scholar]
- [27].Krishnan-Ghosh Y, Liu D, Balasubramanian S. J. Am. Chem. Soc. 2004;126:11009. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- [28].Sessler JL, Jayawickramarajah J, Sherman CL, Brodbelt JS. J. Am. Chem. Soc. 2004;126:11460. doi: 10.1021/ja046773v. [DOI] [PubMed] [Google Scholar]
- [29].Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. J. Am. Soc. Mass Spec. 2006;17:593. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [30].Mazzitelli CL, Chu Y, Reczek JR, Iverson BL, Brodbelt JS. J. Am. Soc. Mass Spec. 2007;18:311. doi: 10.1016/j.jasms.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vairamani M, Gross ML. J. Am. Chem. Soc. 2003;125:42. doi: 10.1021/ja0284299. [DOI] [PubMed] [Google Scholar]
- [32].Sakamoto S, Yamaguchi K. Angew. Chem. Int. Ed. 2003;42:905. doi: 10.1002/anie.200390239. [DOI] [PubMed] [Google Scholar]
- [33].Rosu F, Pirotte S, De Pauw E, Gabelica V. Int. J. Mass Spec. 2006;253:156. [Google Scholar]
- [34].Keller KM, Zhang J, Oehlers L, Brodbelt JS. JMS. 2005;40:1362. doi: 10.1002/jms.927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.