Summary
The mechanisms responsible for maintaining genomic methylation imprints in mouse embryos are not understood. We generated a knockout mouse in the Zfp57 locus encoding a KRAB zinc finger protein. Loss of just the zygotic function of Zfp57 causes partial neonatal lethality, while eliminating both maternal and zygotic functions of Zfp57 results in a highly penetrant embryonic lethality. In oocytes, absence of Zfp57 results in failure to establish maternal methylation imprints at the Snrpn imprinted region. Intriguingly, methylation imprints are re-acquired specifically at the maternally derived Snrpn imprinted region when the zygotic Zfp57 is present in embryos. This suggests that there may be DNA methylation-independent memory for genomic imprints. Zfp57 is also required for the post-fertilization maintenance of maternal and paternal methylation imprints at multiple imprinted domains. The effects on genomic imprinting are consistent with the maternal-zygotic lethality of Zfp57 mutants.
Keywords: ZFP57, KRAB zinc finger, maternal-zygotic effect, genomic imprinting, DNA methylation
Introduction
Parental genetic influence on the development of offspring is evident in both the invertebrate and vertebrate animal kingdoms. Maternal effects are commonly mediated through deposition of the cytoplasmic transcripts or protein products in oocytes during oogenesis in the female germline. These then exert their effects on the fertilized zygotes and affect early embryonic development (De Robertis et al., 2000; Gosden, 2002; Melendez and Greenwald, 2000; Newman-Smith and Rothman, 1998; Nusslein-Volhard, 1991; Priess et al., 1987; Schier, 2001).
Genomic imprinting, the process that causes genes to be expressed according to their parental origin, occurs in plants and mammals and is mediated by epigenetic modifications that differ on the two parental chromosomes (Ferguson-Smith and Surani, 2001; McGrath and Solter, 1984; Reik and Walter, 2001; Surani et al., 1984; Tilghman, 1999; Verona et al., 2003). Over 80 imprinted genes have been discovered in mouse (http://www.mgu.har.mrc.ac.uk/research/imprinting/). Many are known to be localized in clusters regulated by a cis-acting imprinting control region (ICR) that acquires heritable parental origin-specific differential DNA methylation in the male or female germline (Ben-Porath and Cedar, 2000; Lewis and Reik, 2006). The cycle of methylation programming at an ICR begins with erasure of methylation during the development of primordial germ cells. Subsequently, during oogenesis and spermatogenesis de novo methylation is differentially established on the maternal and paternal chromosomes in the germline; these are the so-called, germline imprints. After fertilization, the preimplantation embryonic genome loses much of its methylation through both active and passive demethylation events commencing in the zygote, and subsequently de novo methylation is acquired around the time of implantation (Reik et al., 2001). Germline imprints, however, appear resistant to this post-fertilization methylation reprogramming allowing them to be stably inherited from germline to offspring (Morgan et al., 2005).
Studies in which the de novo and maintenance DNA methyltransferase machinery has been mutated have proven that germline establishment and post-fertilization maintenance of differential methylation is essential for the monoallelic activity of imprinted genes (Bourc’his et al., 2001; Kaneda et al., 2004; Li et al., 1993). In contrast, the mechanisms specifically rendering germline methylation imprints resistant to preimplantation genome-wide demethylation are not understood. Oocyte-derived PGC7/Stella was shown to confer partial protection from demethylation at one class of repetitive sequence and at some imprinted domains (Nakamura et al., 2007). More recently, maternal and zygotic functions of Dnmt1, the maintenance methyltransferase, were found to maintain DNA methylation imprints in preimplantation embryos (Hirasawa et al., 2008).
The majority of germline methylation marks identified to date are established on maternally inherited chromosomes during oogenesis. These include the Snrpn imprinted region implicated in human Prader-Willi and Angelman syndromes. In general, these maternal methylation imprints are located at promoters which, in some cases, regulate large antisense non-coding RNAs expressed from the paternal chromosome that have been implicated in repression of protein coding genes in cis (Pauler et al., 2007). Only three imprinted domains are known to be regulated by a controlling element that is methylated in the paternal germline. Of these, the Igf2-H19 and Dlk1-Dio3 imprinting control elements regulate imprinted genes that are essential for normal prenatal growth and development (Edwards and Ferguson-Smith, 2007; Kawahara et al., 2007; Thorvaldsen and Bartolomei, 2007).
KRAB zinc finger proteins form one of the largest transcription factor families in the mouse and human genome (Looman et al., 2002). They act as potent transcriptional repressors through KRAB box-mediated interaction with KAP-1/TIF-1β co-repressor complexes (Abrink et al., 2001; Friedman et al., 1996; Schultz et al., 2002; Schultz et al., 2001). Mediated by the DNA binding capacity of KRAB zinc-finger proteins, KAP-1 functions to recruit factors associated with DNA methylation (Wiznerowicz et al., 2007) and the formation of repressive chromatin including histone deacetylases and histone methyltransferases (Ayyanathan et al., 2003; Schultz et al., 2002; Schultz et al., 2001). Despite this, there are very few known target genes of the KRAB zinc finger proteins. There is even more limited information as to the function of KRAB zinc finger proteins in vivo except for the variant rsl mouse affecting sexually dimorphic gene expression in the liver (Krebs et al., 2003). We identified Zfp57 during a gene trap-based screen for factors down-regulated upon embryonic stem (ES) cell differentiation (Li and Leder, 2007). We generated a knockout mouse in the Zfp57locus that encodes a putative KRAB zinc finger protein. Our data indicate that Zfp57 is an essential maternal-zygotic effect gene and is required for the establishment and re-acquisition of the maternal methylation imprint at the Snrpn domain. It also maintains both paternal and maternal methylation imprints after fertilization at multiple imprinted regions.
Results
ZFP57 is a KRAB zinc finger protein
Consistent with previous findings (Alonso et al., 2004), we found mouse ZFP57 is a putative KRAB zinc finger protein (Figure 1A). A human homolog was identified indicating conservation of this protein (Figure 1B). To determine whether ZFP57 contains a functional KRAB box, we performed a co-immunoprecipitation (co-IP) interaction assay in which both myc-epitope-tagged mouse ZFP57 and KAP-1/TIF-1β, the obligate co-repressor for KRAB-zinc finger proteins, were overexpressed transiently in COS cells. Antibodies against the myc epitope were used to pull down ZFP57-associated proteins and two different antibodies against two non-overlapping regions of KAP-1/TIF-1β were used to probe the immunoprecipitated material (Figure 1C). KAP-1/TIF-1β was detected when it was co-expressed with ZFP57 (Lane 5 in Figure 1C). These data prove that ZFP57 contains a conserved functional KRAB box that binds KAP-1/TIF-1β. Binding between endogenous ZFP57 and KAP-1/TIFβ was confirmed in ES cells (Supplemental Figure S1A).
Figure 1. ZFP57 is a putative KRAB zinc finger protein.
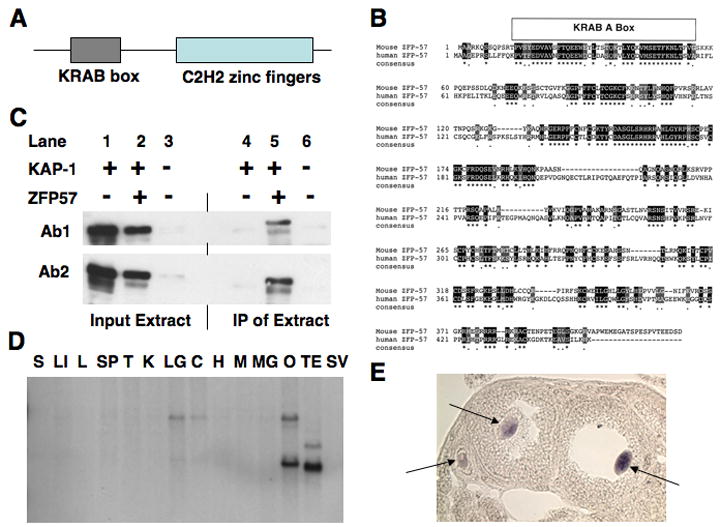
(A) Schematic diagram of the ZFP57 protein.
(B) Sequence alignment between the mouse ZFP57 and the human ZFP57. The identical amino acids are boxed in black and the similar residues are shaded.
(C) Co-IP assays were carried out for KAP-1 and myc epitope-tagged ZFP57. Three left lanes are the western blot of the total cell lysate (Input) from the KAP-1-transfected (lane 1), the KAP-1 and myc epitope-tagged ZFP57 co-transfected (lane 2) and the untransfected cos cells (lane 3). Lane 4, 5, 6 are the western blot of the immunoprecipitate (IP) derived from these three samples when the antibodies against myc epitope were used to pull down ZFP57-associated proteins. The rabbit polyclonal antibodies used for the western blot are Ab1 (anti-KAP1 RBCC) and Ab2 (anti-KAP1 CT) (Schultz et al., 2001).
(D) Adult mouse organ Northern blot of Zfp57. A 1.2kb cDNA fragment encompassing the entire open-reading-frame of Zfp57 was used to probe a Northern blot. Equal amount of polyA RNA was loaded on each well (Chester et al., 1998) and was prepared from the various organs as follows: (S) stomach; (LI) large intestine; (L) liver; (SP) spleen, (T) thymus; (K) kidney; (LG) lung; (C) cerebrum; (H) heart; (M) muscle; (MG) mammary gland; (O) ovary; (TE) testis; (SV) seminal vesicle.
(E) RNA in-situ hybridization reveals that Zfp57 is expressed specifically in the maturing oocytes (purple stain). Frozen sections of the wild-type ovary were probed with an antisense riboprobe derived from a 0.5kb fragment of the 5’ portion of the Zfp57 cDNA. Arrows point to the labelled oocytes inside the cavity of the follicles.
Zfp57 is expressed in oocytes and in a subset of adult tissues
As expected from our original screen, the transcription of Zfp57 is down-regulated when ES cells differentiate (Ahn et al., 2004; Akagi et al., 2005; Li and Leder, 2007). ES cells contain two prominent Zfp57 transcript isoforms (Akagi et al., 2005; Okazaki et al., 1994). Zfp57 also displays a restricted expression pattern in adult mouse organs with the highest level of expression in testis and ovary and at low levels in lung and brain (Figure 1D). The longer transcript appears to be the main product in the lung and brain and the short transcript is the predominant form in ovary. Testis has a third intermediate-sized transcript isoform. Zfp57 transcripts appear to be present at all post-implantation stages (Supplemental Figure S1B).
RNA in-situ hybridization demonstrates specific expression of Zfp57 in oocytes within adult wild-type mouse ovaries (Figure 1E). Interestingly, no other cell types in the ovary, including the follicle cells surrounding the oocytes, express any detectable level of Zfp57 transcripts.
Embryonic and neonatal lethality of Zfp57 mutants indicates distinct zygotic and maternal-zygotic effects
We generated a deleted null allele in the Zfp57 locus (see Supplemental Data and Supplemental Figure S2). From crosses between female and male heterozygous mice, only 7 out of 65 progeny mice (10.8%) at the time of weaning were homozygous mutants, which is less than half of the expected 25%. Likewise, from the crosses between female heterozygous mice and male homozygous mice, only half of the expected homozygotes were obtained (Supplemental Table S1). This loss of homozygous offspring deficient in zygotic, but not maternal Zfp57, mainly occurred at the perinatal or neonatal stage (Table 1). These data indicate that loss of the zygotic function of Zfp57 results in partial lethality (Supplemental Table S1).
Table 1. Embryonic and neonatal lethality of Zfp57 mutant progeny indicates distinct zygotic and maternal-zygotic effects.
Embryos of the different embryonic stages were dissected out from the pregnant female mice in the mixed genetic background. Similar results were obtained with the mice in the 129 Sv/Ev background (Li and Leder, data not shown). Live embryos were confirmed under the dissection microscope by the heartbeat and body movement of the embryos.
| Cross | Stage | % of dead progeny/total | % of dead -/- mutant/total -/- | Type of mutant | ||
|---|---|---|---|---|---|---|
| +/-(f) X +/-(m) | E18.5 | 4.7% (n=85) | 16.7% (n=18) | Zygotic | ||
| P1 pup | 9.8% (n=92)$ | 45% (n=20)$ | ||||
|
| ||||||
| +/-(f) X -/-(m) | E18.5 | 0% (n=32) | 0% (n=12) | Zygotic | ||
| P1 pup | 20.2% (n=84)$ | 39.5% (n=43)$ | ||||
|
| ||||||
| -/-(f) X +/+(m)a | E18.5 | 0% (n=16) | NA | NA | ||
| P1 pup | 0% (n=22) | NA | ||||
|
| ||||||
| +/+(f) X -/-(m)b | E18.5 | 3% (n=33) | NA | NA | ||
| P1 pup | 0% (n=12) | NA | ||||
|
| ||||||
| -/-(f) X +/-(m) | E17.5-18.5 | 34.8% (n=23) | 80% (n=10) | Mat-Zyg | ||
| E11.5-E13.5 | 9.8% (n=34) | 38.1% (n=21) | ||||
|
| ||||||
| -/-(f) X -/-(m) | E17.5-19.5 | 87.5% (n=8) | 87.5% (n=8) | Mat-Zyg | ||
| E14.5-E16.5 | 81.4% (n=59) | 81.4% (n=59) | ||||
| E11.5-E13.5 | 43.4% (n=83) | 43.4% (n=83) | ||||
| E9.5-E10.5 | 7.9% (n=38) | 7.9% (n=38) | ||||
- +/-(f), heterozygous female mouse.
- +/-(m), heterozygous male mouse.
- -/-(f), homozygous mutant female mouse.
- -/-(m), homozygous mutant male mouse.
- +/+(f), wild-type female mouse.
- +/+(m), wild-type male mouse.
- NA, not applicable.
- Mat-Zyg, maternal-zygotic mutant.
total dead P0 or P1 pups.
all progeny from this cross are heterozygous and they lost just the maternal function of Zfp57.
all progeny from this cross are heterozygous and they lost just the paternal function of Zfp57.
In contrast to the crosses generating zygotic mutants for Zfp57, mating homozygous females to heterozygous or homozygous males resulted in no homozygous animals surviving to weaning (Supplemental Table S1). Analysis of prenatal lethality in these crosses indicated that lethality of homozygous embryos commenced around mid-gestation and progressively increased such that by E14.5-E16.5 around 80% of null embryos derived from null female mice, were dead (Table 1). This earlier lethality of homozygotes from null female mice compared to those from heterozygous female mice indicates a maternal effect of Zfp57 on the survival of animals (Supplemental Figure S3). Crosses with oocytes derived from conditionally ablating Zfp57 in female mice using ZP3-driven Cre-recombinase resulted in the same outcome proving the maternal effect of the deletion (Supplemental Table S2). Interestingly, loss of only the maternal or the paternal function of Zfp57 did not cause either embryonic or neonatal lethality indicating zygotic rescue (Table 1).
Defective imprinting in Zfp57 maternal-zygotic mutants
Microarray analysis was conducted comparing transcript expression patterns between the live homozygous E11.5 embryos and heterozygous littermates in a 129 Sv/Ev background derived from the same homozygous (null) female mouse crossed to a heterozygous male. One of the genes most strongly affected was the imprinted Dlk1 gene. Data indicates that expression of Dlk1, a paternally expressed mRNA gene, is dramatically reduced (six-fold decrease) in the homozygous mutant embryos. By contrast, the adjacent Gtl2 gene, a non-coding RNA gene expressed from the maternally inherited chromosome, is expressed at a two-fold increased rate in homozygous mutant embryos. Down-regulation of Dlk1 transcripts was confirmed by Northern blot analysis (Figure 2A) and quantitative RT-PCR (Li and Leder, unpublished data). Dlk1 and Gtl2 are members of a cluster of imprinted genes that also include Rtl1 and Dio3 expressed from the paternally inherited chromosome (Lin et al., 2003). Both Rtl1 and Dio3 (which is over 800kb away from Dlk1) are down-regulated in the homozygous mutant embryo compared with control heterozygous littermates (Figure 2B and Figure 2C). Consistent with the micro-array data, the amount of Gtl2 transcripts in homozygous mutant embryos is about twice that in the heterozygous littermate control embryos (Figure 2D). Because all the active alleles of all the protein-coding genes become repressed on the paternally inherited chromosome, concurrent with an increase in the expression of the normally paternally repressed Gtl2, we deduced that genomic imprinting across the whole Dlk1-Dio3 imprinted domain may be perturbed in homozygous mutant embryos derived from a null female mouse. Like Dlk1-Dio3, the Igf2-H19 domain is regulated by an ICR that acquires methylation during spermatogenesis. Both microarray and Northern blot analysis indicated that expression at Igf2-H19 was not affected in Zfp57 mutants (data not shown).
Figure 2. Loss of Zfp57 affects expression of the genes in the Dlk1-Dio3 imprinted region.
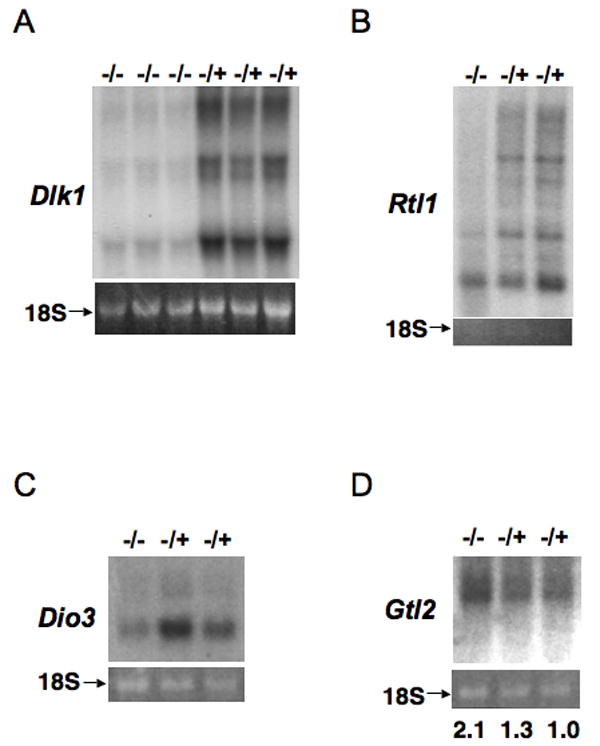
(A) Total RNA samples from three homozygous (-/-) and three littermate heterozygous (-/+) embryos derived from the same null mother were probed with Dlk1 cDNA in Northern blot.
(B), (C), (D) Equal amount of polyA RNA samples from a homozygous (-/-) and two littermate heterozygous (-/+) embryos derived from the same null mother were hybridized with a probe derived from Rtl1, Dio3 or Gtl2, respectively. Numbers in (D) indicate the band intensity of Gtl2 transcripts in each lane based on phosphor image analysis. Similar band intensity of 18S ribosomal RNA shown in the bottom panels of all figures indicates equal loading of the RNA samples.
All embryos are in the pure 129 Sv/Ev background.
Loss of Zfp57 affects the maintenance of DNA methylation imprints
Germline-derived differentially methylated regions (DMR) at imprinting control centers are required for genomic imprinting (Kobayashi et al., 2006; Spahn and Barlow, 2003). At the Dlk1-Dio3 imprinted domain the germline DMR resides in the intergenic region between Dlk1 and Gtl2 (IG-DMR). In the absence of the IG-DMR, imprinting is lost on the maternally inherited chromosome (Lin et al. 2003). Methylation of the IG-DMR on the paternal chromosome has been shown to be required for Dlk1 expression because, in the absence of the maintenance methyltransferase Dnmt1, Dlk1 becomes down-regulated and Gtl2 levels increase (Schmidt et al., 2000). The methylation status of the IG-DMR in Zfp57 mutants was therefore assessed. In the Zfp57 homozygous mutant embryos, DNA methylation at the IG-DMR is indeed affected when genomic DNA samples of different classes of embryos were analyzed (left panel of Figure 3A, Supplemental Figures S4A and S6A). Figure 3A illustrates relative differential DNA methylation levels quantified from Southern blots, an example of which is shown. Consistent with its requirement for Dlk1 activity, differential DNA methylation is completely absent at the IG-DMR in the homozygous mutant embryos derived from a null mother when both the maternal as well as the zygotic Zfp57 is eliminated (Lane 5 of Figure 3A, 0%). This indicates that methylation of the paternal chromosome cannot be maintained in the absence of both maternal and zygotic Zfp57 (maternal-zygotic effect). By contrast, loss of only the maternal function (maternal effect) in the heterozygous embryos derived from a null mother does not appear to cause any significant loss of differential DNA methylation at the IG-DMR (Lane 4 of Figure 3A, 90.9%) indicating that depletion of maternal Zfp57 alone is not sufficient to affect methylation maintenance. We also observed partial loss of differential DNA methylation in the homozygous mutant embryos derived from a heterozygous mother when only the zygotic function of Zfp57 is missing (zygotic effect, Lane 3 of Figure 3A, 67.5%). Differential methylation at the promoter of Gtl2 (Gtl2-DMR) which is regulated by the methylation at the IG-DMR after fertilization, was similarly affected (Supplementary Figure 4B). These results suggest that paternally derived germline methylation cannot be maintained in embryos lacking both maternal and zygotic Zfp57. On the other hand, providing this factor to a null zygote via the oocyte is sufficient to maintain at least some methylation at the domain.
Figure 3. Loss of Zfp57 affects differential DNA methylation at the Dlk1-Dio3 and the Snrpn imprinted regions.
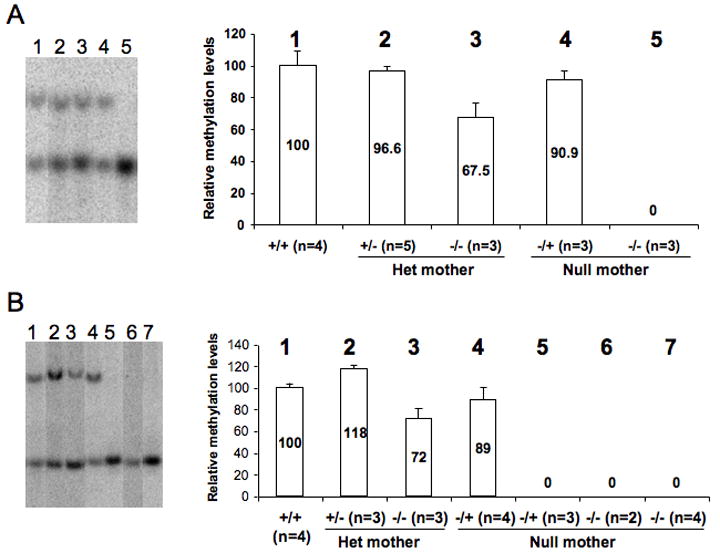
Differential DNA methylation was assessed with the methylation sensitive restriction enzyme HhaI. StuI-digested (Figure 3A) and PstI–digested (Figure 3B) genomic DNA samples from embryos were HhaI digested and hybridized with a probe derived from the differentially methylated regions using Southern blot analysis (left panel). Histograms represent relative differential methylation levels based on quantitative phosphor image analysis of the band intensity of the southern blots shown on the left. The numbers above each bar indicate the representative lane on the Southern blot. Numbers within the bars indicate percentage of differential DNA methylation relative to that of wild-type embryos. Zero indicates absence of methylation. Genotypes and numbers analyzed for each class of embryo are indicated along the horizontal axis. All embryos are E11.5-E13 on a 129 Sv/Ev background.
(A)The IG-DMR from the Dlk1-Dio3 region. Lane 1, wild-type embryos. Lane 2, heterozygous (+/-) embryos from a heterozygous (het) mother. Lane 3, homozygous (-/-) embryos from a het mother. Lane 4, heterozygous (-/+) embryos from a homozygous (null) mother. Lane 5, -/- embryos from a null mother.
(B) The Snrpn DMR. Lane 1, wild-type embryos. Lane 2, +/- embryos from a het mother. Lane 3, -/- embryos from a het mother. Lane 4 and lane 5, -/+ embryos from a null mother. Lane 6, -/- embryos from a null mother and a het father. Lane 7, -/- embryos from a null mother and a null father. This Snrpn Southern data was further confirmed by COBRA analysis on samples including a subset analyzed by Southern blotting. A total of 25 mid-gestation heterozygous embryos from null mothers were assessed with 14 exhibiting methylation acquisition and 11 remaining unmethylated (data not shown).
Other imprinted domains were surveyed to see if differential DNA methylation at ICRs was affected in the Zfp57 homozygous mutant embryos. Differential DNA methylation in the H19 DMR (Tremblay et al., 1997) is normal in the Zfp57 homozygous mutant embryos (Supplemental Figure S4C). Methylation at two families of repeat sequences (Line1 and IAP) was also unaffected (Supplemental Figure S5).
DMRs that acquire their methylation in the maternal germline were assessed. These include Snrpn, Peg1, Peg3, Igf2r and Nnat/Peg5 (Lucifero et al., 2002). Similar to the finding in the Dlk1-Dio3 imprinted region, differential DNA methylation was lost at the Snrpn, Peg1, Peg3 and Peg5/Nnat DMRs in all homozygous mutant embryos (maternal-zygotic) derived from a null mother (Figure 3B and Supplemental Figure S6, and data not shown). Partial loss of DNA methylation at the DMRs was observed when only zygotic Zfp57 is missing and the extent of this varied between individuals (Supplemental Figure S6). Differential DNA methylation is intact in the heterozygous embryos derived from a heterozygous mother. Methylation imprints at the Igf2r imprinting control region were generally not affected in the homozygous embryos except in one embryo lacking both maternal and zygotic Zfp57 (Supplemental Figure S6D).
These findings indicate that, in addition to failing to maintain imprints at Dlk1-Dio3, loss of both maternal and zygotic Zfp57 results in failure to maintain methylation imprints at multiple maternally methylated DMRs. Zygotic Zfp57 is able to rescue loss of maternal Zfp57 to allow imprint maintenance. Absence of zygotic Zfp57 can sometimes compromise maintenance of methylation conferred by maternal Zfp57. Therefore, for effective maintenance of methylation, both maternal and zygotic Zfp57 are required.
Zfp57 is required for the establishment of maternal methylation imprints at Snrpn
Given the absence of methylation at multiple maternally methylated ICRs, it was important to determine whether maternal germline methylation marks were appropriately established. Using both COBRA analysis and bisulphite sequencing, we determined that the Snrpn DMR is not methylated in oocytes derived from homozygous female mice (Figure 4B and Supplemental Figures S7 and S8). In contrast, the Snrpn DMR showed normal methylation in control oocytes from heterozygous females (Figure 4A, Supplemental Figures S7 and S8). Thus, Zfp57 is required for the establishment of the maternal methylation imprint at Snrpn in the female germline.
Figure 4. Differential methylation at the Snrpn DMR is not established in absence of the maternal Zfp57 and can be acquired in the presence of the zygotic Zfp57.
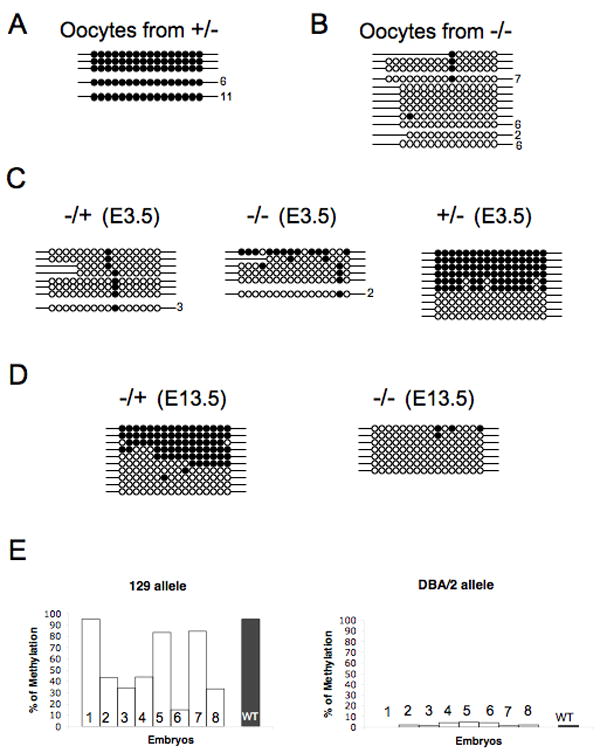
Genomic DNA samples from oocytes or embryos in the 129 Sv/Ev background were subjected to bisulphite sequencing. A total of 16 differentially methylated CpG sites at the Snrpn DMR are shown for Figures A-D. Filled oval, methylated CpG site. Open oval, unmethylated CpG sites. Line with ovals, a unique clone. A unique clone was assigned based on unconverted C residues in the sequences. The number of sequences is shown for a non-unique clone. The high conversion rate fails to distinguish whether these non-unique sequences represent clonal or individual products.
A, Two different pools of unfertilized oocytes isolated from heterozygous female mice.
B, Two different pools of unfertilized oocytes isolated from homozygous female mice.
C, pooled E3.5 embryos. -/+ (E3.5), 29 heterozygous E3.5 embryos from the crosses between homozygous female mice and wild-type male mice. -/- (E3.5), 10 homozygous E3.5 embryos from the crosses between homozygous female mice and homozygous male mice. +/- (E3.5), 16 heterozygous E3.5 embryos from the crosses between wild-type female mice and homozygous male mice. D, genomic DNA samples were made from littermate E13.5 embryos from the cross between a homozygous female mouse and a heterozygous male mouse.
E, Acquisition of DNA methylation imprint occurred on the maternally derived Snrpn DMR. Histograms are shown for the levels of methylation at the CpG sites of this DMR in E11.5-E13 embryos from the cross between a homozygous female mouse in the 129 Sv/Ev background and a wild-type male mouse in the DBA/2 background. Unfilled bars, eight littermate heterozygous embryos (1- 8) from this cross. Filled bar, seven littermate wild-type (WT) embryos from the cross between a wild-type 129 female mouse and a wild-type DBA/2 male mouse. Vertical axis, percentage of methylated CpG sites analyzed. Left panel, maternally derived 129 allele. Right panel, paternally derived DBA/2 allele.
Germline methylation marks were assessed at four other maternally methylated imprinting control regions (Supplemental Figure S7). Unlike Snrpn, germline methylation was found to occur normally at the DMR regions of Peg1, Peg3 and Nnat/Peg5 in unfertilized oocytes derived from either homozygous or heterozygous female mice. Methylation imprints appear to be partially affected at Igf2r in the oocytes produced by homozygous or heterozygous female mice though the significance of this is not clear.
Assessment of paternal germline methylation at the IG-DMR and Igf2-H19 imprinting control regions was conducted in DNA isolated from sperm purified from heterozygous and homozygous mutant males. All samples exhibited normal methylation marks for both regions (Supplemental Figures S9) indicating that Zfp57 does not affect the establishment of these paternal germline methylation imprints. Together, these data suggest that maternal Zfp57 is required for the establishment of the maternal methylation imprint at the Snrpn imprinted region, but is dispensable for Peg1, Peg3 and Nnat/Peg5 imprinting in the female germline. Consistent with the genetic outcome indicating a maternal-zygotic effect for Zfp57, Zfp57 is not required for the establishment of paternal germline methylation. The data also indicate that Zfp57 is not a general maternal germline imprinting factor.
Post-implantation acquisition of differential methylation at the Snrpn DMR
Homozygous embryos generated from homozygous females are not methylated at the Snrpn DMR. This absence of methylation is heritable at preimplantation stages as both heterozygous (-/+) and homozygous (-/-) E3.5 embryos derived null females are unmethylated at the Snrpn DMR (Figure 4C and Supplemental Figure S8C). As expected, the Snrpn DMR was methylated in heterozygous (+/-) E3.5 embryos derived from wild-type female mice (Figure 4C). Surprisingly, however, methylation was evident at Snrpn in approximately half of the heterozygous mid-gestational embryos derived from homozygous female mice (Figures 3B, 4D and 4E). These findings suggest that the acquisition of methylation at the Snrpn DMR in heterozygous mid-gestational embryos derived from null female mice requires zygotic Zfp57 and occurs after E3.5.
Inter-species crosses were carried out to determine whether post-fertilization DNA methylation at Snrpn was acquired specifically on the maternally inherited chromosomes or at random on both maternal and paternal chromosomes. Homozygous female mice (129 Sv/Ev) were mated with wild-type male mice (DBA/2). Polymorphisms exist between these two strains at the Snrpn DMR ((Hiura et al., 2006), Supplemental Figure S10). Based on the bisulphite sequencing results of eight heterozygous embryos from this cross (Figure 4E), methylation was only observed at the 129 Sv/Ev Snrpn DMR, i.e. the maternally derived differentially methylated region, but not at the paternally derived DBA/2-specific region (see Supplemental Figure S11 for detailed information). As expected, differential methylation was maintained on the maternally derived Snrpn DMR in the control cross between 129 Sv/Ev female and DBA/2 male wild-type mice (Supplemental Figure S12).
Endogenous ZFP57 binds to the Snrpn DMR
Affinity purified rabbit polyclonal antibodies against the N-terminal half of ZFP57 protein were generated. These purified antibodies specifically detect exogenous ZFP57 expressed in COS cells and endogenous ZFP57 present in wild-type ES cells. Immunoreactivity is absent in null ES cells (Figure 5A).
Figure 5. Endogenous ZFP57 can bind to the Snrpn DMR region.
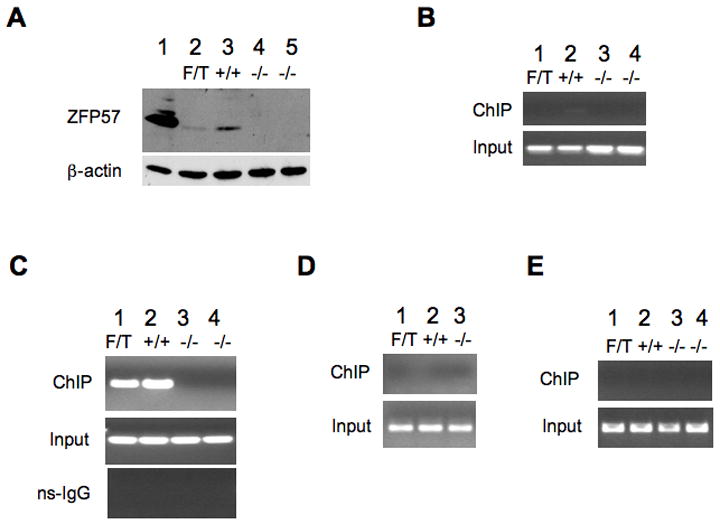
A, Western blot analysis of affinity purified anti-ZFP57 polyclonal antibodies with total cell lysate samples. Lane 1, ZFP57 was over-expressed in COS cells. Lane 2, an ES clone containing one floxed allele (F) and one targeted allele (T) at the Zfp57 locus. Lane 3, wild-type TC1 ES cells. Lanes 4 and 5, two independent Zfp57-null ES clones carrying two deleted alleles. Please see Supplemental Figure S2 and Supplemental Data for the description of the targeted, floxed and deleted alleles.
(B-E) Chromatin immunoprecipitation (ChIP) was performed with approximately one million ES cells. No ChIP PCR product was observed in the negative control samples without the addition of any antibodies (data not shown).
B, the first round PCR product at the Snrpn DMR region.
C, the second round PCR product at the Snrpn DMR region.
D, the second round PCR product at a region about 40kb upstream of the Snrpn DMR region.
E, the second round PCR product at the H19 DMR region.
ChIP, ChIP PCR product. Input, PCR product from the input/starting samples; ns-IgG, ChIP PCR product from the control samples when rabbit non-specific IgG (ns-IgG) antibodies were added during immunoprecipitation. (B-E) Lane 1, an ES clone containing one floxed allele (F) and one targeted allele (T) at the Zfp57 locus. Lane 2, wild-type TC1 ES cells. Lanes 3 and 4, two independent Zfp57-null ES clones carrying two deleted alleles.
To determine whether Zfp57 directly interacts with the Snrpn DMR, chromatin immunoprecipitation (ChIP) was performed to determine whether ZFP57 binds this DMR. Mouse ES cells were used as a model system because they display high de novo methylation activities (Humpherys et al., 2001; Lei et al., 1996; Ooi et al., 2007). Furthermore, trans-acting factors that bind to the control elements within the Snrpn DMR are present in undifferentiated ES cells (Kantor et al., 2004). Analysis of DNA purified after ChIP revealed binding of endogenous ZFP57 to the Snrpn DMR in wild type ES cells and in the ES cells containing one floxed allele and one targeted allele at the Zfp57 locus (Lanes 1 and 2 of Figure 5C) but not to the Snrpn DMR of ES cell clones containing two deleted alleles of Zfp57 (Lanes 3 and 4 of Figure 5C). These experiments which were reproducible for three independent immunoprecipitations indicate that ZFP57 can directly bind to the Snrpn DMR, whereas the control ChIP experiments involving the unaffected H19 DMR or a distant upstream region of the Snrpn DMR did not show any binding activity (Figures 5D and 5E).
Maternal ZFP57 is present in pre-implantation embryos
Embryos isolated from several different crosses were subjected to immunofluorescence staining. As shown in Figures 6A and 6D, there is no detectable ZFP57 present in the homozygous E1.5 or E3.5 embryos derived from a null mother. However, in the embryos derived from the cross between heterozygous female mice and homozygous male mice, ZFP57 was detected in the homozygous E1.5 and E3.5 embryos (Figures 6B and 6G) as well as in the littermate heterozygous E1.5 and E3.5 embryos (Figures 6C and 6F). These data suggest that the maternal gene product of Zfp57 is carried over from oocytes and deposited in early embryos, as illustrated in Supplemental Figure S3. Zygotic ZFP57 was detectable in the heterozygous E3.5 embryos from the cross between homozygous female mice and wild-type male mice (Figure 6H).
Figure 6. Maternal ZFP57 is present in pre-implantation embryos.
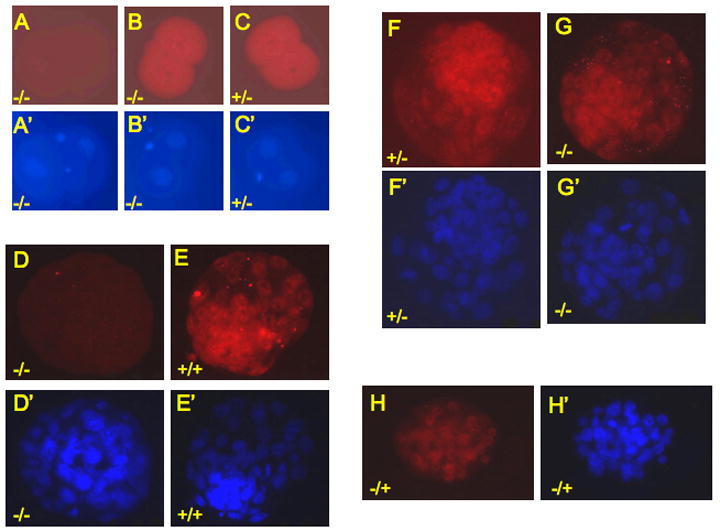
Pre-implantation embryos were stained with affinity-purified polyclonal antibodies against ZFP57 (Figures A-H). They were also incubated with Hoechst dye to illuminate genomic DNA (Figures A’-H’).
A homozygous E1.5 embryo (A or A’) and a homozygous E3.5 embryo (D or D’) were derived from the cross between homozygous female mice and homozygous male mice. Littermate E1.5 embryos (B and C) and E3.5 embryos (F and G) were derived from the crosses between heterozygous female mice and homozygous male mice.
B (or G), a homozygote (-/-).
C (or F), a heterozygote (+/-).
The wild-type E3.5 embryo (E) was derived from the cross between a wild-type female mouse and a wild-type male mouse. The heterozygous (-/+) E3.5 embryo (H) was from the cross between a homozygous female mouse and a wild-type male mouse.
Discussion
Mechanisms of establishment and maintenance of germline-derived methylation imprints are not fully understood. In a screen for genes down-regulated upon ES cell differentiation, we identified a maternal-zygotic effect gene, Zfp57, that contributes to the stable maintenance of methylation imprints during development.
Consistent with its expression in the female germline, Zfp57 is required for the establishment of the germline methylation imprint at the Snrpn imprinted region. More detailed biochemical analysis needs to be undertaken to determine whether ZFP57 is directly involved in the acquisition or maintenance of this methylation in the female germline. In contrast, maternal Zfp57 is not essential for the establishment of the genomic imprints at other examined maternally methylated regions (Peg1, Peg3 and Nnat/Peg5) in oocytes, or in the male germline. Consistent with the maternal-zygotic effect evident from the genetic data, a role for Zfp57 in imprinting establishment appears to be specific to the female germline.
In contrast, maternal Zfp57 plays a much broader role in the maintenance of both paternal and maternal methylation imprints in embryos. Genomic imprints are not maintained at the paternally methylated IG-DMR of the Dlk1-Dio3 domain and at maternally methylated Peg1, Peg3 and Nnat/Peg5 in the absence of both maternal and zygotic functions of Zfp57 in embryos. These data indicate that ZFP57 plays a role in the post-fertilization maintenance of genomic imprints at a large subset of the imprinted regions. Indeed, it was just published online that zygotic mutations in human Zfp57 also affect differential DNA methylation at a subset of imprinted regions (Mackay et al., 2008). Variable effects of this mutation in different individuals are consistent with the defects we observe in the zygotic mutant mice. Another maternal effect gene, Stella/PGC7, appears to play a partially protective role in the maintenance of the other two paternally-derived methylation imprints at H19 and Rasgrf1, and three maternally-derived methylation imprints at Peg1, Peg3 and Peg10 (Nakamura et al., 2007). In contrast to Zfp57, Stella/PGC7 does not affect Dlk1-Dio3, Snrpn and Peg5 or play a role in the germline establishment of imprints. The overlapping effect of Zfp57 and Stella at Peg1 and Peg3 suggests that their maintenance functions may not be mutually exclusive.
Interestingly, differential DNA methylation at the Snrpn DMR was established in the presence of the zygotic Zfp57 in post-implantation embryos even though germline methylation was either not established or lost in oocytes and remained absent at E3.5 (Supplemental Figure S13). Since ZFP57 can bind to the Snrpn DMR in ES cells, it is likely that ZFP57 can bind to the unmethylated Snrpn DMR directly during methylation acquisition in embryos. This acquisition of methylation only occurs at the maternally derived Snrpn DMR. This suggests that there may be a DNA methylation-independent genomic imprint that can act as imprinting memory at the Snrpn imprinting control region. Previously it has been shown that the imprinting of some placental genes occurs in the absence of maintenance methylation, likely involving repressive histone modifications (Lewis et al., 2004; Umlauf et al., 2004). It will be interesting to determine whether repressive histone modifications play a role in the heritable methylation-independent germline mark at Snrpn. Future experiments will address the many intriguing questions arising from this finding including the nature of the heritable memory and whether there is a bias for the grandmaternal over the grandpaternal maternal allele. Since some heterozygous embryos derived from null female mice had no or only partial DNA methylation around midgestation whereas others displayed a fully methylated maternal allele, it is possible that differential methylation at the Snrpn DMR in the presence of the zygotic Zfp57 occurs over time in embryos and all progeny from this cross may acquire full differential methylation at the maternal allele in late stages of development.
The effects of ZFP57 on imprint methylation are consistent with the lethality of embryos described for the range of genetic crosses: (1) Loss of just zygotic Zfp57 causes partial neonatal lethality as well as partial loss of differential DNA methylation; (2) loss of only the maternal Zfp57 does not appear to cause any lethality or any loss of differential DNA methylation due to the rescue of the maternal effect by zygotic Zfp57; (3) eliminating both the maternal as well as zygotic Zfp57 results in complete loss of differential DNA methylation as well as a highly penetrant embryonic lethality around mid-gestation. This lethality is consistent with phenotypes expected from the cumulative effects of loss of imprinting of a subset of imprinted genes although Zfp57 may also play a role at some unidentified non-imprinted loci.
How might this KRAB zinc finger protein be influencing the epigenetic state of imprinted domains? Interestingly, it was recently reported that the KRAB domain can trigger heritable de novo DNA methylation when targeted to a reporter transgene specifically in early mouse embryos (Wiznerowicz et al., 2007). This is consistent with an endogenous role for the KRAB zinc finger protein ZFP57 in mediating DNA methylation at some imprinted domains. We therefore suggest a model in which ZFP57 might be directly involved in targeting methylation to certain imprinting control regions; in the female germline at Snrpn and at several imprinted ICRs after fertilization. This implies that the maintenance of imprints during early development is a regulated event and more than a mere protection against active and passive demethylation. It is possible that ZFP57 can also bind to the methylated Snrpn DMR and other imprinted regions. Future experiments focusing on the functions of ZFP57 and associated complexes may elucidate a novel pathway of epigenetic control involving zinc-finger proteins and lead to a better understanding of the underlying mechanisms of the establishment and maintenance of DNA methylation.
Experimental Procedures
Targeting at the Zfp57 locus
A genomic fragment of Zfp57 was used to make the targeting construct (see Supplemental Data). The linearized construct was electroporated into ES cells and candidate targeted ES clones following G-418 drug selection were screened by Southern blot. A confirmed targeted ES clone was used to generate germline-transmissible chimera mice and corresponding floxed and deleted alleles at the Zfp57 locus were obtained.
Bisulphite mutagenesis and PCR primers
The protocol for bisulphite mutagenesis is described in Arnaud et al., 2003. PCR primers used for the Snrpn DMR and the IG-DMR are the same as those used in Lucifero et al., 2002. For other primers, please see the Supplemental Data. Prior to cloning for bisulphite sequencing, sample aliquots were tested by combined bisulphite restriction analysis (COBRA) for later comparison with sequenced products to rule out cloning bias. For the oocyte-specific Snrpn bisulphite analysis, two biological replicates on pools of oocytes derived from heterozygote (n=311 from 12 females and n=174 from 8 females) and homozygote females (n=312 from 7 females and n=185 from 8 females) were subjected to both bisulphite sequencing and COBRA analysis. For E3.5 embryos numbers are indicated in the legend to Figure 4 with additional pooled embryos assessed by COBRA analysis as described in Figure S8.
Methylation-sensitive Southern blot analysis
DNA was isolated, double digested, blotted and hybridized as described in (Takada et al., 2002), including probes for the Gtl2 promoter and IG-DMR. For the probe of the Snrpn DMR, please see the Supplemental Data. For Southern blotting, a total of 17 heterozygotes and 13 homozygotes from heterozygous female mice plus 16 heterozygotes and 16 homozygotes from null female mice were generated. DNA was used for multiple probe hybridizations with sample overlap between the Gtl2-DMR and IG-DMR, and Snrpn and H19 DMRs. In addition there was overlap between samples used for Southern blot analysis and for COBRA analysis. For example, of 7 heterozygotes from null female mice analyzed for the Snrpn DMR by Southern blotting, 4 plus an additional 18 were assessed by COBRA. All genotype-specific methylation analysis by Southern blotting was confirmed by COBRA analysis though not necessarily on all the same samples. Genotype-specific methylation was consistent for each genotype using these two different approaches.
Northern blot analysis
The polyA+ Northern blot used in Figure 1D was generated with RNA isolated from adult organs and used previously (Chester et al., 1998) and repeated with fresh tissues which gave the same results (not shown). Either total RNA or polyA+ RNA derived from midgestation embryos were used for Northern blots on imprinted genes shown in Figure 2. Probes were derived from the corresponding cDNAs.
Generation and purification of anti-ZFP57 antibodies and immunostaining
A portion of Zfp57 cDNA encoding the N-terminal portion of ZFP57 protein (Met1 to Arg 240) was amplified by PCR and cloned into the NcoI and BamHI sites of the expression vector pQE60 (Qiagen). Six Histidine-tagged protein was induced by the addition of IPTG to the transfected bacterial clones and purified under denaturing conditions according to the manufacturer’s manual. The purified denatured protein was directly injected into the rabbits and anti-ZFP57 sera were generated with a standard protocol (Cocalico Biologicals, Inc., USA). Rabbit anti-ZFP57 polyclonal antibodies were purified with the Histidine-tagged ZFP57 recombinant protein attached to a Nickel-NTA affinity column (Gu et al., 1994). After elution with 4M MgCl2, the purified polyclonal antibodies were dialyzed against water for one hour and then against Tris-buffered saline solution (25mM Tris, pH 8.0) exhaustively at 4°C.
Immunofluorescence staining in embryos
Embryos were harvested from superovulated female mice and fixed in 4% paraformaldehyde. They were stained with the purified anti-ZFP57 antibodies following a previously published protocol (Payer et al., 2003).
Co-immunoprecipitation
Mouse monoclonal antibodies against the Myc epitope were used to immunoprecipitate the exogenously transfected Myc epitope-tagged ZFP57 in COS cells. Purified rabbit polyclonal antibodies against ZFP57 were used to pull down the endogenous ZFP57 in ES cells.
Chromatin immunoprecipitation (ChIP)
ChIP was conducted according to the protocol recommended by Upstate (USA). For PCR amplification of immunoprecipitated DNA, please see PCR primers in the Supplemental Data.
Supplementary Material
Acknowledgments
The authors want to thank Montserrat Michelman for help with ES cells. We are grateful to Lina Du and Arlene Sharpe at Brigham and Women’s Hospital transgenic mouse facility for generating the Zfp57 conditional knockout mouse. Special thanks go to Morgan Fleishman for the help with isolation of oocytes and early embryos. We also appreciate the generosity of William J. Fredericks and Frank J. Rauscher III at the Wistar Institute for the anti-KAP-1 antibodies and the KAP-1 expression construct, Nicholas Chester for the polyA mouse organ blot and Boris Reizis for reagents. We thank Alex Bishop, Nicholas Chester and Holger Babbe for the comments on the manuscript and members of the Ferguson-Smith lab for helpful discussions. X. Li was a postdoctoral fellow of the Helen Hay Whitney Foundation and M. Ito and N Youngson’s experiments were supported by the BBSRC, NIH and MRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrink M, Ortiz JA, Mark C, Sanchez C, Looman C, Hellman L, Chambon P, Losson R. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A. 2001;98:1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JI, Lee KH, Shin DM, Shim JW, Lee JS, Chang SY, Lee YS, Brownstein MJ, Lee SH. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev Biol. 2004;265:491–501. doi: 10.1016/j.ydbio.2003.09.041. [DOI] [PubMed] [Google Scholar]
- Akagi T, Usuda M, Matsuda T, Ko MS, Niwa H, Asano M, Koide H, Yokota T. Identification of Zfp-57 as a downstream molecule of STAT3 and Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun. 2005;331:23–30. doi: 10.1016/j.bbrc.2005.03.118. [DOI] [PubMed] [Google Scholar]
- Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, Peters J, Craigen W, Preece M, Stanier P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Human molecular genetics. 2003;12:1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes & development. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Cedar H. Imprinting: focusing on the center. Curr Opin Genet Dev. 2000;10:550–554. doi: 10.1016/s0959-437x(00)00126-x. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Chester N, Kuo F, Kozak C, O’Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom’s syndrome gene. Genes & development. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Larrain J, Oelgeschlager M, Wessely O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat Rev Genet. 2000;1:171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & development. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gosden RG. Oogenesis as a foundation for embryogenesis. Mol Cell Endocrinol. 2002;186:149–153. doi: 10.1016/s0303-7207(01)00683-9. [DOI] [PubMed] [Google Scholar]
- Gu J, Stephenson CG, Iadarola MJ. Recombinant proteins attached to a nickel-NTA column: use in affinity purification of antibodies. Biotechniques. 1994;17:257, 260–262. [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes & development. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kantor B, Makedonski K, Green-Finberg Y, Shemer R, Razin A. Control elements within the PWS/AS imprinting box and their function in the imprinting process. Human molecular genetics. 2004;13:751–762. doi: 10.1093/hmg/ddh085. [DOI] [PubMed] [Google Scholar]
- Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, Ferguson-Smith AC, Kono T. High-frequency generation of viable mice from engineered bi-maternal embryos. Nature biotechnology. 2007;25:1045–1050. doi: 10.1038/nbt1331. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- Krebs CJ, Larkins LK, Price R, Tullis KM, Miller RD, Robins DM. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes & development. 2003;17:2664–2674. doi: 10.1101/gad.1135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nature genetics. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li X, Leder P. Identifying genes preferentially expressed in undifferentiated embryonic stem cells. BMC Cell Biol. 2007;8:37. doi: 10.1186/1471-2121-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nature genetics. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Looman C, Abrink M, Mark C, Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nature genetics. 2008 doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Melendez A, Greenwald I. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics. 2000;155:1127–1137. doi: 10.1093/genetics/155.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Human molecular genetics. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Newman-Smith ED, Rothman JH. The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr Opin Genet Dev. 1998;8:472–480. doi: 10.1016/s0959-437x(98)80120-2. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C. Determination of the embryonic axes of Drosophila. Development (Cambridge, England) 1991;1:1–10. [PubMed] [Google Scholar]
- Okazaki S, Tanase S, Choudhury BK, Setoyama K, Miura R, Ogawa M, Setoyama C. A novel nuclear protein with zinc fingers down-regulated during early mammalian cell differentiation. J Biol Chem. 1994;269:6900–6907. [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Schier AF. Axis formation and patterning in zebrafish. Curr Opin Genet Dev. 2001;11:393–404. doi: 10.1016/s0959-437x(00)00209-4. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes & development. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes & development. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn L, Barlow DP. An ICE pattern crystallizes. Nature genetics. 2003;35:11–12. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, Ferguson-Smith AC. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Human molecular genetics. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Bartolomei MS. SnapShot: imprinted gene clusters. Cell. 2007;130:958. doi: 10.1016/j.cell.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS. A 5’ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nature genetics. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated Box Repressor Domain Can Trigger de Novo Promoter Methylation during Mouse Early Embryogenesis. J Biol Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


