Abstract
Posttranslational modification of histones by biotinylation can be catalyzed by both biotinidase (BTD) and holocarboxylase synthetase (HCS). Biotinylation of histones is an important epigenetic mechanism to regulate gene expression, DNA repair, and chromatin remodeling. The role of BTD in histone biotinylation is somewhat ambiguous, given that BTD also catalyzes removal of the biotin tag from histones. Here, we sought to develop BTD inhibitors for future studies of the role of BTD in altering chromatin structure. We adopted an existing colorimetric BTD assay for use in a novel 96-well plate format to permit high-throughput screening of potential inhibitors. Biotin analogs were chemically synthesized and tested for their ability to inhibit human BTD. Seven of these compounds inhibited BTD by 26% to 80%. Biotinyl-methyl 4-(amidomethyl) benzoate had the largest effect on BTD, causing an 80% inhibition at 1 mM concentration. Enzyme kinetics studies were conducted to determine Vmax, Km, and Ki for the seven inhibitors; kinetics were consistent with the hypothesis that biotinyl-methyl 4-(amidomethyl) benzoate and the other compounds acted by competitive inhibition of BTD. Finally, biotinyl-methyl 4-(amidomethyl) benzoate did not affect biotin transport in human cells, suggesting specificity in regard to biotin-related processes.
Keywords: biotinidase, biotinyl-methyl 4-amidomethyl benzoate, histones, inhibitors
1. Introduction
Mammals contain two biotin-metabolizing enzymes, namely holocarboxylase synthetase (HCS) and biotinidase (BTD). HCS is a cytoplasmic, mitochondrial, and nuclear protein [1–3] whereas BTD is predominantly secreted into serum [4,5] but can also be found in the cell nucleus [2,4]. HCS catalyzes the covalent binding of biotin to a specific lysine residue in apocarboxylases to produce holocarboxylases [6]. The classical function of BTD is to recycle biotin by hydrolyzing the amide bond between biotin and lysine residues (biocytin) in breakdown products of holocarboxylases [7].
Recently it was proposed that both BTD and HCS also catalyze the biotinylation of ε-amino groups in specific lysine residues in histones [1,8–11]. Posttranslational modifications of histones have distinct functions in chromatin metabolism. For example, trimethylation of K4 in histone H3 is associated with transcriptional activation of genes, whereas dimethylation of K9 in histone H3 is associated with transcriptional silencing [12,13].
All five major classes of histones are targets for biotinylation in human cells [10]. While the physiological roles of histone biotinylation are under investigation, there is increasing evidence that histone biotinylation plays a role in DNA repair [14], and gene silencing and heterochromatin structures in human cells [15], and stress resistance and lifespan in Drosophila [16,17]. Biotinylation of histones is mediated by both HCS [1,16] and BTD [8], but evidence has been provided that HCS is the dominant histone-biotinyl ligase [16].
Biotinylation of histones is a reversible modification. Ballard et al. suggested that debiotinylation of histones might be mediated by BTD [18]. The regulation of BTD to favor debiotinylation of histones over biotinylation of histones by the same enzyme is unknown. A number of variables may regulate the catalytic activity of BTD. First, the availability of substrate might favor either biotinylation or debiotinylation of histones. For example, locally high concentrations of biocytin might shift the reaction equilibrium towards biotinylation of histones [8,19]. Second, proteins may interact with BTD at the chromatin level, favoring either biotinylation or debiotinylation of histones. Third, three alternatively spliced variants of BTD have been identified [20]. Theoretically, these variants may have unique functions with regard to histone biotinylation. Fourth, BTD possesses six glycosylation sites [21,22]; glycosylation of BTD might affect its cellular location [23].
Our long-term goal is to identify the roles of BTD in biotinylation and debiotinylation of histones. As a first step towards this goal, we generated a first generation of synthetic inhibitors of BTD, and we developed a 96-well plate assay for high-throughput screening of putative BTD inhibitors.
Previous studies have proposed using biotin, di-isopropylfluorophosphate, and thiol reagents such as p-chloromercuribenzoate as inhibitors of BTD [4]. These compounds are of limited use for the following two reasons. First, di-isopropylfluorophosphate and p-chloromercuribenzoate are general inhibitors of enzymes as opposed to being specific inhibitors of BTD. Second, simultaneous inhibition of BTD in both cytoplasm and nucleus make it impossible to link potential effects of low BTD activity to altered histone debiotinylation in the nucleus as opposed to impaired biotin recycling in the cytoplasm. The studies presented here lay the groundwork for the development of second-generation inhibitors that can be targeted directly to the nucleus to interfere with histone debiotinylation without affecting cytoplasmic events.
2. Materials and methods
2.1. BTD purification
Human plasma, which is known to contain high levels of BTD activity [4], served as a source of BTD in this study. Approximately 150 mL of human blood was collected through venipuncture into heparinized containers. This study was approved by the Institutional Review Board for Human Subjects at the University of Nebraska-Lincoln. Plasma was separated from blood cells by centrifugation [870 g for 30 min]. Previous studies indicated that BTD precipitates between 30% to 50% saturation with ammonium sulfate [24]. Thus, for all experiments described here plasma BTD was precipitated using 30% to 50% saturation with ammonium sulfate. Precipitated proteins, usually from 10 mL of plasma were re-suspended in 1.5 mL of potassium phosphate buffer (0.1 M, pH 6) and dialyzed for 24 to 36 hours against three changes of 1 L of potassium phosphate buffer at 4°C. Dialyzed samples were subdivided into 1-mL aliquots and stored at −20°C. Please note that these crude preparations of BTD might contain proteins that affect analysis of Vmax, Km, and Ki. Individual aliquots were thawed and used as needed for BTD assays as described below. The amount of protein was quantified by using the Nanodrop 1000 spectrophotometer (Wilmington, DE). Plasma BTD activity is known to vary by about 25% among healthy individuals [25], explaining the sample-to-sample variation described below.
2.2. BTD inhibitors
Putative inhibitors of BTD were synthesized by conjugating biotin to various allylamines using carbodiimide as the coupling reagent. Briefly, equimolar amounts (0.21 mmol) of d-biotin (50 mg), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (39 mg), 4-(dimethylamino)pyridine (25 mg), allylamine derivatives (Table 1) were dissolved in 5 mL of N,N-dimethylformamide in an 8-mL vial. The solution was stirred under nitrogen atmosphere overnight. After the reaction was complete [checked by thin-layer chromatography and stained with iodine or phosphomolybdic acid solution], DMF was evaporated under vacuum. The crude product was column chromatographed on silica gel (Silica gel 60, 40–63 µm, EMD Chemical Inc.; Gibbstown, NJ). The column was first washed with 50 mL dichloromethane and the product eluted with 150 mL dichloromethane:methanol (10:1, by volume). Solvents were removed by vacuum drying to obtain a white solid powder product at a 40% to 90% yield. 1H and 13NMR was used to verify the identity of the product.
Table 1.
Inhibitors of BTD
| Inhibitors | % inhibitiona | Structure |
|---|---|---|
| Biotinyl anilide | 55±0.1 | 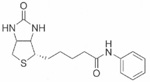 |
| Biotinyl allylamide | 37±0.09 | 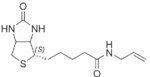 |
| Biotinyl N-methylanilide | 26±0.05 | 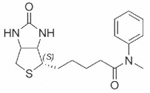 |
| Biotinyl methyl 4-(amidomethyl) benzoate | 80±0.02 | 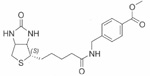 |
| Biotinyl 2-amido-pyridine | 40±0.05 | 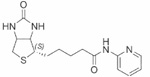 |
| Biotinyl 4-amidophenylboronic acid | 53±0.02 | 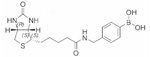 |
| Biotinyl benzylamide | 47±0.01 | 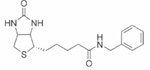 |
Percent inhibition compared with inhibitor-free control after 2 hours of incubation of BTD + inhibitor + substrate. Values are means ± SD (n = 6; P < 0.05 compared with inhibitor-free control).
2.3. BTD assay
BTD activity was measured as the rate of hydrolysis of N-biotinyl-4-aminobenzoic acid to release 4-aminobenzoic acid (PABA). The latter was quantified using N-1-naphthylethylenediamine dihydrochloride as described by Knappe et al. [26] and Backman-Gullers et al. [27], and modified by Nilsson & Ronge [28]. These protocols were adapted for microtiter plates as follows. In a 96-well microtiter plate, 500 µg of partially purified BTD from plasma and 12 µL of 10 mM putative BTD inhibitors (120 nmoles/well; 1.0 mM final concentration) were mixed with 88 µL of 54 mM sodium phosphate buffer (pH 6.0), containing 1.08 mM disodium EDTA and 4.3 mM cysteamine hydrochloride (prepared fresh); samples were preincubated at 37°C for 60 min. Then, 10 µL of 6mM N-(+)-biotinyl-PABA (60 nmoles/well; 0.5 mM final concentration) were added to each well and plates were incubated for 120 min at 37°C. Reactions were stopped by adding 30 µL of 6 M hydrochloric acid and the following compounds were added at 3-min intervals at room temperature: 55 µL water, 15 µL of 14.5 mM sodium nitrite (prepared fresh), 15 µL of 43.8 mM ammonium sulfamate, and 15 µL of 3.86 mM N-1-naphtylethylenediamine dihydrochloride. Incubation was continued for 10 min. Precipitated proteins were removed by centrifugation (1260 g for 10 min) and the supernatant was transferred to a new plate and the absorbance was measured at 546 nm. Previous studies suggested that BTD activity is maximal at 37°C and pH 6.0 [24] and, thus, all tests were run under these conditions. One unit of BTD activity is defined as the amount of protein required to release 1 nanomole of PABA 120 min−1 under the conditions of the assay.
2.4. Enzyme kinetics
Km, Vmax, and Ki [29] were determined as follows. The concentration of inhibitors was kept constant (0.5 mM) in enzyme assays as described above, while the concentration of the substrate N-(+)-biotinyl-PABA was varied from 0.05 mM to 1 mM. The enzyme kinetics module of Sigmaplot 10.0 was used for calculations [30].
2.5. Biotin transport
Theoretically, the biotin analogs tested here might affect both biotin transport into human cells and BTD activity. Here, biotin transport was quantified using a physiological concentration of [3H]biotin (475 pM) in the presence or absence of putative BTD inhibitors (0.5 mM) as described [31]; the Km of biotin transporters is in the low micromolar range [32]. Human Jurkat cells were used for biotin transport studies [33].
2.6. Statistical analysis
Heterogeneous variances were identified by using Bartlett’s test, and data were log transformed where applicable [34]. Significance of differences was tested by one-way ANOVA. Fisher’s Protected Least Significant Difference procedure was used for posthoc testing. StatView 5.0.1 (SAS Institute; Cary, NC) was used to perform all calculations. Differences were considered significant if P < 0.05. Data are expressed as mean ± SD.
3. Results
3.1. Calibration and linearity of the BTD assay
BTD activity was dependent on substrate concentration, enzyme abundance and time. First, known amounts (0 to 40 nanomoles) of synthetic PABA were used to generate a standard curve (Fig. 1). This assay was linear up to 40 nanomoles PABA. Second, dependence of BTD activity on substrate concentration was quantified by incubating BTD with 0 (control), 0.1, 0.25, 0.5, and 0.75 mM of N-(+)-biotinyl-PABA per well in a 96-well plate. PABA released from N-(+)-biotinyl-PABA reached a plateau at 0.5 mM of N-(+)-biotinyl-PABA per well as judged by absorbance at 546 nm (Fig. 2); all subsequent reactions were carried out using 0.5 mM of N-(+)-biotinyl-PABA per well. Third, partially purified BTD from plasma was added in increasing concentration: 0, 0.1, 0.2, 0.4, 0.5, 0.75, and 1 mg of protein per well. The assay was linear up to a concentration of 1 mg of partially purified BTD per well (Fig. 3). Fourth, a time course was conducted to determine whether the hydrolysis of N-(+)-biotinyl-PABA was linear with time. BTD and N-(+)-biotinyl-PABA were incubated in assay buffer for up to 4 hours after the addition of 0.5 mM of N-(+)-biotinyl-PABA at 37°C. The hydrolysis of N-(+)-biotinyl-PABA was linear up to 4 hours (Fig. 4); all subsequent incubations with BTD were carried out for 2 hours following the addition of 60 nanomoles (0.5 mM final concentration/well) N-(+)-biotinyl-PABA unless otherwise noted.
Fig. 1.
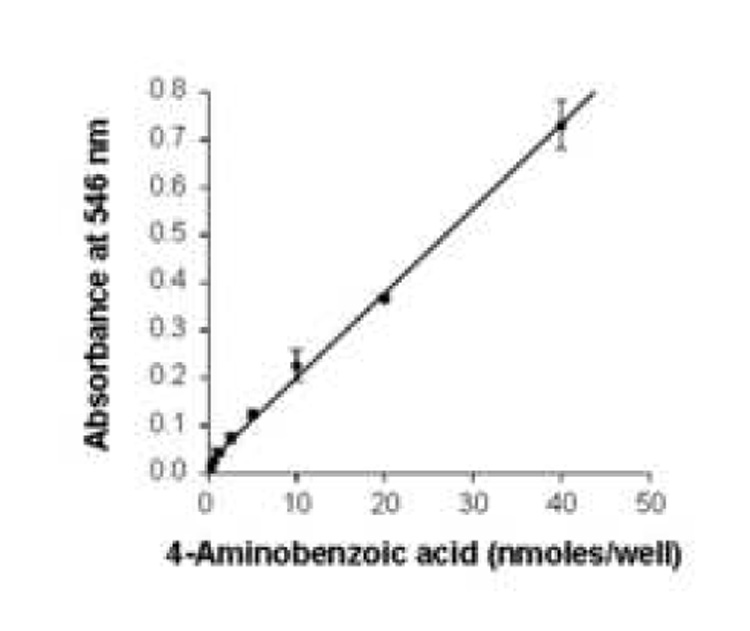
Standard curve for colorimetric quantification of PABA. Absorbance of derivatives of PABA was measured at 546 nm. The standard deviations are too small to be visible at low amounts of PABA (y = 0.0013x + 0.021; r = 0.998).
Fig. 2.
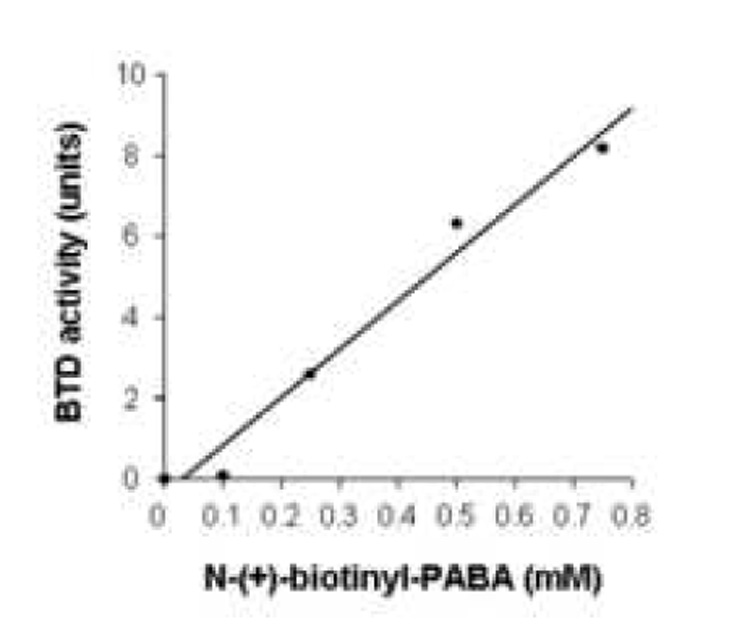
BTD activity depends on the concentration of its substrate, N-(+)-biotinyl-4-amidobenzoic acid. BTD activity was monitored by the release of PABA from N-(+)-biotinyl-PABA. The standard deviations are too small to be visible (y = 0.11x − 1.19; r = 0.989).
Fig. 3.
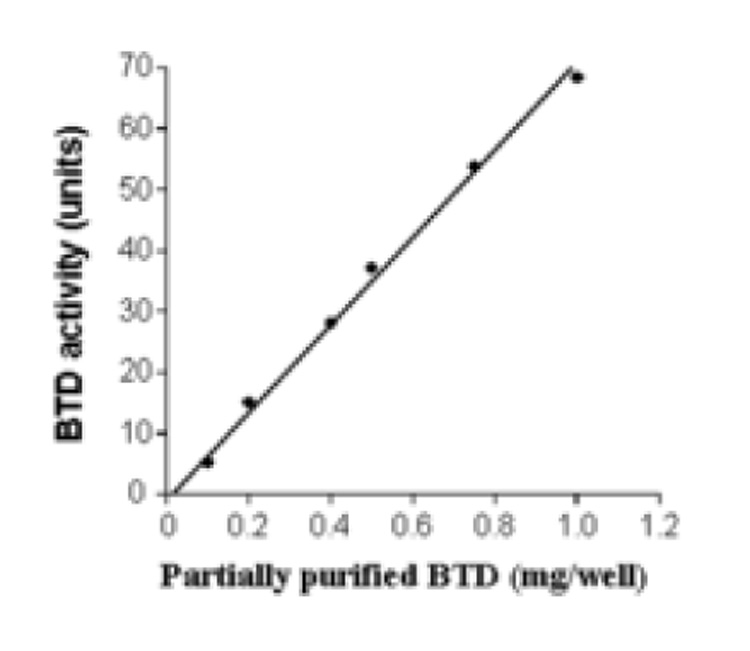
Hydrolysis of N-(+)-biotinyl-4-amidobenzoic acid (60 nmoles/well) depends on the amount of partially purified BTD. The standard deviations are too small to be visible (y = 0.072x − 1.03; r = 0.997).
Fig. 4.
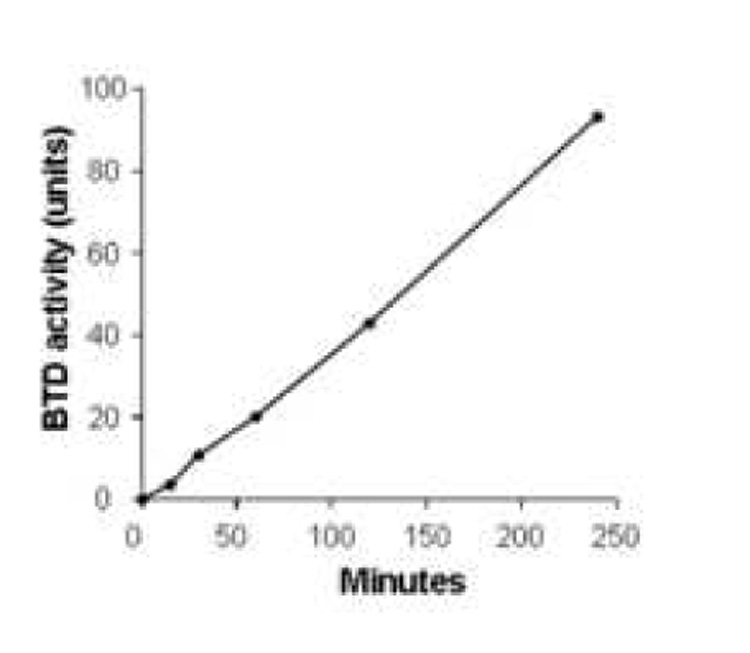
Hydrolysis of N-(+)-biotinyl-4-amidobenzoic acid by partially purified BTD is linear for up to 4 hours. Values are means ± SD (n = 6). The standard deviations are too small to be visible.
3.2. Inhibitors of BTD
Seven out of the thirteen compounds tested here inhibited BTD. In contrast, the following compounds did not significantly affect BTD activity and were used as negative controls in selected experiments: biotinyl diphenylamide; biotinyl dibenzylamide; biotinyl (R)-1-(1-naphthyl)ethylamide; biotinyl 2,4,6-trimethylbenzenamide; biotinyl 3,4-dimethanilide; and biotinyl 4-amidobenzonitrile. These compounds had no meaningful effects on BTD activity even if concentrations as high as 0.5 mM were tested. The compounds tested here are all biotin analogs and it is highly unlikely that concentrations greater than 0.5 mM can be achieved in biological fluids in vivo [35]. Figure 5 depicts compound biotinyl 2,4,6-trimethylbenzenamide as a representative example.
Fig. 5.
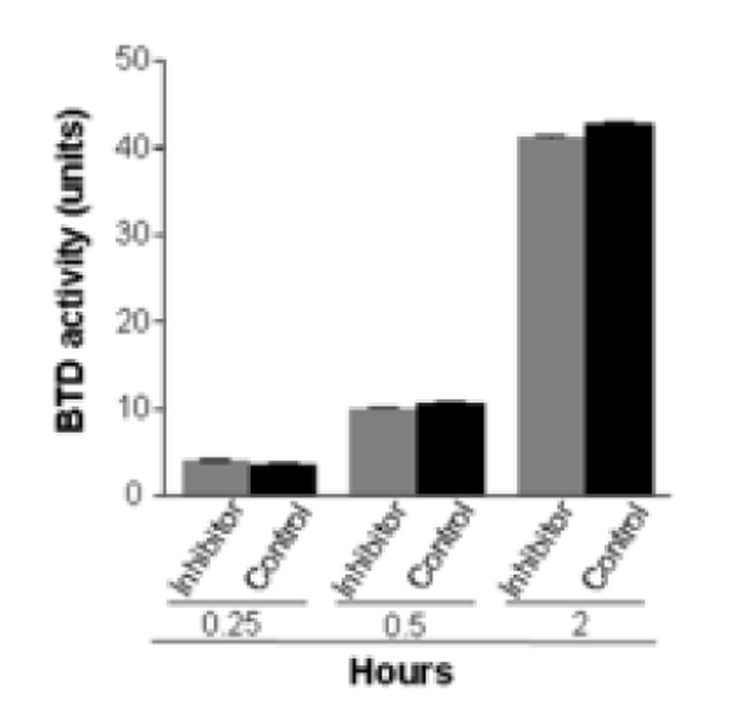
Biotinyl 2,4,6-trimethylbenzenamide (1 mM) does not inhibit BTD. The effects of the treatment are not significantly different compared with inhibitor-free controls. The incubation times denote the hours of incubation of partially purified BTD, substrate, and inhibitor. Values are means ± SD (n = 6; P > 0.05).
In contrast, the following compounds inhibited BTD activity by more than 25%: biotinyl anilide; biotinyl allylamide; biotinyl N-methylanilide; biotinyl-methyl 4-(amidomethyl) benzoate; biotinyl 2-amido-pyridine; biotinyl 4-amidophenylboronic acid; and biotinyl benzylamide. Biotinyl-methyl 4-(amidomethyl) benzoate was the most effective compound of all the inhibitors tested. For example, if BTD was incubated with N-(+)-biotinyl-PABA in the presence of biotinyl methyl 4-(amidomethyl) benzoate for 30, 60, and 120 min, the hydrolysis of N-(+)-biotinyl-PABA was 80%, 59%, and 30%, respectively, of inhibitor-free controls containing only DMSO. A greater percentage inhibition at earlier time points is an event frequently seen with competitive inhibitors [29]. Importantly, the percent inhibition of BTD increased if concentrations of inhibitors were increased, consistent with competitive inhibition of BTD by biotinyl methyl 4-(amidomethyl) benzoate (Fig. 7). For example, if BTD was incubated with N-(+)-biotinyl-PABA in the presence of 0.01 mM, 0.1 mM, and 1 mM biotinyl methyl 4-(amidomethyl) benzoate, the inhibition of hydrolysis of N-(+)-biotinyl-PABA was 16%, 50%, and 80%, respectively, compared to inhibitor-free controls containing only DMSO (Fig. 7).
Fig. 7.
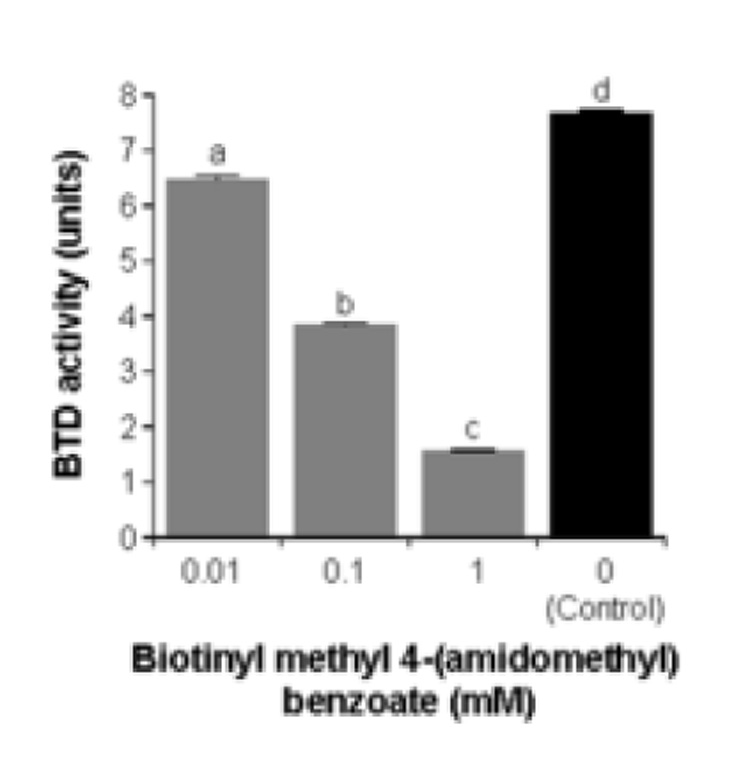
Biotinyl-methyl 4-(amidomethyl) benzoate inhibits BTD activity in a concentration-dependent fashion. Values are means ± SD (n = 6). a,b,c,d Bars not sharing the same letter are significantly different from the inhibitor-free control (P < 0.05).
Enzyme kinetics studies were conducted to confirm that the biotin analogs tested here decreased BTD activity by competitive inhibition. In these experiments we kept the concentrations of inhibitors constant (0 vs. 0.5 mM) and varied the concentrations of substrate, N-(+)-biotinyl-PABA (0.05 mM to 1 mM). For all the inhibitors tested, the percent inhibition of BTD decreased if the concentration of N-(+)-biotinyl-PABA was increased. Km and Vmax values were consistent competitive inhibition (Table 2). The Ki value for biotinyl methyl 4-(amidomethyl) benzoate was smaller than Ki for all the other inhibitors, suggesting that this compound has a greater affinity for BTD than the other compounds tested.
Table 2.
Kinetics of BTD inhibitorsa
| Kinetic variables | |||
|---|---|---|---|
| Inhibitor name | Vmax | Km | Ki |
| nmol × min−1 × mg−1 | mM | mM | |
| Biotinyl methyl 4-(amidomethyl) | |||
| benzoate | 0.29 ± 0.001 | 0.04 ± 0.0004 | 0.06 ± 0.008 |
| Biotinyl anilide | 0.26 ± 0.006 | 0.07 ± 0.048 | 0.26 ± 0.183 |
| Biotinyl 2-amido-pyridine | 0.28 ± 0.002 | 0.01 ± 0.003 | 0.27 ± 0.011 |
| Biotinyl allylamide | 0.35 ± 0.008 | 0.06 ± 0.002 | 0.09 ± 0.011 |
Data are means ± S.D.
Finally, we determined whether biotinyl methyl 4-(amidomethyl) benzoate affected cellular biotin transport, potentially producing artifacts in future studies of BTD-mediated chromatin biology at the cellular level. Biotin uptake into Jurkat cells was quantified in the presence of excess biotinyl methyl 4-(amidomethyl) benzoate, biotinyl anilide (specificity control) and DMSO (negative control). Biotinyl methyl 4-(amidomethyl) benzoate did not affect biotin uptake into Jurkat cells compared with DMSO controls (13 ± 0.6 vs. 13 ± 0.9 amol × 5×10−6 cells × 30min−1; P > 0.05; n=3) whereas biotinyl anilide caused a significant decrease of biotin uptake (5.4 ± 0.3 amol × 5×10−6 cells × 30min−1; P > 0.05; n=3).
4. Discussion
Previous studies of BTD depended on using time-consuming cuvette-based assays [26–28]; making high-throughput screening of BTD inhibitors a tedious task. Here, we present a 96-well plate assay that for the first time offers the means for high-throughput screening of BTD activity and putative BTD inhibitors.
Using this assay and an array of synthetic biotin derivatives, we succeeded in identifying seven putative BTD inhibitors. The most efficient compound was biotinyl-methyl 4-(amidomethyl) benzoate which inhibited BTD up to 80% at a concentration of 1 mM. Our kinetics studies suggest that biotinyl-methyl 4-(amidomethyl) benzoate (and other compounds tested here) act by reversible, competitive inhibition of BTD.
The inhibitors presented here are biotin derivatives and are likely to primarily affect proteins involved in biotin metabolism such as BTD, HCS, and biotin transporters, as opposed to affecting enzyme activity globally. Unspecific inhibitors such as para-hydroxymercuribenzoate, hydroxylamine, and mercaptoethanol do not offer the degree of specificity needed for cell biology studies [8]. For example, mercaptoethanol reduces sulfhydryl groups in enzymes, causing conformational changes that affect enzyme activity. Our studies on biotin transport provide evidence that some BTD inhibitors do not affect biotin uptake, suggesting specificity for BTD. We are currently in the process of investigating affects of BTD inhibitors on other histone biotinyl ligases such as HCS.
Please note, however, that even if an inhibitor is specific for BTD, it will abolish biological functions of this enzyme to the same extent in various cell compartments. For example, loss of BTD activity may result in biological changes that could either be a result of decreased recycling of biotin from breakdown products of carboxylases in the cytoplasm [36], or of remodeling of chromatin through histone debiotinylation in the nucleus [2]. Our first-generation inhibitors of BTD do not overcome the problem of specifically targeting cytoplasmic versus nuclear BTD. A second generation of BTD inhibitors is currently being developed in our laboratory. The goal of these ongoing studies is to create an inhibitor that specifically targets nuclear BTD. Also, we will seek to modify these second generation inhibitors so that they do not undergo significant metabolism and loss of activity. Once these second-generation inhibitors become available, we hope to specifically characterize roles of BTD in chromatin remodeling.
Fig. 6.
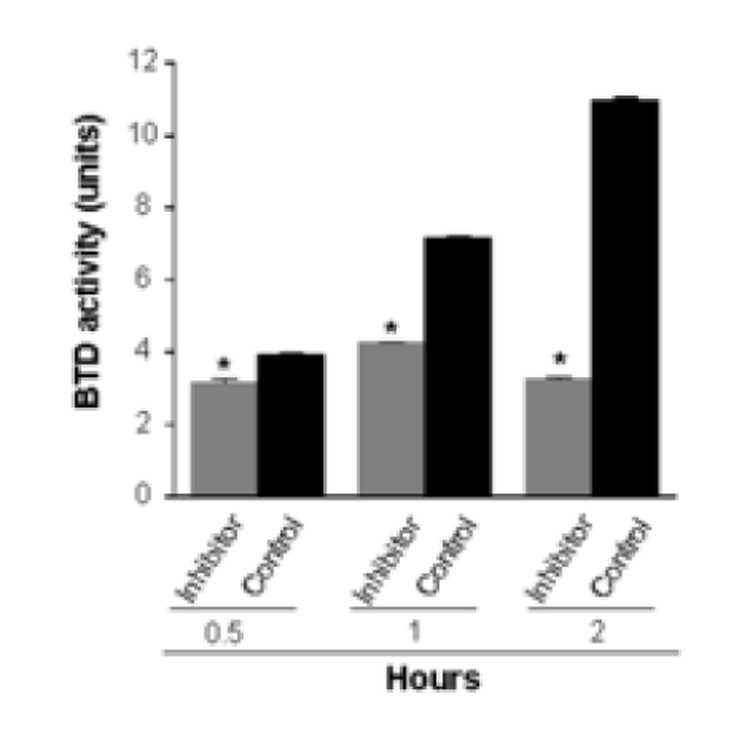
Biotinyl-methyl 4-(amidomethyl) benzoate (1 mM) inhibits BTD. Values are means ± SD (n = 6; *P < 0.05 compared with inhibitor-free control). The incubation times denote the hours of incubation of partially purified BTD, substrate, and inhibitor.
Fig. 8.
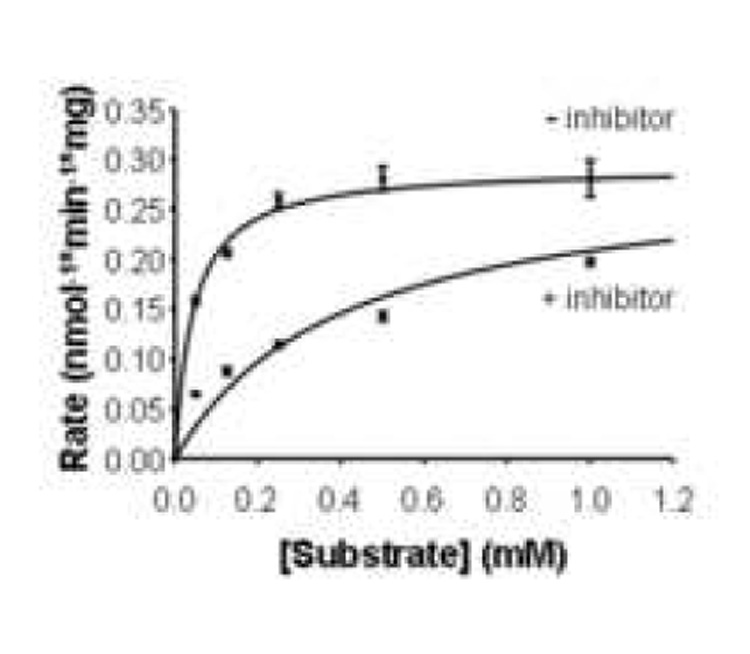
BTD activity is reduced in a substrate-concentration dependent manner in the presence or absence of 0.5 mM of the inhibitor biotinyl-methyl 4-(amidomethyl) benzoate.
Fig. 9.
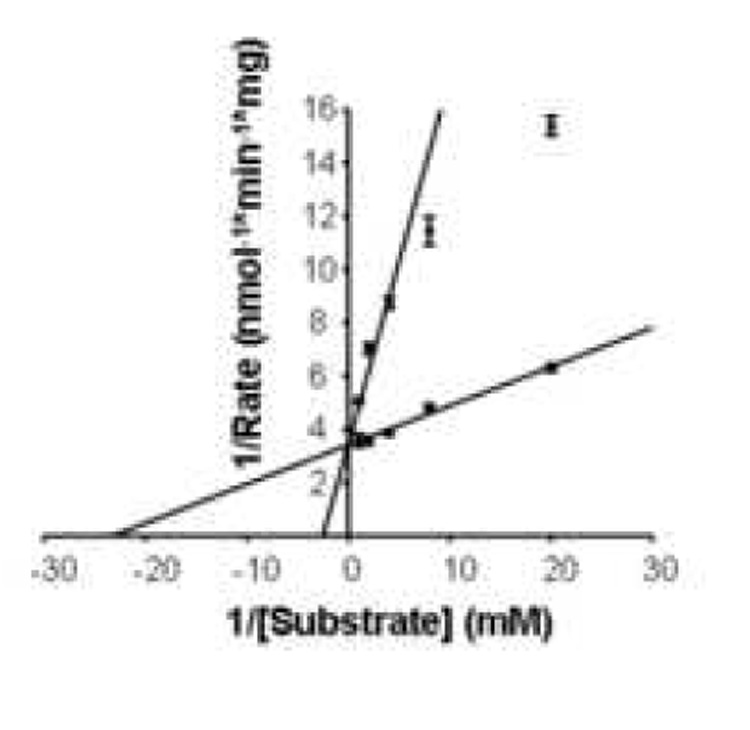
Lineweaver-Burk plot demonstrating competitive inhibition of BTD by biotinyl methyl 4-(amidomethyl) benzoate. Vmax = 0.29 ± 0.009 nmol/min−1/mg−1; Km = 0.04 ± 0.007 mM; Ki = 0.06 ± 0.01 mM; n = 3.
Acknowledgment
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK 063945 and ES 015206, USDA grant 2006-35200-17138, and by NSF EPSCoR grants EPS-0346476 and EPS-0701892.
Abbreviations
- BTD
biotinidase
- DMSO
dimethyl sulfoxide
- HCS
holocarboxylase synthetase
- MCD
multiple carboxylase deficiency
- PABA
para-aminobenzoic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum. Mol. Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 2.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J. Nutr. Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiba Y, Suzuki Y, Aoki Y, Ishida Y, Narisawa K. Purification and properties of bovine liver holocarboxylase synthetase. Arch. Biochem. Biophys. 1994;313:8–14. doi: 10.1006/abbi.1994.1351. [DOI] [PubMed] [Google Scholar]
- 4.Pispa J. Animal biotinidase. Ann. Med. Exp. Biol. Fenniae. 1965;43:4–39. [PubMed] [Google Scholar]
- 5.Wolf B, Grier RE, McVoy JRS, Heard GS. Biotinidase deficiency: a novel vitamin recycling defect. J. Inherit. Metab. Dis. 1985;8:53–58. doi: 10.1007/BF01800660. [DOI] [PubMed] [Google Scholar]
- 6.Dakshinamurti K, Chauhan J. Biotin-binding proteins. In: Dakshinamurti K, editor. Vitamin Receptors: Vitamins as Ligands in Cell Communication. Cambridge, UK: Cambridge University Press; 1994. [Google Scholar]
- 7.Wolf B, Heard GS. Biotinidase deficiency. In: Barness L, Oski F, editors. Advances in Pediatrics. Chicago, IL: Medical Book Publishers; 1991. [PubMed] [Google Scholar]
- 8.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem. Mol. Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 9.Hymes J, Wolf B. Human biotinidase isn't just for recycling biotin. J. Nutr. 1999;129:485S–489S. doi: 10.1093/jn/129.2.485S. [DOI] [PubMed] [Google Scholar]
- 10.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur. J. Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 11.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur. J. Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 12.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 13.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 14.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double strand breaks in human JAr choriocarcinoma cells. J. Nutr. 2005;135:2337–2342. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. doi: 10.1016/j.jnutbio.2006.12.014. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J. Nutr. 2006;136:2735–2742. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J. Nutr. 2007;137:885–889. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballard TD, Wolff J, Griffin JB, Stanley JS, Calcar Sv, Zempleni J. Biotinidase catalyzes debiotinylation of histones. Eur. J. Nutr. 2002;41:78–84. doi: 10.1007/s003940200011. [DOI] [PubMed] [Google Scholar]
- 19.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu. Rev. Nutr. 2005;25:175–196. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 20.Stanley CM, Hymes J, Wolf B. Identification of alternatively spliced human biotinidase mRNAs and putative localization of endogenous biotinidase. Mol. Genet. Metab. 2004;81:300–312. doi: 10.1016/j.ymgme.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Cole H, Reynolds TR, Lockyer JM, Buck GA, Denson T, Spence JE, Hymes J, Wolf B. Human serum biotinidase cDNA cloning, sequence, and characterization. J. Biol. Chem. 1994;269:6566–6570. [PubMed] [Google Scholar]
- 22.Knight HC, Reynolds TR, Meyers GA, Pomponio RJ, Buck GA, Wolf B. Structure of the human biotinidase gene. Mamm. Genome. 1998;9:327–330. doi: 10.1007/s003359900760. [DOI] [PubMed] [Google Scholar]
- 23.McMahon RJ. Biotin in metabolism and molecular biology. Annu. Rev. Nutr. 2002;22:221–239. doi: 10.1146/annurev.nutr.22.121101.112819. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan J, Dakshinamurti K. Purification and characterization of human serum biotinidase. J. Biol. Chem. 1986;261:4268–4275. [PubMed] [Google Scholar]
- 25.Wolf B, Grier RE, Allen RJ, Goodman SI, Kien CL. Biotinidase deficiency: An enzymatic defect in late-onset multiple carboxylase deficiency. Clin. Chim. Acta. 1983;131:273–281. doi: 10.1016/0009-8981(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 26.Knappe J, Brümmer W, Biederbick K. Reinigung und Eigenschaften der Biotinidase aus Schweinenieren und Lactobacillus Casei. Biochem. Z. 1963;338:599–613. [PubMed] [Google Scholar]
- 27.Backman-Gullers B, Hannestad U, Nilsson L, Sörbo B. Studies on lipoamidase: characterization of the enzyme in human serum and breast milk. Clin. Chim. Acta. 1990;191:49–60. doi: 10.1016/0009-8981(90)90057-y. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson L, Ronge E. Lipoamidase and biotinidase deficiency: Evidence that lipoamidase and biotinidase are the same enzyme in human serum. Eur. J. Clin. Chem. Clin. Biochem. 1992;30:119–126. [PubMed] [Google Scholar]
- 29.Segel IR. Biochemical Calculations. 2nd Edition. New York: John Wiley & Sons, Inc.; 1968. p. 430. [Google Scholar]
- 30.Systat Software, Inc. [accessed April 11, 2007];Sigmaplot 10.0. accessed www.systat.com.
- 31.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am. J. Physiol. Cell Physiol. 1998;275:C382–C388. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 32.Prasad PD, Wang H, Kekuda R, Fujita T, Fei Y-J, Devoe LD, Leibach FH, Ganapathy V. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J. Biol. Chem. 1998;273:7501–7506. doi: 10.1074/jbc.273.13.7501. [DOI] [PubMed] [Google Scholar]
- 33.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J. Nutr. 2002;132:887–892. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 34.SAS Institute. StatView Reference SAS Publishing. StatView Reference. 1999 [Google Scholar]
- 35.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J. Nutr. 1995;125:941–946. doi: 10.1093/jn/125.4.941. [DOI] [PubMed] [Google Scholar]
- 36.Wolf B, Heard GS, McVoy JRS, Grier RE. Biotinidase Deficiency. Ann. NY Acad. Sci. 1985;447:252–262. doi: 10.1111/j.1749-6632.1985.tb18443.x. [DOI] [PubMed] [Google Scholar]


