Abstract
Here we identify the pharmacophore in a peptoid that antagonizes Vascular Endothelial Growth Factor Receptor-2 (VEGFR2) in vitro and in vivo. Only three of the side chains in the peptoid are required for activity. Surprisingly however, main chain atoms also form critical interactions with the receptor.
Rapid identification of the ‘minimum pharmacophore’ of a lead compound is a vital step in the drug development process since it sets the stage for subsequent optimization. With peptide-based agents, this exercise is simplified by the regular structure of the molecule. A common practice is to evaluate a series of derivatives in which each residue in turn is replaced with a glycine or alanine (alanine scanning).1, 2 Recently, we reported the effective application of glycine scanning to a peptoid (N-substituted oligoglycine) inhibitor of the 19S regulatory particle of the proteasome. This allowed us to create a minimal derivative of the original hit with about half the mass and thus increased cell permeability and potency.3 We have also reported the isolation of highly specific peptoid ligands for the extracellular domain of the Vascular Endothelial Growth Factor Receptor-2 (VEGFR2),4 an integral membrane receptor that triggers angiogenesis when bound by its cognate hormone VEGF5. A dimerized derivative (GU40C4) of one of these nine residue peptoids (GU40C; see Fig. 1) is a low nM ligand for the receptor’s extracellular domain and is a potent antagonist of angiogenesis in vivo.4 Inhibition of VEGFR2-mediated angiogenesis is a validated strategy to slow the growth of tumors as well as to treat “wet” macular degeneration.6–14 Thus, this peptoid is of potential therapeutic interest and its optimization is an important goal. Therefore, we sought to identify the minimal pharmacophore in GU40C as the initial step in this effort.
Figure 1.
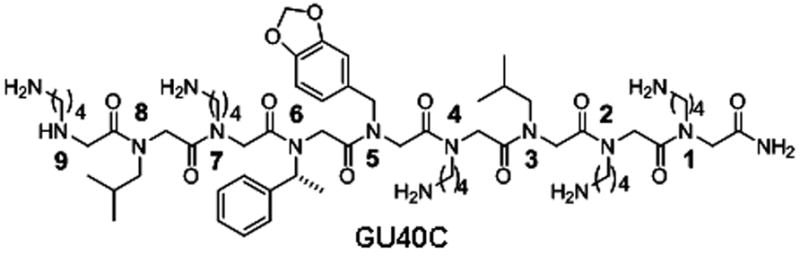
GU40C Structure of GU40C. Residues are numbered starting from C-terminus.
First, nine derivatives of GU40C were synthesized in which each of the nine residues in the parent peptoid was replaced with a glycine. All these derivatives were synthesized with a C-terminal cysteine to facilitate fluorescein attachment via maleimide chemistry. The affinity of each of these derivatives for the extracellular domain (ECD) of VEGFR2 was then determined using an ELISA-like binding assay described in our previous report4. The results are shown in Fig. 2 (black bars). Only two side chains (the 6th and 8th from the C-terminus) appeared to be important for binding of GU40C to the VEGFR2 ECD.
Figure 2.
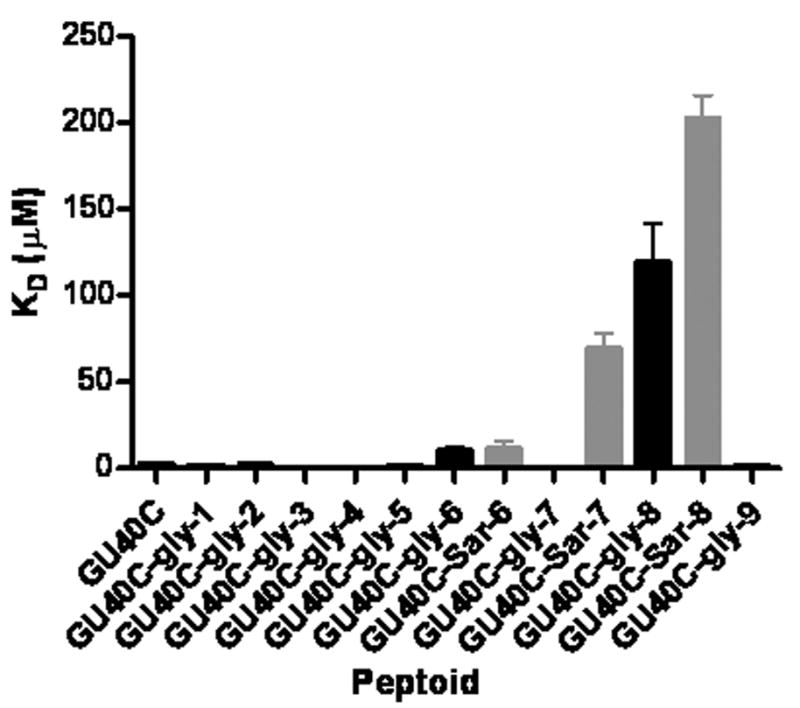
Glycine (black bars) sarcosine (grey bars) scan binding results of GU40C. Please refer Figure 1 for residue numbers.
To buttress these data, we repeated the analysis, but replaced each monomer in the peptoid with sarcosine rather than glycine. Since secondary amides have a strong preference for a transoid configuration about the peptide bond, while tertiary amides do not, it is possible that glycine substitution could introduce conformational constraints not present in the parent peptoid and thus the comparison of the derivative to the parent molecule might reflect issues other than simply deleting the side chain. For example, if the preferred binding conformation of a peptoid involved a cisoid conformation about a particular peptide bond in the molecule, then replacement of the side chain with a hydrogen would discriminate against this conformation and presumably inhibit binding, even though the side chain was not involved directly. A sarcosine scan has the effect of replacing each of the side chains in turn with a methyl group rather than a hydrogen, preserving the tertiary amide bond, but removing the bulk of the side chain. Therefore, we decided to conduct a sarcosine scan in the region of the molecule identified as being critical for binding by the glycine scan.
As shown in Fig. 2 (grey bars), substitution of the methyl group for isobutyl moiety at position 8 or the α-methylbenzyl group at position 6 weakened binding of the peptoid for the VEGFR2 ECD significantly, consistent with the glycine scan results. However, in contrast with the glycine scanning result, substitution of the lysine-like side chain at position 7 with methyl also reduced binding affinity. This result was confirmed by competition binding assays that compared directly the relative affinities of the peptoids with glycine and sarcosine substitution at position 7 (see supplementary figure 5). We do not fully understand the basis of the different results obtained using the two scanning methods at position 7. One possibility might be that a polar substituent capable of donating a hydrogen bond to solvent might be favorable there. In any case, the combined data from the glycine and sarcosine scans indicate that the N-terminal region of GU40C, specifically positions 6–8 (see Fig. 1), are important for binding of the peptoid to VEGFR2.
Based on these data, it seemed reasonable to speculate that a trimeric peptoid including positions 6–8 would be a good ligand for the VEGFR2 ECD, an interesting possibility since this molecule would have a mass of less than 450 Daltons. To test this idea, we synthesized a fluorescein-conjugated tetramer peptoid GU40C(1) that contains the three original residues at 6–8 positions along with an N-terminal glycine (Fig. 3A).
Figure 3.
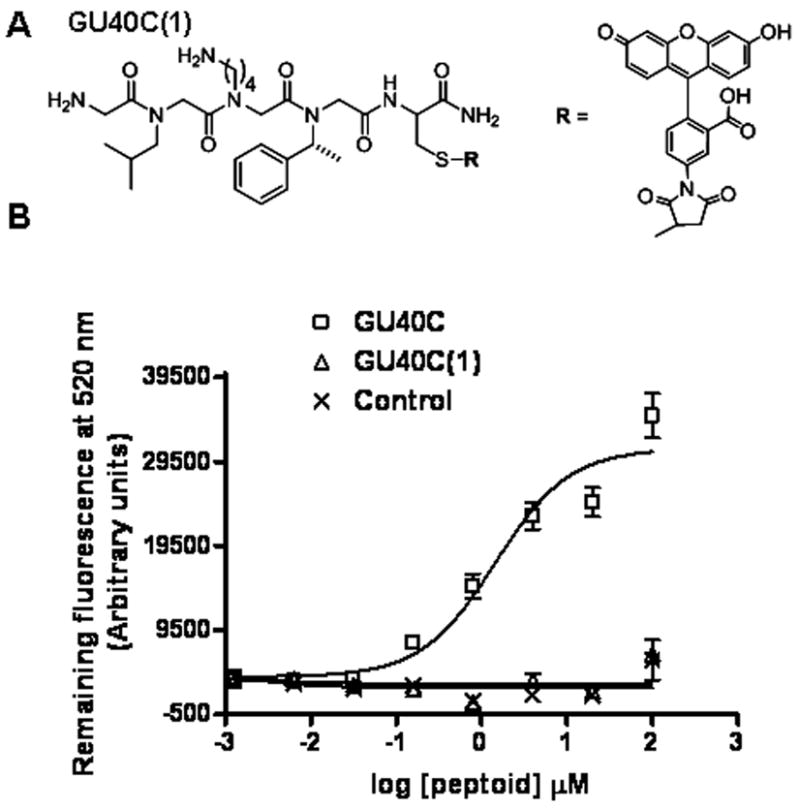
Shortest derivative of GU40C which contains only the important side chains and it’s binding isotherm. (A) Structure of GU40C(1) (B) Binding isotherms of GU40C, GU40(1) and a control peptoid that does not bind selectively to VEGFR2 ECD.
The affinity of this minimized GU40C derivative for the receptor ECD was then tested, again using the ELISA-like binding assay. Somewhat surprisingly, this molecule showed no detectable binding to the receptor ECD at any of the concentrations tested (Figure 3B). Combined with the glycine scanning data shown in Figure 2, this result suggested the possibility that some of the main chain atoms in the parent peptoid might be involved in receptor binding.
Therefore, we decided to reintroduce the full backbone, but leave out the side chains, except those important residues at positions 6–8 [GU40C(2) –Figure 4A, first structure]. Interestingly, this compound recognized the receptor ECD with an affinity similar to that of the GU40C parent peptoid (Figure 4B, square and circle data points), confirming the conclusion derived from the scanning experiments that the side chains at positions 1–5 are not involved in receptor binding. This observation, combined with the sarcosine scanning data, confirm that some of the backbone amide bonds within the first five residues participate in the binding event. We also synthesized and tested GU40C(3) (Fig. 4A), a compound containing the essential side chains and two amide groups C-terminal to these residues, but which are out of register with the amides in the parent peptoid. This compound had no detectable affinity for the receptor ECD (Fig. 4B).
Figure 4.
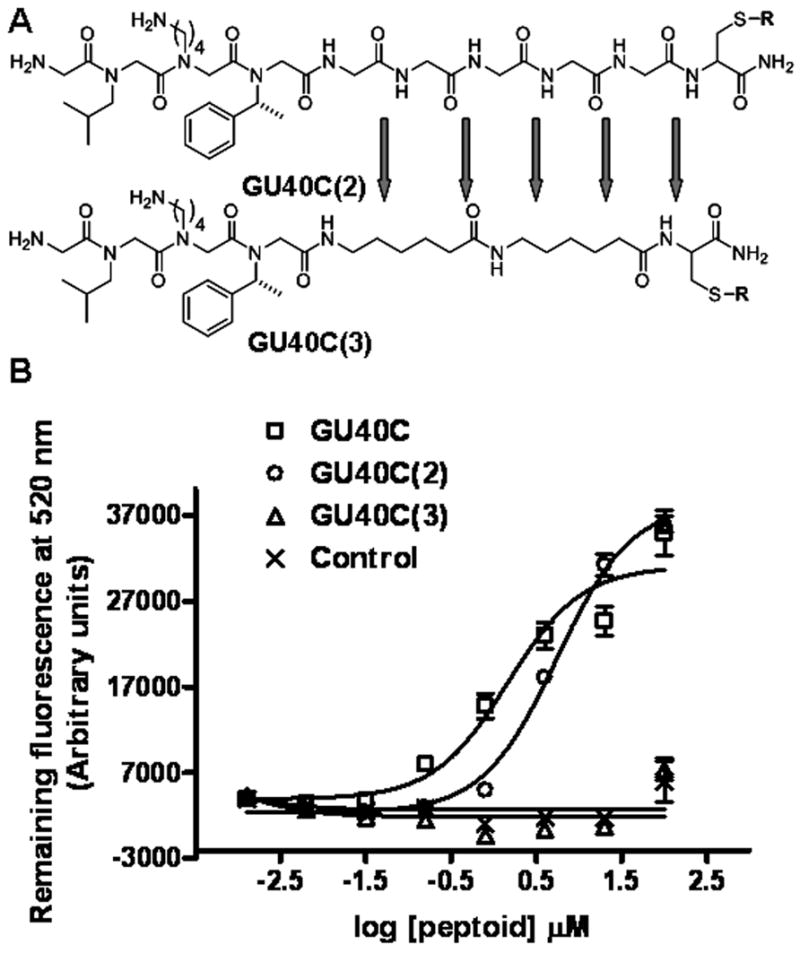
Structures and binding isotherms of GU40C and its derivatives. (A) Structures of GU40C(2) and GU40C(3). R: please see figure 3. (B) Binding isotherms of GU40C and its derivatives. Only GU40C and GU40C(2) are able to display binding to VEGFR2 ECD.
To identify the main chain amides important for binding, we synthesized five different truncated versions of GU40C, each containing all four of the N-terminal residues (Figure 5A). Increasing concentrations of these truncated versions were competed with a constant amount of fluorescein-conjugated GU40C for binding to the VEGR2 ECD. The results are shown in Figure 5B. Unlabeled GU40C competed efficiently with its labeled counterpart as expected (Figure 5B). Elimination of the first C-terminal residue weakened binding about ten-fold (Fig. 5B). Further elimination of the next three C-terminal residues further diminished binding only slightly. However, deletion of the next residue essentially abolished binding of the peptoid to the VEGFR2 ECD.
Figure 5.
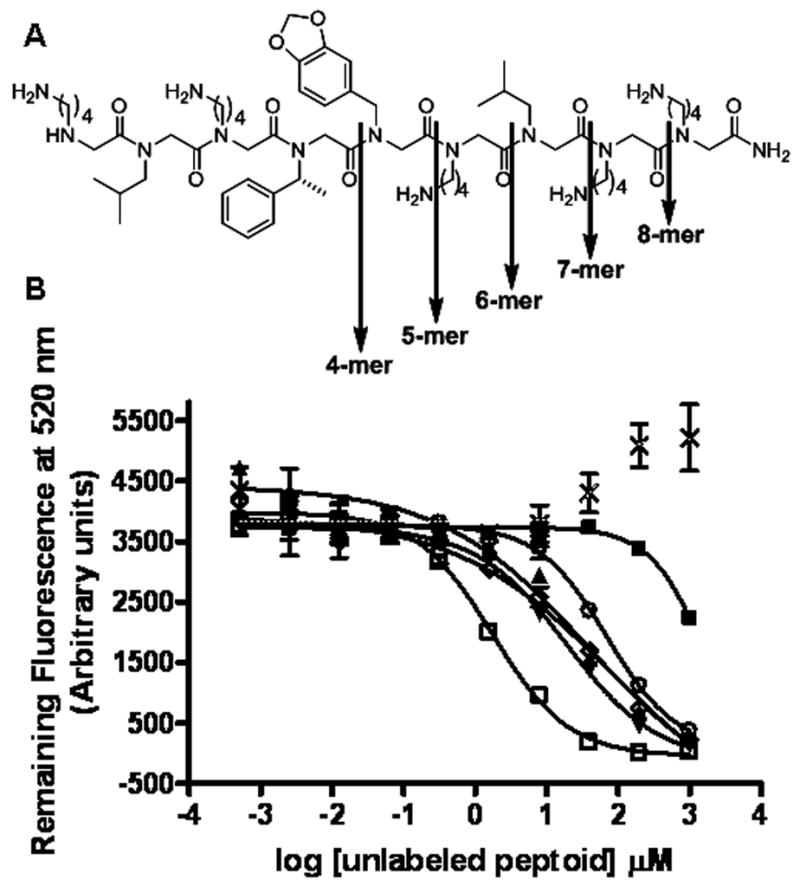
GU40C truncation study results. (A) Downward arrows show GU40C truncation positions (B) Competitive binding assay results; increasing concentrations of unlabelled GU40C and truncated derivatives were competed with a constant amount of fluoraceinated GU40C. Symbols represent; GU40C (□), 8-mer (▲), 7-mer (▼), 6-mer (◇), 5-mer (○), 4-mer (■), control (×).
In summary, the data described above have defined the minimal pharmacophore of the peptoid VEGFR2 antagonist GU40C4. Only three side chains are important for peptoid-receptor binding (the 6th through 8th counting from the C-terminus; Fig. 6) as shown by glycine and sarcosine scanning (Fig. 2). However, a smaller peptoid containing only these residues is inactive (Fig. 3). Furthermore, an analysis of truncated derivatives of GU40C showed that elimination of the first C-terminal residue reduced the affinity of the peptoid by about ten-fold and removal of the fifth residue essentially abolished binding. Combined with the insensitivity of the binding affinity to the removal of the side chains at these residues, these data argue that it is the main chain residues at positions 1 and 5 that contact the receptor ECD. This model is further supported by the fact that the nine residue peptoid GU40C(2), which contains only the side chains at positions 6–8 but is otherwise comprised of glycines, binds the receptor ECD about as well as the GU40C parent.
Figure 6.
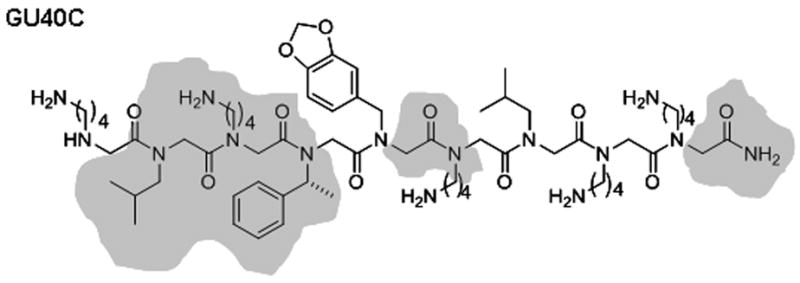
Structure of the GU40C with highlighted residues that are proposed to constitute the minimal pharmacophore.
Fig. 6 highlights the residues that are proposed to constitute the minimal pharmacophore of GU40C based on these results. This study also highlights how we can take advantage of the regular structure of peptoids to rapidly identify the pharmacophore of a bioactive molecule. Current efforts are focused on optimizing the fit of the important side chains with the receptor ECD to improve the potency of the compound.
Supplementary Material
Experimental details, structures of glycine, sarcosine and truncated derivatives of GU40C and their MALDI-TOF confirmation data. Competitive binding assay data.
Acknowledgments
This work was supported by a contract from the National Heart, Lung, and Blood Institute (NO1-HV-28185) for the UT Southwestern Center for Proteomics Research, a grant from the Welch Foundation (I-1299), and the Effie Marie Cain Scholarship for Angiogenesis Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El Kasmi KC, Theisen D, Brons NH, Muller CP. Molecular immunology. 1998;35:905. doi: 10.1016/s0161-5890(98)00087-x. [DOI] [PubMed] [Google Scholar]
- 2.Slon-Usakiewicz JJ, Purisima E, Tsuda Y, Sulea T, Pedyczak A, Fethiere J, Cygler M, Konishi Y. Biochemistry. 1997;36:13494. doi: 10.1021/bi970857h. [DOI] [PubMed] [Google Scholar]
- 3.Lim HS, Archer CT, Kim YC, Hutchens T, Kodadek T. Chemical communications (Cambridge, England) 2008:1064. doi: 10.1039/b717861a. [DOI] [PubMed] [Google Scholar]
- 4.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. Journal of the American Chemical Society. 2008;130:5744. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N. Endocrine reviews. 2004;25:581. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 6.Ambresin A, Mantel I. Rev Med Suisse. 2007;3:137. [PubMed] [Google Scholar]
- 7.Brekken RA, Overholser JP, Stastny VA, Waltenberger J, Minna JD, Thorpe PE. Cancer Res. 2000;60:5117. [PubMed] [Google Scholar]
- 8.D’Andrea LD, Del Gatto A, Pedone C, Benedetti E. Chem Biol Drug Des. 2006;67:115. doi: 10.1111/j.1747-0285.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 9.Gerber HP, Kowalski J, Sherman D, Eberhard DA, Ferrara N. Cancer Res. 2000;60:6253. [PubMed] [Google Scholar]
- 10.Getmanova EV, Chen Y, Bloom L, Gokemeijer J, Shamah S, Warikoo V, Wang J, Ling V, Sun L. Chem Biol. 2006;13:549. doi: 10.1016/j.chembiol.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Hicklin DJ, Ellis LM. J Clin Oncol. 2005;23:1011. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 12.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F. N Engl J Med. 2004;350:2335. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 13.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Nature. 1993;362:841. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 14.Klohs WD, Hamby JM. Curr Opin Biotechnol. 1999;10:544. doi: 10.1016/s0958-1669(99)00033-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental details, structures of glycine, sarcosine and truncated derivatives of GU40C and their MALDI-TOF confirmation data. Competitive binding assay data.


