Abstract
The dsDNA bacteriophage P22 has a ring shaped dodecameric complex composed of the 84 kDa portal protein subunit which forms the central channel of the phage’s DNA packaging motor. The overall morphology of the P22 portal complex is similar to that of the portal complexes of Phi29, SPP1, T3, T7 phages and herpes simplex virus. Secondary structure prediction of P22 portal protein and its threading onto the crystal structure of the Phi29 portal complexes suggested that P22 portal protein complex shares conserved helical modules which were found in the dodecameric interfaces of the Phi29 portal complex.
To identify the amino acids involved in inter-subunit contacts in the P22 portal ring complexes and validate the threading model, we performed comparative hydrogen/deuterium exchange analysis of monomeric and in vitro assembled portal proteins of P22 and the dodecameric Phi29 portal. Hydrogen/deuterium exchange experiments provided evidence of inter-subunit interactions in the P22 portal complex similar to those in the Phi29 portal which map to the regions predicted to be conserved helical modules.
Introduction
Virus assembly is a multi-step process driven by precise protein/protein or protein/nucleic acid interactions. Formation of an infectious virus often proceeds through the assembly of sub-structures or non-infectious precursor particles. For example, the tailed dsDNA bacteriophages and herpesviruses first form a precursor to the capsid termed either a procapsid or a prohead. During procapsid assembly twelve molecules of the portal protein form a ring-like channel which occupies one of the twelve pentameric vertices of the procapsid. Genomic DNA is packaged into the procapsid through the unique portal vertex using a two subunit terminase protein complex fueled by ATP hydrolysis. Shortly after the encapsidation of DNA, the portal channel is closed by the binding of head completion proteins. Tail attachment follows resulting in infectious virions.1,2
Based on their morphological similarity and the fact that they share common morphogenic features such as multi-step assembly and DNA packaging into preformed procapsids it has been proposed that there are evolutionary relationships among the tailed dsDNA bacteriophages and herpesviruses,1,2 In support of this it has been determined that the structural proteins such as coat proteins and scaffolding proteins share a common fold.3–11 For example, a common fold, termed the HK97 or Johnson fold, was found in the major coat proteins of bacteriophages HK975, T46, P227, Phi298, T79, epsilon 1511 and herpes simplex virus (HSV-1) despite a lack of sequence conservation.10 Although they display no detectable sequence similarity as well as large variations in subunit size (from 36 kDa in Phi29 to 84 kDa in P22), the portal complexes of dsDNA bacteriophages and herpesviruses all share a dodecameric ring-like structure.11–18 Bacteriophage Phi29 was the first dsDNA phage whose portal complex structure was solved at the atomic level.19 The Phi29 portal protein is composed of three distinct domains: the wide-end domain, the central domain, and the narrow-end domain.19 The wide-end and narrow-end domains are predominantly composed of short β-sheets and extended coils whereas the central domain which connects the wide- and narrow-end domains consists of three long α-helices. These three long helices run anti-parallel to one other and a central channel is formed through the lateral interactions of twelve three α-helical bundles.19 The inside of the central channel is predominantly negatively charged presumably facilitating passage of DNA during packaging and ejection.19 In the prohead, the α-helical bundles of the central domain penetrate the capsid at an angle of approximately 40° with respect to the central 12-fold axis.19 The wide-end domains are located inside of the prohead contacting the surrounding coat proteins whereas the narrow-end domains are exposed outside presumably allowing interactions of packaging components such as pRNA and terminase complexes.19
Recently, the crystal structure of the bacteriophage SPP1 portal complex has been solved.20 Although the in vitro form is composed of 13 subunits rather than 12, its overall shape and protein fold is strikingly similar to the Phi29 dodecameric ring.18,20,21 The stem domain and clip (residues 250–350) of SPP1 form the central channel of the portal ring and match well with the central and narrow-end domains of the Phi29 portal complexes, respectively.18–20 Since the SPP1 portal subunit has approximately 200 more residues than does Phi29, additional structural features were observed in its crystal structure. The SPP1 portal complexes present a crown domain not observed in the Phi29 ring and a thicker wing domain than is seen in Phi29.20 The crown domain of the SPP1 portal complex is composed of three short α-helices (residues 421–467) located near the C-terminus of the SPP1 portal protein. Mutations in the crown domain altered formation and stability of the ring20 suggesting the crown domain is involved in inter-subunit interactions upon assembly.
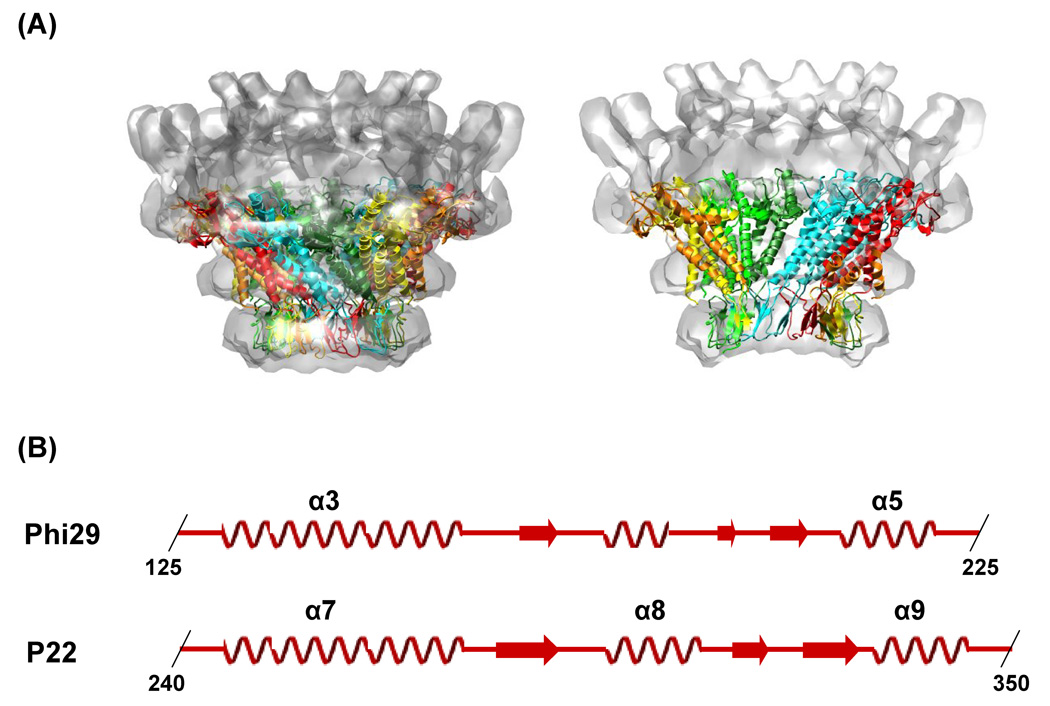
There is no atomic structure of the bacteriophage P22 portal protein available. Cryo-EM studies of the P22 portal complex assembled in vitro and in situ have determined that they have a ring-like morphology similar to that observed for Phi29, SPP1, T3, T7 and herpes simplex virus (Figure 1A).11–19,22,23 Although these cryo-EM reconstructions provided global structural information they did not reach atomic resolution. The predicted secondary structure of the P22 portal protein presented a regular arrangement of long helical components in the middle of the amino acid sequence (Figure 1B) and threading the secondary structure prediction onto the crystal structure of the Phi29 portal complex suggested that the P22 portal protein shares the helical modules found at the dodecameric interface of the Phi29 portal protein complex (Figure 1).18,24
Figure 1.
Comparison between the Phi29 and the P22 portal complexes. (A) Overlay of the crystal structure of Phi29 (PDB: 1IJG) onto the P22 virion’s portal density. The Phi29 crystal structure fits well into the electron density of the distal half of the P22 portal complex (B) Putative conserved secondary structure modules in the portal proteins of P22 (residues 240–350) and Phi29 (residues 125–225). The secondary structure of the Phi29 portal complex was obtained from the crystal structure of the Phi29 portal complex (PDB: 1IJG). The secondary structure of the P22 portal protein was predicted using the program PredictProtein (www.predictprotein.org). The regions of α-helix (coil) and β-strand (arrow) are indicated and the boundary residues are numbered.
To identify the amino acids involved in subunit-subunit contacts in the P22 portal ring and validate the threading model we performed mass spectrometry based comparative hydrogen/deuterium exchange analyses of the Phi29 portal complex and the P22 portal protein in both the monomeric and oligomeric forms. Hydrogen exchange experiments are sensitive to the dynamics and solvent accessibility of the protein backbone.25–27 Ligand binding can result in increased exchange protection by direct occlusion of the binding site or through distant conformational changes.28 However, it has been demonstrated that increased protection in the rapidly exchanging residues generally correlates with the occlusion mechanism rather than with alterations in global dynamics.28 The results demonstrate that the helices that modulate the inter-subunit contacts in the Phi29 portal complex are well protected, and that the regions which were predicted to form similar helical interactions in the P22 portal protein undergo a substantial increase in exchange protection upon ring formation.
Results
Hydrogen/deuterium exchange of the Phi29 portal complex
To characterize the conserved helical modules in the portal complexes of the related bacteriophage Phi29 and P22, we performed an analysis of the hydrogen/deuterium exchange kinetics by mass spectrometry. We initiated investigation with the Phi29 portal complex, since its X-ray crystal structure is available and therefore its hydrogen/deuterium exchange behavior can be directly mapped onto the crystal structure.19 Phi29 portal protein subunits spontaneously form dodecameric rings and dissociation of the rings results in protein aggregation. Therefore, all the hydrogen/deuterium exchange experiments were performed with the dodecameric ring.
To initiate exchange the Phi29 portal rings were diluted 10-fold into deuterated buffer (50mM Tris, 400mM NaCl, pH 7.9) and incubated at room temperature. The exchange reactions were sampled at various times (30 sec to 8 hours), quenched at low pH (pH 2.5) and then flash frozen in liquid nitrogen for subsequent analysis by LC ESI hybrid ion trap-FT ICR mass spectrometry (LTQ-FT, Thermo Finnigan). The quenched samples were digested with pepsin and the peptic fragments were identified by a combination of exact mass matching and MS/MS sequencing. Approximately 90% of the Phi29 portal protein primary sequence, including some overlapping fragments, was identified and used in the analysis of the exchange kinetics (Figure 2A).
Figure 2.
Amino acid sequences of the Phi29 portal protein and hydrogen/deuterium exchange profile for peptide residues 136–159 and 160–185. (A) Amino acid sequence of the Phi29 portal protein. The observed peptic fragments of the Phi29 portal protein are underlined (blue lines). The peptic fragments, which were identified by a combination of exact mass matching and MS/MS sequencing, span ~90% of the protein primary sequence. The secondary structure was determined and mapped as in Figure 1. (B) Mass spectra of the peptic fragment corresponding to residues 136–159 (left panel) and 160–185 (right panel) from the Phi29 portal complexes at various exchange times. In each panel, the bottom spectrum represents the un-exchanged control.
Electrospray ionization (ESI) generally produces multiply charged ions and mass analyzers detect mass-to-charge ratio (m/z). In any given peptide, the random incorporation of naturally occurring isotopes (primarily 13C) results in a cluster of related peaks with a spacing of ~1 Da/z, termed the isotopic distribution. Replacement of an amide proton with a deuteron results in a 1 Da mass increase and a shift in the isotopic distribution towards higher m/z (Figure 2B, C, 5B, and C). The raw mass spectra in figure 2B and C demonstrate the time dependent incorporation of deuterium into peptides spanning residues 136–159 and 160–185 of the Phi29 portal ring. Although similar in length (24 and 26 residues) and contiguous in the primary sequence, they have very different exchange profiles. While the peptide spanning residues 136–159 was well protected from exchange, the amide protons of the peptide 160–185 exchanged rapidly. These exchange patterns imply that residues 160–185 are flexible and exposed to solvent without forming inter-subunit interactions whereas residues 136–159 are buried either within the subunit or at the inter-subunit interface. In the crystal structure, residues 160–185 lie in the narrow-end domain which is predominantly composed of extended coils whereas residues 136–159 are located in helix 3 which is one of the three long helices in the central domain (Figure 4).
Figure 5.
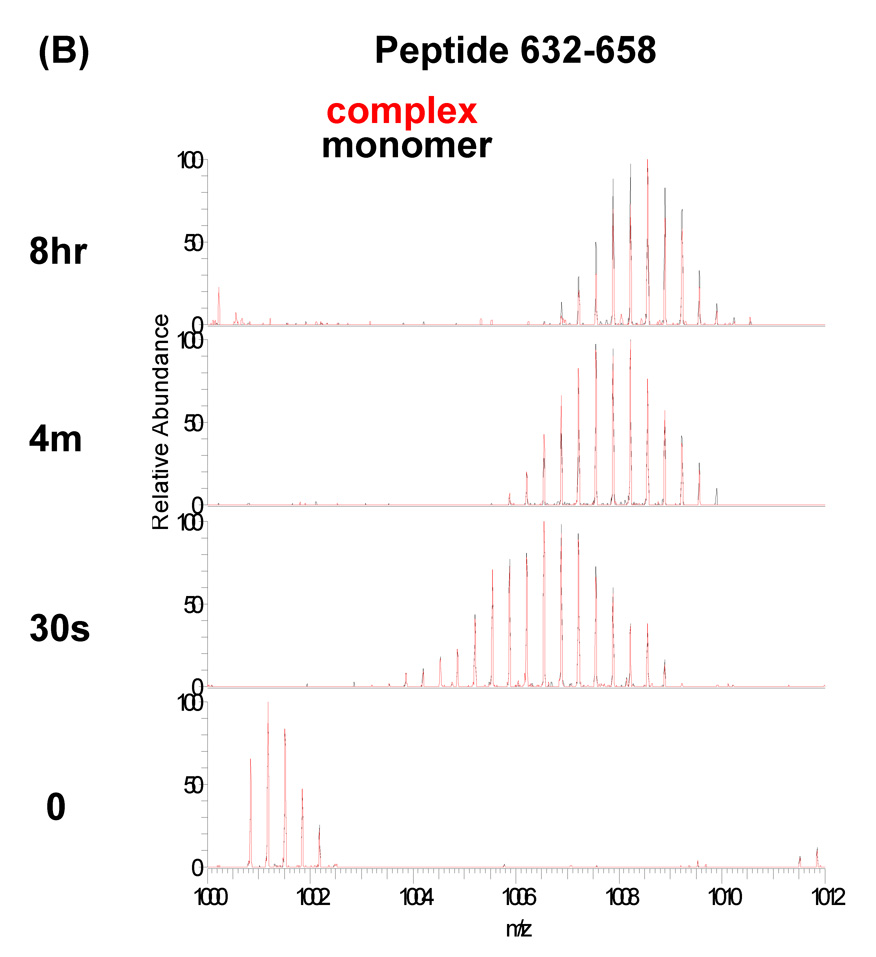
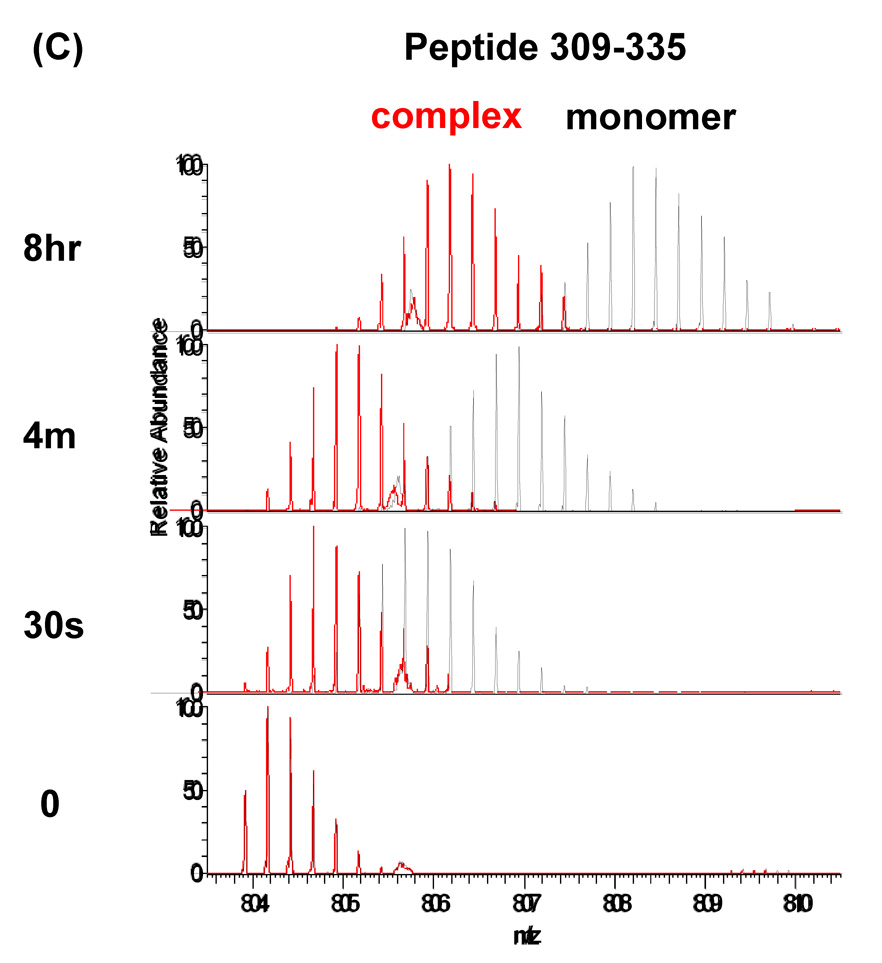
Amino acid sequences of the P22 portal protein and hydrogen/deuterium exchange profile for peptide residues 632–658 and 309–335. (A) Amino acid sequence of the P22 portal protein. The observed peptic fragments of the P22 portal protein are underlined (blue lines). The peptic fragments, which were identified by a combination of exact mass matching and MS/MS sequencing, span ~90% of the protein primary sequence. The predicted elements of secondary structure were calculated and mapped onto the sequence as described in Figure 1. (B and C) Overlaid mass spectra of the peptic fragment corresponding to residues 632–658 and 309–335, respectively, from P22 portal protein monomer (black) and complex (red) at various exchange times. In each panel, the bottom spectrum represents the un-exchanged control.
Figure 4.
Hydrogen/deuterium exchange profile map of the Phi29 portal complex. Hydrogen/deuterium exchange rates are categorized three groups, fast (red), intermediate (yellow), and slow (blue), based on the curve fitting results from a three component exponential model. Top (left panel) and side views (right panel) of the Phi29 portal complex crystal structure (PDB: 1IJG) are mapped according to exchange rates using PyMol. Regions where are not covered by hydrogen/deuterium exchange are colored grey.
Helical bundles in the central domain of the Phi29 portal complex were well protected from exchange
To quantify the rate of exchange of the peptic fragments, we calculated the centroids of the isotopic distribution at each time point and plotted them against the exchange times (Figure 3, 6). The progress curves for each individual peptide were fit to a three component exponential model (Figure 3 and Figure 6). The average back exchange was estimated to be 20% based on the extent of exchange averaged across rapidly exchanging peptides after eight hours. Based upon the exponential fit, the amide protons in each peptide were assigned to one of three groups, a (fast), b (intermediate) and c (slow), with exchange rate constants k1 (>1min−1), k2 (0.01min−1– 1min−1) and k3 (<0.01min−1), respectively.25
Figure 3.
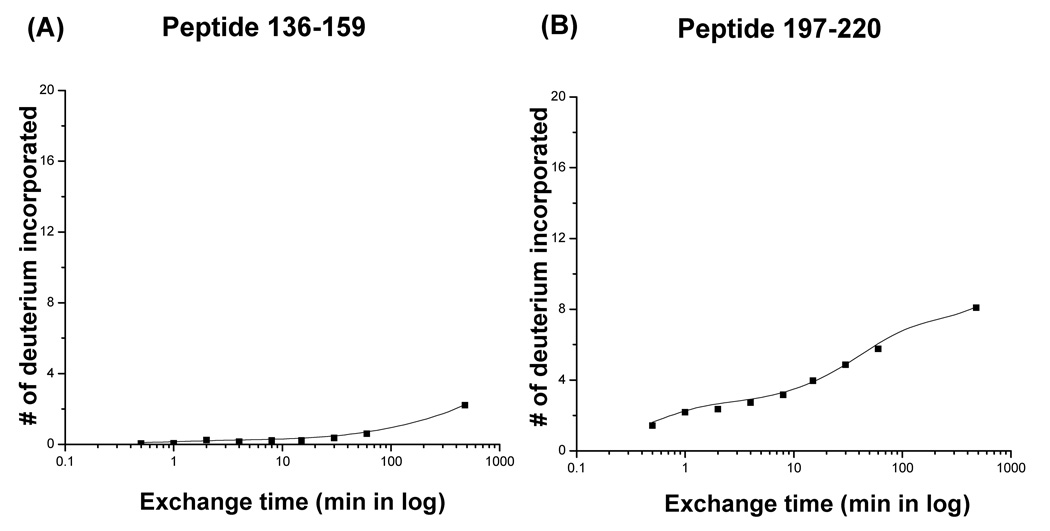
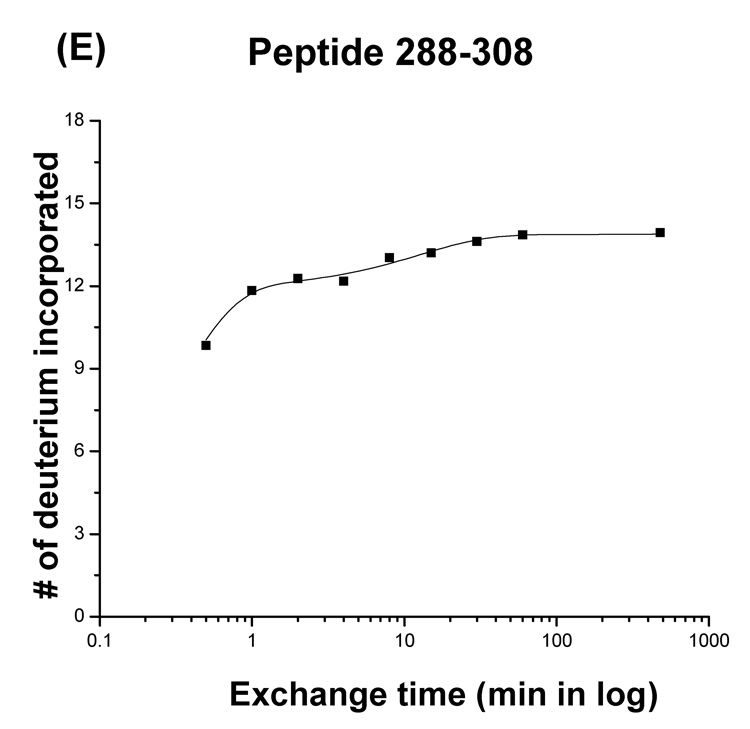
Plots of the number of deuterium atoms incorporated according to the exchange period for peptic fragments from the Phi29 portal complexes (dots). The solid line represents the fit obtained by three component exponential fitting the exchange data (see materials and methods). Peptic fragments (A) 136–159, (B) 197–220, (C) 160–185, (D) 1–13, and (E) 288–308. Highest number on the y-axis represents the theoretical number of exchangeable amide protons.
Figure 6.
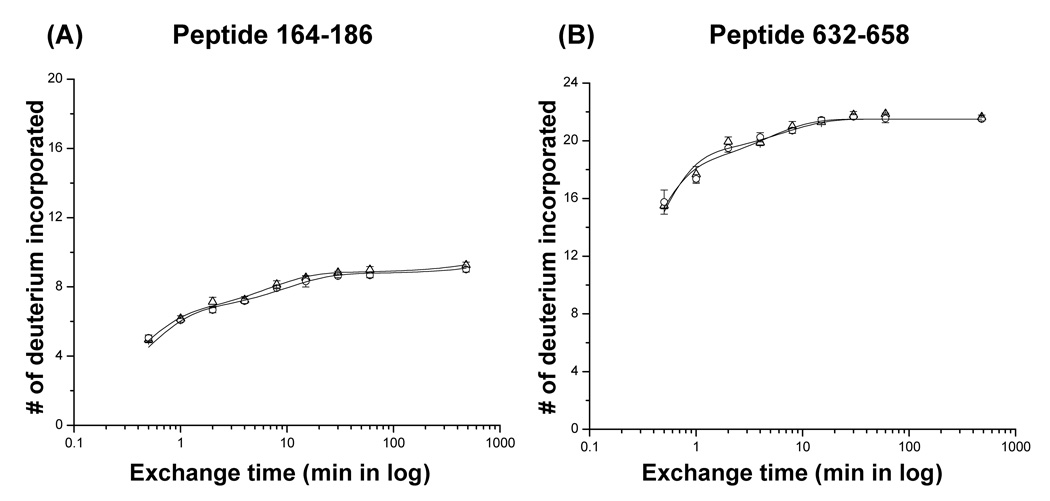
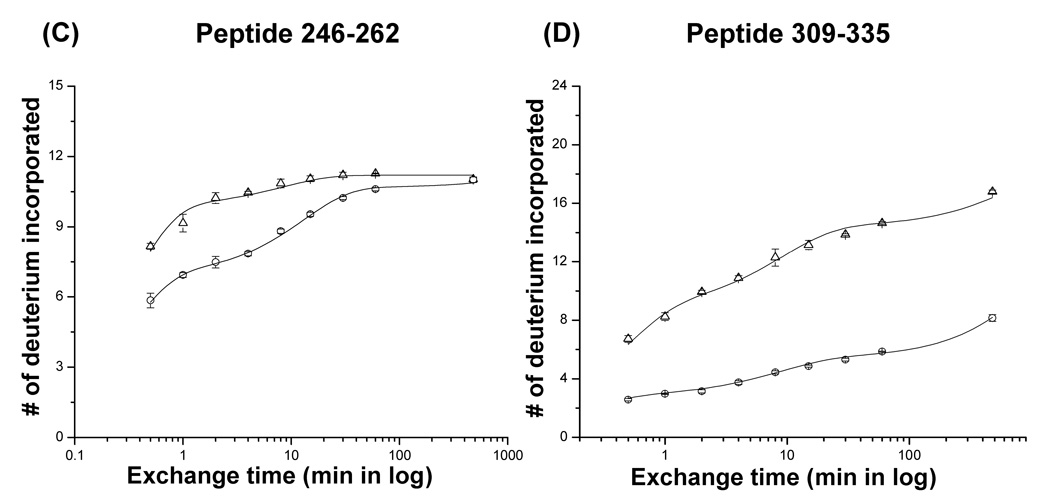
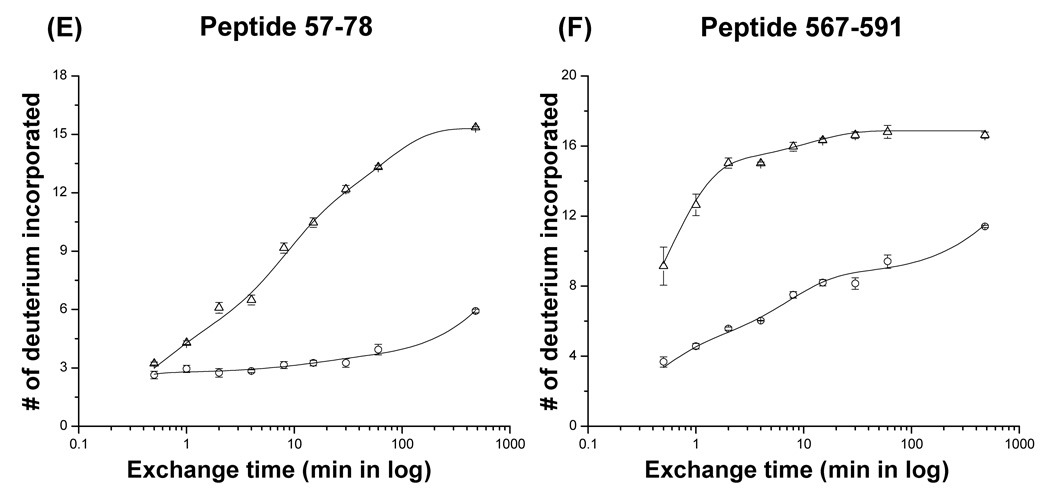
Plots of the number of deuterium atoms incorporated according to the exchange period for peptic fragments from the P22 portal monomers (open triangles) and complexes (open circles). The error bars represent the standard deviation for three independent measurements. The solid line represents the fit obtained by three component exponential fitting the exchange data (see materials and methods). Peptic fragments (A) 164–186, (B) 632–658, (C) 246–262, (D) 309–335, (E) 57–78, and (F) 567–591. Highest number on the y-axis represents the theoretical number of exchangeable amide protons.
The centroids of the isotopic distribution of representative peptides were plotted against exchange time (Figure 3). As was evident from the raw mass spectra, residues 136–159 remained un-exchanged even at late times (less than 5% exchanged in 8 hours), whereas the peptide spanning residues 160–185 exchanged rapidly (50% of exchangeable amide protons were exchanged within 1 min) and reached completion within 8 hours (Figure 3A, C).
The helices in the central domain of the Phi29 portal complexes, fragments 136–159 and 197–220 corresponding to helix 3 (α3) and helix 5 (α5), were well protected from exchange over the full exchange time (8 hours) suggesting that the helical bundles form very strong inter-subunit contacts consistent with their role in stabilizing the ring implied by the crystal structure (Figure 3A, B, and 4).19 The Phi29 portal complex has an additional long helix (helix 1, α1) in the central domain which presumably has similar interactions to the other helices (helices 3 and 5). Although we could not follow the exchange kinetics of this particular region, as will be discussed later the putative homolog was detected in the P22 portal complex.
In the crystal structure of the Phi29 portal complex, the N- and C-termini (residues 1–10 and 285–308, respectively) are unstructured.19 Peptides corresponding to residues 1–13 and 288–308 were detected in the digests and exchanged rapidly consistent with the X-ray crystallographic data (Figure 3D, E).
To map the exchange data onto the crystal structure we categorized each peptide as fast (red), intermediate (yellow), and slow (blue) exchanging based upon the exchange rate of the dominant component (Figure 4 and Supplement 1). For example, the peptide spanning residues 136–159 was categorized as a slow peptide because more than 90% of the exchangeable protons exchanged slowly. The outer edge of the wide-end domain and the entire narrow-end domain exchanged rapidly. In contrast, the helical bundles in the central domain and the inner face of the wide-end domain were well protected from exchange suggesting the helical bundles in the central domain interact strongly to form the stable ring like structure (Figure 4 and Supplement 1).
Hydrogen/deuterium exchange of the P22 portal monomers and ring complexes
There is no atomic structure of the P22 portal complex available. However, monomeric P22 portal protein subunits can be purified and assembled into rings in vitro.29 In vitro assembly was driven by adding 10 mM CaCl2 to 10 µM of monomeric portal proteins. Calcium induced assembly produces mixed populations of 11- and 12- subunit rings at approximately a 3:1 ratio.30 While electron microscopy studies demonstrated that portal proteins exclusively form dodecameric ring-like structures in situ,11–19,22,23 in vitro assembly studies of the portal proteins demonstrated that they can form ring-like structures with different numbers of subunits (for example, 11, 12, and 13 subunits) suggesting plasticity of inter-subunit interactions.30–35 However, crystal structures and cryo-EM data suggests that there are only slight morphological and inter-subunit interfacial differences between the various oligomers.17,20,21
We performed comparative hydrogen/deuterium exchange analysis of purified monomers and in vitro assembled ring complexes using the same procedures employed for the Phi29 portal complex. Because of the large size of the P22 portal protein (84 kDa) in comparison to the Phi29 portal protein (36 kDa) it generated three times as many peptic fragments. Digests of large proteins produce fragments of similar mass resulting in overlapping spectral peaks. Typically, peptic fragments are first separated by reverse phase liquid chromatography in front of the ESI-MS to reduce spectral complexity. However, in exchange experiments it is crucial to minimize the analysis time to limit back exchange. The high resolution and mass accuracy of the FT-ICR MS allowed us to resolve most of the peptides we were interested in without extensive chromatographic separation. The 100,000 resolving power allows us to distinguish peaks that lie within 0.05 m/z at 1,000 m/z and the 2 ppm mass accuracy allows us to unambiguously determine the peak to peak spacing and therefore which peaks belong to which isotopic distribution (Supplement 2).
Although FT-ICR is capable of determining the mass of peptic fragments to within 2 ppm, nonspecific digestion (using pepsin) of large proteins like P22 portal protein can in principle generate a large number of isobaric fragments or fragments within 2 ppm of one another. In the case of the P22 portal proteins, we used 43 peptides to follow the exchange kinetics. Among these 43, there were 7 isobaric candidates found in the amino acid sequence and 13 peptides which have two or more candidates within 2 ppm of one another. Therefore, it was necessary to unambiguously identify the fragments using a combination of accurate mass measurement and MS/MS sequencing (Supplement 3). By taking advantage of the hybrid ion trap-FT ICR MS, we could follow exchange kinetics of peptic fragments which covered approximately 90% of 732 residue P22 portal primary sequence (Figure 5A).
Predicted helical modules of the P22 portal proteins were protected from exchange upon ring assembly
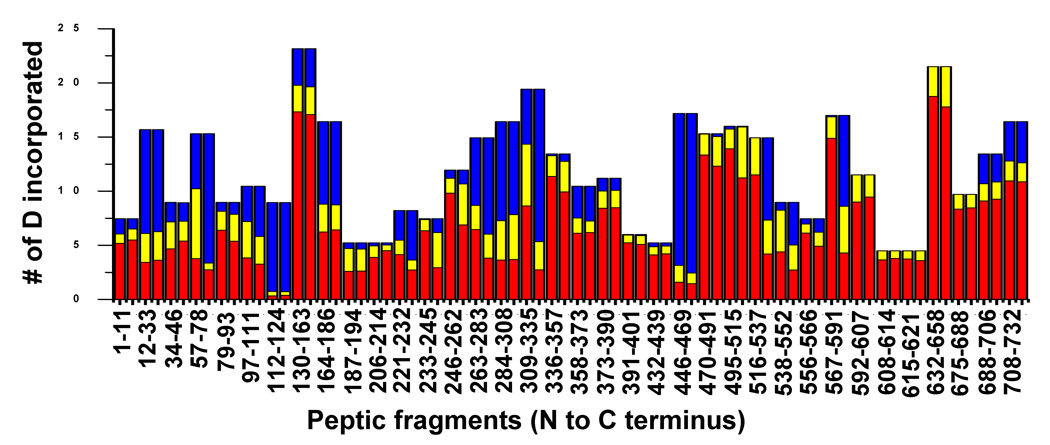
The mass spectra of the peptic fragment spanning residues 632–658 and 309–335 in both the monomers and ring are overlayed in Figure 5B and C, respectively. While the peptide derived from residues 632–658 exchanged rapidly and identically in both forms, the peptide derived from residues 309–335 exchanged rapidly in the monomers but was well protected in the rings (Figure 5B, C). To identify the peptides whose protection changed upon assembly the centroid of the isotopic distribution was plotted against the exchange time for each peptide found as a monomer/complex pair, and fitted with a three component exponential model as described above (Figure 6, 7). Most of the fragments showed very similar exchange patterns in both forms (Figure 7). For example, the progress curves for peptides 164–184 and 632–658 were unchanged upon assembly (Figure 6A, B). Previous genetic and cryo-EM image reconstruction studies demonstrated that deletion of the C-terminal 130 residues does not affect ring formation.16 All the fragments belonging to the C-terminal 140 residues (592–732) of the P22 portal protein showed exactly the same exchange profiles in both monomers and the ring (Figure 6B, 7) consistent with the suggestion that they do not participate in inter-subunit interactions in the ring. The fact that so many peptides were unchanged between the two forms suggests that there was no significant global conformational change upon assembly.
Figure 7.
Overall pair-wise hydrogen/deuterium exchange comparison between the P22 portal monomers and complexes. Each bar represents the relative contributions of the three components fast (red), intermediate (yellow) and slow (blue) with exchange rate constants k1 (>1 min−1), k2 (0.01 min−1 – 1 min−1) and k3 (<0.01 min−1), respectively.
Despite the overall similarity of the exchange profile several regions, including the predicted helical modules, did show significant changes upon ring assembly (Figure 6C, D, 7). Approximately one hundred residues (240–340) of the P22 portal protein are predicted to have a very similar secondary structure arrangement (Figure 1B and Figure 8) to the helical bundles in the central domain of the Phi29 portal complexes.18,24 The peptides, 246–262 and 309–335, which lie within the predicted helical elements showed a pronounced increase in protection upon ring formation. In the monomer, 90% of the exchangeable amide protons in the peptide 246–262 exchanged within 1 min and exchange was completed in 4 min. In the ring, a smaller fraction exchanged rapidly and the rest were well protected, exchanging gradually and only converging after for about 8 hours (Figure 6C). The peptide spanning residues 309–335 also showed protection in the ring. In the monomer, this peptide reached nearly complete exchange after 8 hours while in the it was only approximately 30% exchanged after 8 hours (Figure 6D).
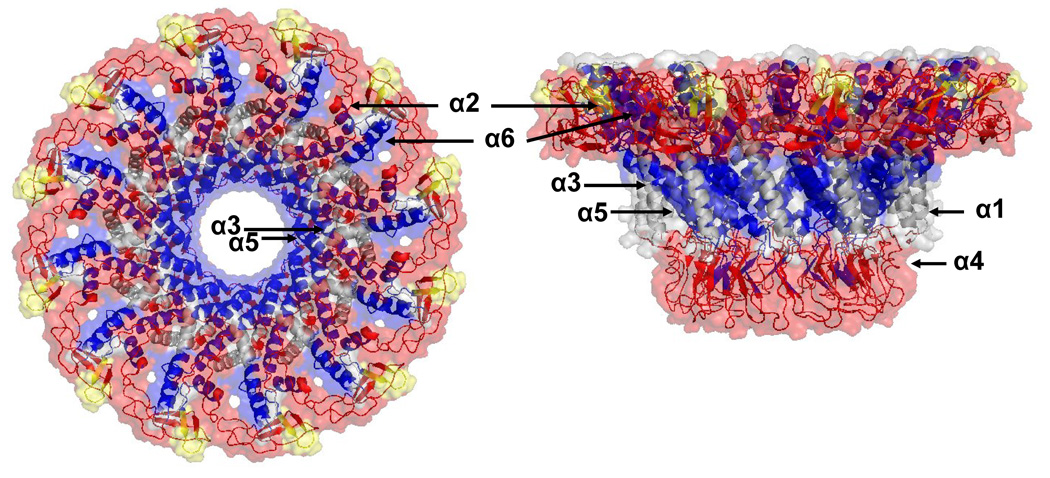
Figure 8.
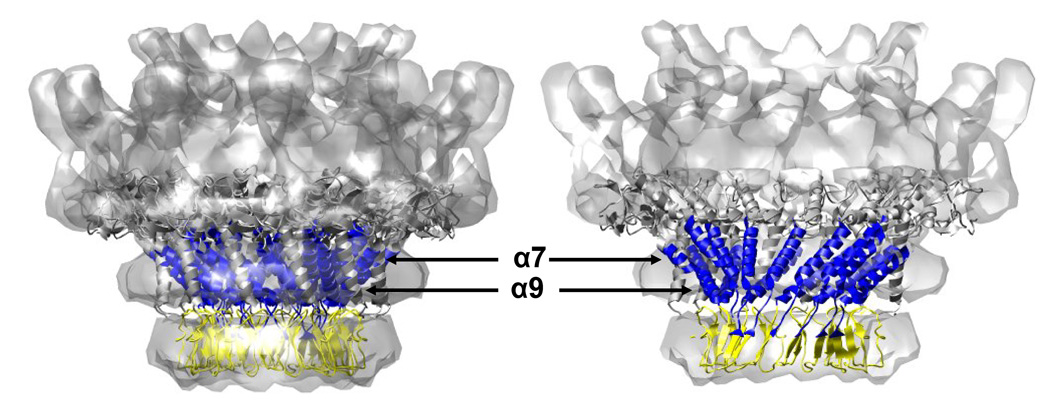
The crystal structure of the Phi29 connector was docked into the cryoEM reconstruction of the P22 portal as in Figure 1. Using the alignment of predicted helical regions of P22 with the known helical regions of Phi29 (as shown in Figure 1) the extent of exchange protection observed upon P22 portal assembly was mapped onto the structure. Blue represents peptides showing protection upon assembly, yellow represents no appreciable change upon assembly, and grey represents regions that lie outside of the putative conserved domain.
The central domain of the Phi29 portal complex consists of three long helices (helices 1, 3, and 5). Helix 1 is located at the very end of the N-terminus of the protein (residues 17–35) and this peptide was not detected in our experiments. In the crystal structure it makes close contacts with helices 3 and 5 forming a helical bundle in the central domain of the Phi29 portal complex. In the P22 portal hydrogen/deuterium exchange experiments, a significantly different exchange pattern was observed in the N-terminal region (Figure 6E, 7). The peptide 57–78 which is also predicted to be long helix exchanged gradually in the monomer reaching completion after 8 hours. In contrast, it was well protected in the ring reaching only approximately 40% exchanged after 8 hours (Figure 6E, 7). This result suggests that residues 57–78 in the P22 portal protein may correspond to helix 1 of Phi29.
The exchange data for peptides 57–78, 246–262, and 309–335 in the monomers and the rings suggests that the threading model is essentially correct, and that the helices are stable in the monomer, but upon assembly interactions between these helices further increases their stability. This interpretation is consistent with spectroscopic data that indicates that a small increase in helicity accompanies assembly.24,29
Additional protected regions were observed in the P22 portal complexes
When the crystal structure of the Phi29 portal ring is positioned in the cryo-EM reconstruction of the P22 portal complex, the density corresponds well with the bottom half of the P22 portal ring (Figure 1A). In the P22 ring the wing domain (or wide end domain) is twice as thick as in Phi29 and presents an additional density on the top termed the crown domain. The P22 portal protein has 424 more residues than does Phi29 (732 vs. 308 AA) (Figure 1) and these additional residues were proposed to comprise the additional wing and crown domain density.16,17 As the 3-D alignment and hydrogen/deuterium exchange data suggested that the N-terminus and the predicted conserved helical modules (up to 400 residues) likely correspond to the narrow end domain and the lower half of the wing domain, it is reasonable to consider that the C-terminal 350 residues correspond to the top of the wing and crown domains observed in the P22 cryo-EM reconstructions.
While the very C-terminal 140 residues (592–732) of the P22 portal protein showed rapid and unchanged exchange in both forms the upstream 100 residues (495–591) showed increased protection in the ring (Figure 7). For example, the peptide spanning residues 567–591 in the monomer was more than 90% exchanged within 2 min whereas in the ring it was well protected at early times and exchanged only gradually (Figure 6F). This result suggests that residues 567–591 make new contacts upon assembly and may contribute to the electron density of either the crown domain or the top half of the wing domain in the P22 portal complexes.
Discussion
Although comparisons of genome or protein sequences have been widely used to compare and classify lineages of viruses, close relationships have been detected among viruses even in the absence of sequence homology based on structural similarity.3–18 While structural data from spectroscopic techniques such as circular dichroism and Raman spectroscopy is available, the structures of relatively few viral proteins have been solved at the atomic level. For example, the only portal structures solved at atomic resolution are those of Phi2919 and SPP1.20 However, cryo-EM structures of portal complexes are available for several other family members and their structures demonstrate that they share a similar ring-like morphology.11–18 While cryo-EM reconstruction can provide information on the global morphology and topology of protein complexes, even in the most favorable cases the achievable resolution makes it difficult to directly identify the amino acids involved in subunit-subunit contacts. Mass spectrometry based hydrogen/deuterium exchange represents a good complement to cryo-EM because of its ability provide region specific dynamic information of supramolecular complexes such as viruses.36–40
In this study the exchange profiles of Phi29 and P22 portal complexes were monitored by mass spectrometry and compared to identify the amino acids involved in subunit-subunit contacts. For Phi29, significant exchange protection was seen in helices 3 and 5 which form the stable central domain of the Phi29 portal complex (Figure 3A, B, and 4). A similar degree of protection was observed in the regions of the P22 portal protein predicted to form helices and suggested to form the conserved helical bundles (Figure 8). The fact that the protection in this region increased upon the association of the subunit into rings further supports the hypothesis that these regions lie at the site of inter-subunit contacts (Figure 6C, D). The presumptive regions of intersubunit contact implicated by hydrogen/deuterium exchange experiments agree well with biochemical data obtained by Raman spectroscopy, proteolytic digestion, and mutational analysis. Replacement of cysteine 283 with serine results in a decreased ability of the portal to oligomerize and decreased infectivity whereas similar mutations as cysteines 153 and 173 do not. Similarly, Raman spectroscopy studies suggested that the central region of the P22 portal sequence is involved in inter-subunit interactions upon assembly because the most significant changes accompanying assembly were detected in the tyrosine markers arising from the central region (residues 223–571), rather than the C-terminal region (residues 572–732).24
The P22 portal protein which has 424 more residues than Phi-29 also presents with a crown domain and thickened wing domain similar to the SPP1 portal complex.16,17 The crown domain of SPP1 is composed of three short α-helices located near the C-terminus of the subunit. Although the C-terminal 140 residues (592–732) in the P22 portal protein were predicted to be α-helical, no increase in exchange protection upon assembly was observed for these residues suggesting this region does not participate in inter-subunit contacts (Figure 6B, 7). This suggestion is consistent with deletion studies16 that demonstrate that the C-terminal 140 residues are not critical for ring assembly. Two additional clusters of four consecutive short α-helices (residues 390–470 and 520–590) were predicted to lie near the C-terminus. Residues 390–470 showed no increase in protection upon assembly while residues 520–590 did (Figure 6F, 7). Therefore, we tentatively assign residues 520–590 as contributing to the crown domain. As shown in the SPP1 portal crystal structure, the rest of additional residues may contribute to the extra electron density of the wing domain.16,17,20
Although understanding the protein-protein interactions and conformational changes in large macromolecular complexes is crucial to understanding their biological function in the cell, doing so represents a substantial challenge for structural biology due to the difficulties of obtaining high resolution structures. In this report, we used hydrogen/deuterium exchange to obtain experimental evidence in support of a structural model for P22 portal derived from threading the predicted secondary structure onto the crystal structure of a related protein complex. This is a logical extension of the use of H/D exchange to identify protein/protein interaction sites and the use of H/D exchange to constrain threading models as pioneered by the Komives lab to the case of a supramolecular structures which display similar gross morphology and putative common elements of secondary structure in the absence of identifiable sequence similarity.
Materials and methods
Preparation of Phi29 portal complexes
The Phi29 portal complexes were over-expressed in E. Coli BL-21 from the plasmid pPLc28D inducing by heat shock. The proteins were purified as described previously with several modifications.41 Briefly, the cells were collected at four hour post induction and resuspended in cell lysis buffer (0.3 M NaCl, 50 mM Tris, pH 7.7). Resuspended cell pellets were treated with 200 ng/ml lysozyme and 50 ng/ml DNase and RNase on ice. The cell debris was removed by centrifugation at 12,000g for 45min and the supernatant was further purified with HiTrap SP HP column (Amersham) with a NaCl gradient of 0.3 – 1 M in 50 mM Tris, pH 7.7. The proteins were eluted at 0.75 M NaCl and dialyzed against buffer containing 0.3 M NaCl and 50 mM Tris, pH 7.7 to reduce ionic strength. Dialysed samples were applied on the HiTrap Q HP column and eluted with 0.3 – 1 M NaCl gradient. The proteins were eluted around at 0.43 M NaCl and equilibrated with reaction buffer (0.4 M NaCl, 50 mM Tris, pH 7.7) through dialysis. Molecular mass of purified Phi29 portal protein was measured by ESI time-of-flight (TOF) equipped with Z-spray (Micromass LCT, Waters). Mass of 35,746.0 Da was observed in good agreement with the theoretical mass of 35,747.2 Da.
Preparation of P22 portal proteins
The C-terminally his-tagged P22 full-length portal protein was over-expressed and purified as described previously with several modifications.29 Briefly, the P22 portal protein was over-expressed in E.coli BL-21 and purified by IMAC using 5 ml HiTrap chelating Sepharose column (Amersham) as described previously. Eluate from HiTrap column was dialyzed against 50 mM Tris, 50 mM NaCl, 2 mM DTT, 10 mM EDTA overnight at 4 °C. The monomeric P22 portal proteins were separated from other oligomeric forms on the 1 ml anion exchange HiTrap Q Sepharose column (Amersham) by applying linear gradient of 10 column volumes of buffer containing 1 M NaCl and 1 mM EDTA. Fractions containing monomers were collected, concentrated by ultrafiltration, and frozen at −80°C. ESI-TOF analysis of the P22 portal protein provided protein mass of 83,664.0 Da in good agreement with the value of 83,666.6 Da predicted for P22 portal protein.
Oligomerization of the P22 portal proteins into the ring complexes was induced by addition of CaCl2 to the final concentration of 10 mM.30 The reaction was left at room temperature for 1 hour and oligomeric forms were separated and collected by using 1 ml HiTrap Q Sepharose column (Amersham) as described above except buffer where 1 mM EDTA was replaced by 10 mM CaCl2. The purified P22 portal complexes were concentrated by ultrafiltration and frozen at −80°C in aliquots. Presence of portal rings in the final prep was confirmed by size exclusion chromatography, electron microscopy, and native ESI MS.30
Hydrogen/deuterium exchange experiments
Hydrogen/deuterium exchange experiments were performed as described previously for various viral capsids with slight modifications.37 The Phi29 portal complexes and the P22 portal proteins were exchanged by 10 fold dilution into each deuterated buffer (0.4 M NaCl, 50 mM Tris, pH 7.7 for Phi29 and 50 mM Tris, 50 mM NaCl, 2 mM DTT, with or without 10 mM CaCl2 for P22 portal proteins) to reach 90% final concentration at 21 °C and sampled at zero time, 30 seconds, one, two, four, eight, 15 and 30 minutes, one, and eight hours. The exchange reactions were quenched by the addition of formic acid to 1%, immediately flash frozen in liquid nitrogen, and stored at −80 °C until analysis. The samples were thawed rapidly, mixed with an equal volume of pepsin (~10 µM finally), injected into 20 µl loop, and digested for one minute on ice. Digested samples (~50 pmol) were desalted and separated onto a C4 trap (Michrom BioResources, Inc.) and introduced into the ESI source. The injection valve, C4 trap and tubing were submerged completely in an ice bath. The peptides were rapidly eluted with a 5–95% acetonitrile gradient (50 µl/min Agilent 1100 capillary pump). Exchange mass analyses were performed on an ESI hybrid linear quadrupole ion trap (LTQ)-FT ICR mass spectrometer (LTQ-FT, Thermo Finnigan, San Jose CA.). The LTQ-FT mass spectrometer was set to collect FT-ICR MS scans only (300 – 2000 m/z) with FT-ICR only mode. The amount of deuterium incorporated was determined by calculating the centroid of the isotopic distributions. The eluted peptides were identified by combination of exact mass measurement (< 2 ppm) with 7T FT-ICR MS scans and collision induced dissociation (CID) tandem mass spectrometry (MS/MS) sequencing with the LTQ‥
Progress curves for individual peptides were fitted to a sum of three exponentials derived from the exchange rate expression.25
Where D is the observed number of deuterium atoms incorporated at time t and N is the corrected total number of exchangeable protons. N for each peptide is corrected by multiplying theoretical number of exchangeable amide protons by 0.8 which accounts for back-exchange (20% back-exchange, see below). The theoretical number of exchangeable amide protons of each peptide was calculated by multiplying number of exchangeable amide protons (total number of amino acid of the peptide – 1 – number of proline) by 0.9, because deuteration is achieved by 10-fold dilution of samples into deuterated buffer and maximized deuteration reached 90% in the buffer. The exchange extent of rapidly exchanging peptides at the final time point (8 hrs) was evaluated, compared with the theoretical number of exchangeable amide protons, and the average back exchange was estimated as 20%. a, b and c represent the number of amino acids that exchange with rate constant k1 (fast, >1.0 min−1), k2 (intermediate, 0.01–1.0 min−1) and k3 (slow, <0.01 min−1) respectively.
Supplementary Material
S1. Hydrogen/deuterium exchange rates of individual peptic fragments of the Phi29 portal complexes. Each bar represents the relative contributions of the three components fast (red), intermediate (yellow) and slow (blue) with exchange rate constants k1 (>1 min−1), k2 (0.01 min−1 – 1 min−1) and k3 (<0.01 min−1), respectively.
S2. High resolution FT-ICR mass spectrum of two peptides (34–46 and 538–552) generated from peptic digests of P22 portal proteins. Overlapped isotopic distributions (left panel) and enlarged view of mass spectrum showing the capability of FT-ICR to resolve peaks separated by as little as 65 × 10−3 u (right panel).
S3. Identification of the peptic fragments using accurate mass measurement and MS/MS mass tagging. (A) Comparison between the observed mass spectrum (top panel) and simulated mass spectra of candidate peptides (residues 446–469 (middle) and 499–523(bottom)). Monoisotopic mass is observed at m/z = 940.12037 and two candidate peptides have same chemical composition with monoisotopic mass at m/z = 940.12034. Mass errors between the observed and the candidates are less than 1 ppm. (B) Ion trap MS/MS of precursor [940.12037]3+ ion. Corresponding fragment peaks are assigned as indicated on the top sequence. Identity of ion is unambiguously determined by MS/MS mass tagging.
Acknowledgement
This work was supported by NIH grant GM47980 (P.E.P) and SS RR-17261. Funds for the operation of the UAB Biomedical FT-ICR MS Laboratory were provided in part by the Supporters of the UAB Comprehensive Cancer Center and the Department of Biochemistry and Molecular Genetics. We wish to thank Gabe Lander and Jack Johnson for supplying the EM based density of P22 portal dodecamers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Casjens S, King J. Virus assembly. Ann. Rev. Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- 2.Chiu W, Burnett R, Garcea R. Structural biology of viruses. Oxford: University Press, New York; 1997. [Google Scholar]
- 3.Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PE, Jr, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 4.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage Phi29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 5.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 6.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Nat. Acad. Sci. U.S.A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct.l Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 8.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol. Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SHW, Carazo JM, Carrascosa JL. Quasi-Atomic Model of Bacteriophage T7 Procapsid Shell: Insights into the Structure and Evolution of a Basic Fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Baker ML, Jiang W, Rixon FJ, Chiu W. Common Ancestry of Herpesviruses and Tailed DNA Bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 2001;310:1027–1037. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- 13.Carrascosa JL, Carazo JM, Ibanez C, Santisteban A. Structure of phage phi 29 connector protein assembled in vivo. Virology. 1985;141:190–200. doi: 10.1016/0042-6822(85)90251-x. [DOI] [PubMed] [Google Scholar]
- 14.Donate LE, Herranz L, Secilla JP, Carazo JM, Fujisawa H, Carrascosa JL. Bacteriophage T3 connector: three-dimensional structure and comparison with other viral head-tail connecting regions. J. Mol. Biol. 1988;201:91–100. doi: 10.1016/0022-2836(88)90441-x. [DOI] [PubMed] [Google Scholar]
- 15.Kochan J, Carrascosa JL, Murialdo H. Bacteriophage lambda preconnectors : Purification and structure. J. Mol. Biol. 1984;174:433–447. doi: 10.1016/0022-2836(84)90330-9. [DOI] [PubMed] [Google Scholar]
- 16.Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. The EMBO J. 2005;24:2087–2095. doi: 10.1038/sj.emboj.7600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The Structure of an Infectious P22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 18.Agirrezabala A, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. The EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage Phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron Cryotomography Reveals the Portal in the Herpesvirus Capsid. J. Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Casado A, Moore SD, PE P, Thomas GJ. Structure of bacteriophage P22 portal protein in relation to assembly: investigation by Raman spectroscopy. Biochemstry. 2001;40:13583–13591. doi: 10.1021/bi0110488. [DOI] [PubMed] [Google Scholar]
- 25.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Quart. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 26.Truhlar SME, Croy CH, Torpey JW, Koeppe JR, Komives EA. Solvent Accessibility of Protein Surfaces by Amide H/2H Exchange MALDI-TOF Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2006;17:1490–1497. doi: 10.1016/j.jasms.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truhlar SME, Torpey JW, Komives EA. Regions of I{kappa}B{alpha} that are critical for its inhibition of NF-{kappa}B{middle dot}DNA interaction fold upon binding to NF-{kappa}B. Proc. Nat. Acad. Sci. U.S.A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore SD, Prevelige PE., Jr Structural Transformations Accompanying the Assembly of Bacteriophage P22 Portal Protein Rings in Vitro. J. Biol. Chem. 2001;276:6779–6788. doi: 10.1074/jbc.M007702200. [DOI] [PubMed] [Google Scholar]
- 30.Poliakov A, Duijn Ev, Lander G, Fu C-y, Johnson JE, Prevelige J, Peter E, Heck AJR. Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J. Struct. Biol. 2007;157:371–383. doi: 10.1016/j.jsb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the Connector of Bacteriophage T7 at 8 A Resolution: Structural Homologies of a Basic Component of a DNA Translocating Machinery. J. Mol. Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Jekow P, Behlke J, Tichelaar W, Lurz R, Regalla M, Hinrichs W, Tavares P. Effect of the ionic environment on the molecular structure of bacteriophage SPP1 portal protein. Eur. J. Biochem. 1999;264:724–735. doi: 10.1046/j.1432-1327.1999.00601.x. [DOI] [PubMed] [Google Scholar]
- 33.Orlova EV, Dube P, Beckmann E, Zemlin F, Lurz R, Trautner TA, Tavares T, van HeeL M. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 1999;6:842–846. doi: 10.1038/12303. [DOI] [PubMed] [Google Scholar]
- 34.Valpuesta JM, Sousa N, Barthelemy I, Fernandez JJ, Fujisawa H, Ibarra B, Carrascosa JL. Structural Analysis of the Bacteriophage T3 Head-to-Tail Connector. J. Struct. Biol. 2000;131:146–155. doi: 10.1006/jsbi.2000.4281. [DOI] [PubMed] [Google Scholar]
- 35.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and Polymorphism of the UL6 Portal Protein of Herpes Simplex Virus Type 1. J. Virol. 2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mo. Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 37.Kang S, Prevelige PE., Jr Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J. Mol. Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Lisal J, Lam TT, Kainov DE, Emmett MR, Marshall AG, Tuma R. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat. Struct. Mol. Biol. 2005;12:460–466. doi: 10.1038/nsmb927. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Lane LC, Smith DL. Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001;10:1234–1243. doi: 10.1110/ps.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu C, Prevelige P. Dynamic motions of free and bound phi29 scaffolding protein identified by hydrogen deuterium exchange mass spectrometry. Protein Sci. 2006;15:731–743. doi: 10.1110/ps.051921606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibanez C, Garcia JA, Carrascosa JL, Salas M. Overproduction and purification of the connector protein of Bacillus subtilis phage Phi29. Nucleic Acid Res. 1984;12:2351–2365. doi: 10.1093/nar/12.5.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Hydrogen/deuterium exchange rates of individual peptic fragments of the Phi29 portal complexes. Each bar represents the relative contributions of the three components fast (red), intermediate (yellow) and slow (blue) with exchange rate constants k1 (>1 min−1), k2 (0.01 min−1 – 1 min−1) and k3 (<0.01 min−1), respectively.
S2. High resolution FT-ICR mass spectrum of two peptides (34–46 and 538–552) generated from peptic digests of P22 portal proteins. Overlapped isotopic distributions (left panel) and enlarged view of mass spectrum showing the capability of FT-ICR to resolve peaks separated by as little as 65 × 10−3 u (right panel).
S3. Identification of the peptic fragments using accurate mass measurement and MS/MS mass tagging. (A) Comparison between the observed mass spectrum (top panel) and simulated mass spectra of candidate peptides (residues 446–469 (middle) and 499–523(bottom)). Monoisotopic mass is observed at m/z = 940.12037 and two candidate peptides have same chemical composition with monoisotopic mass at m/z = 940.12034. Mass errors between the observed and the candidates are less than 1 ppm. (B) Ion trap MS/MS of precursor [940.12037]3+ ion. Corresponding fragment peaks are assigned as indicated on the top sequence. Identity of ion is unambiguously determined by MS/MS mass tagging.