Abstract
“RNA bandages” are composed of two 6-12-mer 2′-OMe RNA strands complementary to a mRNA target that are joined by a photocleavable linker. These tandem oligonucleotides typically exhibit much higher affinity for the mRNA than the individual strands. An RNA bandage with binding arms of different lengths and a 4-base gap blocked translation in vitro of GFP mRNA; subsequent near-UV irradiation restored translation. This provides a general method of photomodulating hybridization for a variety of oligonucleotide-based technologies.
Photochemical methods for controlling oligonucleotide function have become increasingly important in biological research.1 Light-activated (“caged”) oligonucleotides have been used to regulate DNA hybridization,2–4 polymerase,5–9 ribozyme,10, 11 DNAzyme,12, 13 aptamer,14 and RNase H activity,4, 15 RNA folding,16 and RNA interference,17, 18 as well as gene expression in cells and embryos.17, 19–23 Most caging groups for biological use involve the nitrobenzyl (NB) moiety and its derivatives, which allow for removal at relatively long wavelengths (~365 nm) without harmful side products. Previously, our lab used a NB-containing photocleavable linker to join an antisense oligodeoxynucleotide (asODN) to a much shorter complementary strand, which controls binding of the asODN to an mRNA target. For example, in human leukemia cells, photoactivation of a caged asODN initiated the degradation of c-myb mRNA by an RNase H-dependent mechanism.21 And in zebrafish embryos, caged asODNs were shown to block expression of chordin and bozozok, upon irradiation.20
While these caged asODNs succeeded in turning gene expression “off” after photolysis, a related technique for turning gene expression “on” would be equally useful. The timing and location of protein expression within the cell has profound consequences for proper cellular development, and light-activated control of mRNA would allow temporal and spatial analysis of protein function. Herein, we describe the synthesis of an “RNA bandage”, in which tandem oligonucleotides are joined by a photocleavable linker. The bandage binds and protects the target mRNA from translation, until UV irradiation cuts the bandage (Fig. 1).
Figure 1.
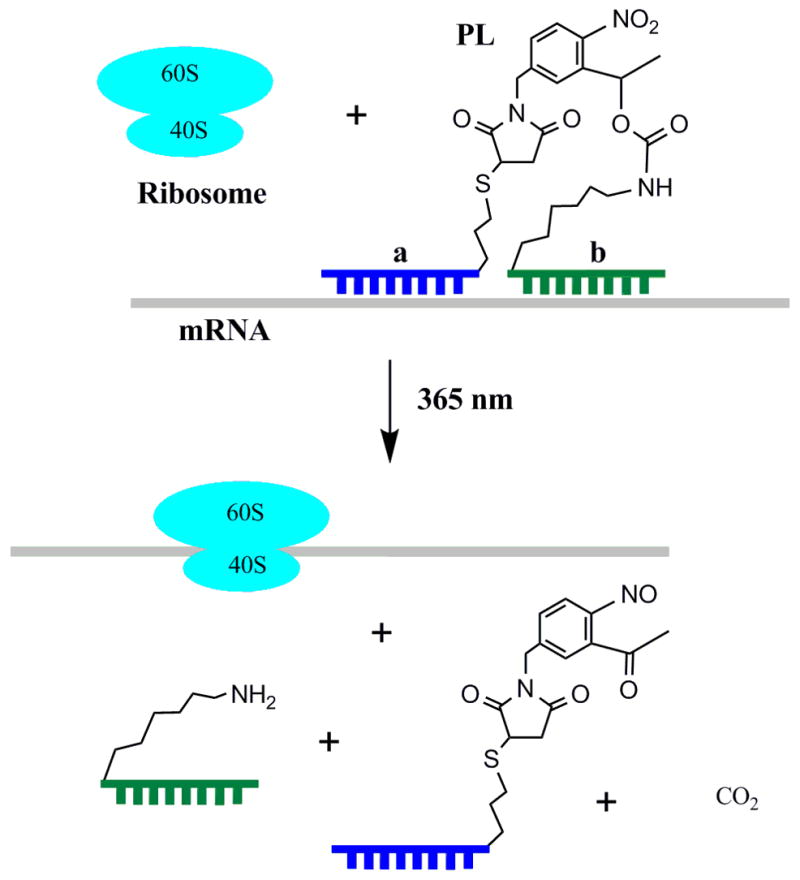
RNA bandage (a-PL-b) binds mRNA and blocks protein synthesis until cut with UV light.
Ando et al. generated the first caged mRNA by statistically labeling the phosphodiester backbone with a large number of coumarin photoactive blocking groups, thereby perturbing mRNA structure and preventing protein synthesis in zebrafish.22 However, UV photolysis yielded relatively little “active” mRNA, due to the low quantum efficiency of removing dozens of blocking groups on a single oligonucleotide. In contrast, RNA bandages seek to cage mRNA function by employing a single, site-specific photoactive group. This strategy has several advantages: shorter irradiation times, more efficient synthesis, and purification of a light-activated oligonucleotide that can target the 5′ untranslated region (UTR) of any complementary mRNA molecule. This method has the potential to allow photomodulation of endogenous cellular mRNA, as well as exogenously supplied mRNA.
RNA bandages consist of two short antisense oligonucleotides that are both complementary to a target mRNA (Fig. 1). Many structural modifications of antisense oligonucleotides have been identified that improve mRNA binding affinity and specificity.24 For example, 2′-O-alkylation of the RNA ribose ring improves hybridization and nuclease resistance. Specifically, 2′-O-methyl (2′-OMe) RNA has advantages of ease and low cost of synthesis and has been used extensively for antisense targeting.24
Previously, 2′-OMe RNA antisense molecules as short as 14–16 bases in length were shown to inhibit completely the translation of a complementary mRNA in rabbit reticulocyte lysate.25 The antisense oligonucleotides were only effective when directed towards the 5′ UTR and the first 20 bases of the coding region. It was hypothesized that the 2′-OMe RNA-mRNA duplex acted to block the ribosome in a mechanism similar to that of intramolecular mRNA hairpins.26 These hairpins located in the 5′ UTR affect the early stages of translation initiation. Therefore, our antisense sequences were designed to target the start codon and the Kozak sequence in the 5′ UTR, which are important for efficient translation.
The synthesis of RNA bandages was carried out in two steps using the 1-(5-(N-maleimidomethyl)-2-nitrophenyl)ethanol N-hydroxysuccinimide ester photocleavable linker (PL).4 First, the b strand, a 2′-OMe RNA oligonucleotide with 5′ amine modification, was reacted with PL in DMSO and TEA for 1 h and then purified by reverse-phase HPLC. The PL-b conjugate was then reacted in phosphate buffer for at least 4 h with 2-fold excess of a strand, a 2′-OMe RNA oligonucleotide containing 3′ thiol modification. Afterwards, the reaction was treated for 2 h with 0.1 M DTT to reduce any possible disulfides. The pure product (a-PL-b) was isolated by RP-HPLC. Each step occurred in 50% yield, with HPLC traces generally showing complete consumption of starting material.
Conjugates 1–4 (Table 1) were synthesized, which were varied in sequence to generate large differences in melting temperature (ΔTm) between the bandages (a-PL-b) and their individual a and b strands in binding an RNA target (Tg). Tg comprises the bases 1–26 of a transcribed full-length GFP mRNA targeted by the bandages in subsequent in vitro translation assays. Conjugates 1–3 showed ΔTm’s of 32, 20, and 24 °C, indicating greater photomodulation than is typical for light-activated oligonucleotides that employ multiple caging moieties: ΔTm = 11.0–21.5 °C, in duplexes with targets of similar length as Tg.2, 13, 15
Table 1.
Bandage sequences and melting temperatures in °C, hybridized to target RNA (Tg).
| Bandage | Sequence 5′-(a-PL-b)– 3′ | Bases Targeted | Tm | Tm(a-PL-b) | ΔTma | ||
|---|---|---|---|---|---|---|---|
| A | b | a | b | ||||
| 1 | CAUGGUGUCUAGCCAG b | 24 ← 17 | 16 ← 9 | 28.2 | 30.1 | 62.2 | 32 |
| 2 | GCCAUGGUGUCUAGCCAGCU | 26 ← 17 | 16 ← 7 | 38.2 | 52.8 | 72.4 | 20 |
| 3 | CAUGGUGUCUAGCCAGCUUGGGUC | 24 ← 13 | 12 ← 1 | 42.4 | 57.3 | 81.2 | 24 |
| 4 | UGGUGU_ _ _ _CCAGCUUGGGUC | 22 ← 17 | 12 ← 1 | < 20 | 57.3 | 63.4 | 6 |
| Tg | 3′ – CGGUACCACAGAUCGGUCGAACCCAG – 5′ | ||||||
ΔTm represents the difference between Tm (a-PL-b) and the Tm of the a or b strand, whichever has the higher Tm when hybridized to Tg.
The a strand is in regular font and the b strand is in bold, with the photocleavable linker (PL) connecting the 3′ end of a with the 5′ end of b.
Initially, the individual a and b strands of each RNA bandage were tested for their ability to block translation of a capped GFP mRNA transcript in rabbit reticulocyte lysate using the radiolabel S-35 methionine to monitor protein production. The antisense strands were used in moderate excess over the target mRNA (less than 20:1), in order to limit nonspecific binding. The only strand found to block translation in these studies was the 12-mer oligonucleotide 3a covering the start codon and nine bases upstream (bases 13–24). Secondary structure prediction by RNAstructure 4.527 showed that the 5′ region of the mRNA forms a stem-loop with the a and b strands mostly targeting the loop and stem, respectively (Fig. 2). Based on this prediction, the stem bases 8–15 and 23–30 of the mRNA should be less accessible than bases 16–22 of the loop or the unstructured regions at the 5′ end, 1–7. Loop structures have been previously shown to be good targets for antisense inhibition.28
Figure 2.
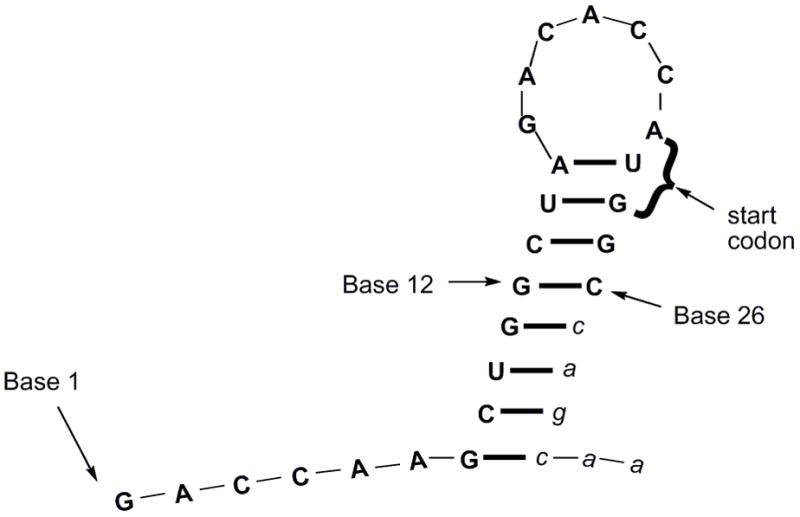
Secondary structure prediction of first 32 bases of GFP mRNA target sequence. Bases 27–32 (lowercase letters) extend beyond Tg and were not targeted by the RNA bandage, but likely contribute to the secondary structure of the full-length mRNA.
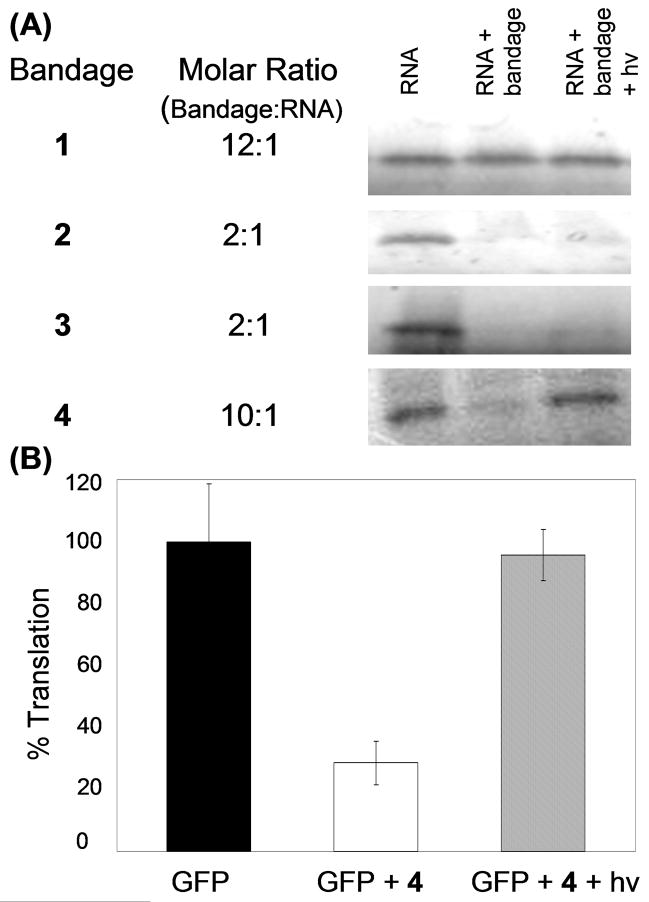
The conjugates were subsequently tested, and 1, which targeted the same bases as 3a plus bases 9–12, only reduced translation by 10% at 12-fold excess over GFP mRNA (Fig. 3A). UV irradiation was performed with a transilluminator (5 min, 9 mW/cm2 at peak intensity, 365 nm) as previously optimized for this PL,4 but translation did not return to original levels. Notably, the Tm of 1 was 20 °C higher than that of 3a (Table 1). These results indicated that strands 1a and 1b work cooperatively when joined by PL to increase the bandage-mRNA duplex stability, yet the ribosome readily displaces 1. Thus, longer and more stable 20-and 24-base bandages (2 and 3) covering different regions of Tg were designed, which succeeded in blocking in vitro protein synthesis at 2-fold excess over the GFP mRNA. However, UV irradiation failed to restore translation with either bandage (Fig. 3A).
Figure 3.
(A) Analysis of S-35 methionine-labeled translation products from in vitro translation assay by denaturing polyacrylamide gel electrophoresis. Lane 1 is GFP mRNA transcript only. Lane 2 is the GFP mRNA and the bandage in the indicated excess over mRNA. Lane 3 is the GFP mRNA and bandage after 5-min UV irradiation. (B) Quantification of average percent GFP translation for bandage 4 with and without UV irradiation, normalized to no-bandage control. Error bars indicate standard deviation from three trials.
To address the restoration problem, a bandage 4 was designed that incorporated 6- and 12-base strands with a 4-base gap in the middle. This bandage targeted a 22-base region of the mRNA, and created similar steric bulk to 2 and 3, while incorporating a 6-mer a strand of much lower thermal stability. This asymmetric bandage was designed to promote strand dissociation after photoactivation. At a 10-fold excess over mRNA, 4 reduced translation to 30% of original protein expression levels (Fig. 3B). Most importantly, upon irradiation, activity was restored to 95% of original values. Inclusion of a control luciferase mRNA in the reaction confirmed that the bandage’s effects on translation were specifically targeted to the GFP mRNA. Other controls showed that UV irradiation of the GFP mRNA did not affect translation efficiency.
The ability of an RNA bandage to block the ribosome is clearly related to thermodynamics, as the conjugates with highest melting temperature (2 and 3, Table 1) are effective at only a 2:1 ratio with mRNA. The structures of Tg, the GFP mRNA, and bandage play more subtle roles, as 4 (Tm = 63.4 °C) was much more effective at blocking translation than 1 (Tm = 62.2 °C). Furthermore, there is no simple correlation between an RNA bandage’s ability to modulate translation and the difference in melting temperature (Δ Tm) between a-PL-b and the more stable a or b strand (Table 1). In fact, bandage 4 with the lowest Δ Tm proved most effective at photoregulating translation. With this GFP mRNA as the target, the best bandage design was to position a very low Tm a strand at the start codon and a high Tm b strand several bases away. We infer that the a strand serves to bind the start codon and inhibit translation initiation, as long as the attached b strand provides high affinity for the target mRNA. This asymmetric tandem structure promotes the dissociation of the bandage from the mRNA target after photoactivation.
We have shown that tandem a-PL-b oligonucleotides can be designed to have higher affinity for a target RNA sequence than the individual a and b strands. However, uncertainties about RNA secondary structure and the mechanisms by which antisense oligonucleotides block translation make it difficult to identify targets and design RNA bandages simply from duplex melting temperatures. The bandage strategy for modulating RNA activity should work most predictably in systems where an accessible sequence of mRNA is already known, or for shorter, less-structured targets such as PCR primers, microRNAs, or siRNAs. Photolabile bonds have previously been incorporated within oligonucleotides using phosphoramidite chemistry.29–32 PL offers additional possibilities to vary the gap between a and b strands, and link nonstandard oligos (e.g., phosphorothioate, peptide, or locked nucleic acids) that may improve targeting in vivo.
In conclusion, an efficient synthesis was developed for joining two noncomplementary 2′-OMe RNA strands by a photocleavable linker to generate tandem oligos 1–4 that bind tightly to a RNA target Tg. In mRNA translation assays, 1–3 provided the limiting cases for poor photomodulation: 1 was too short and unstable, with only 10% blockage prior to irradiation, even at 10-fold excess over RNA; 2 and 3 were too stable, with 100% blockage, even after UV irradiation, and at only 2-fold excess over RNA. Based on these results, an extended and more asymmetric RNA bandage 4 was designed, which proved successful at photomodulating in vitro protein translation levels more than 3-fold. This strategy can likely be generalized to other oligonucleotides and intracellular targets, in order to control gene function with light.
Supplementary Material
Details about experimental procedures can be found in the online version.
Acknowledgments
We thank Eric Meggers, Barry Cooperman, Ronen Marmorstein, and Jim Eberwine for access to equipment and helpful discussions. We thank NIH (1R01GM083030, GM071339) and the Camille and Henry Dreyfus Foundation for support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.(a) Tang XJ, Dmochowski IJ. Mol Biosys. 2007;3:100. doi: 10.1039/b614349k. [DOI] [PubMed] [Google Scholar]; (b) Mayer G, Heckel A. Angew Chem Intl Ed. 2006:4900. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]; (c) Young DD, Deiters A. Org Biomol Chem. 2007;5:999. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 2.Ghosn B, Haselton FR, Gee KR, Monroe WT. Photochem Photobiol. 2005;81:953. doi: 10.1562/2004-11-15-RA-373. [DOI] [PubMed] [Google Scholar]
- 3.Krock L, Heckel A. Angew Chem Int Ed. 2005;44:471. doi: 10.1002/anie.200461779. [DOI] [PubMed] [Google Scholar]
- 4.Tang XJ, Dmochowski IJ. Angew Chem Int Ed. 2006;45:3523. doi: 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]
- 5.Young DD, Edwards WF, Lusic H, Lively MO, Deiters A. Chem Commun. 2008:462. doi: 10.1039/b715152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asanuma H, Tamaru D, Yamazawa A, Liu MZ, Komiyama M. Chembiochem. 2002;3:786. doi: 10.1002/1439-7633(20020802)3:8<786::AID-CBIC786>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 7.Patnaik S, Kumar P, Garg BS, Gandhi RP, Gupta KC. Bioorg Med Chem. 2007;15:7840. doi: 10.1016/j.bmc.2007.08.042. [DOI] [PubMed] [Google Scholar]
- 8.Yamazawa A, Liang XG, Asanuma H, Komiyama M. Angew Chem Int Ed. 2000;39:2356. doi: 10.1002/1521-3773(20000703)39:13<2356::aid-anie2356>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Tang XJ, Richards JL, Peritz AE, Dmochowski IJ. Bioorg Med Chem Lett. 2005;15:5303. doi: 10.1016/j.bmcl.2005.08.058. [DOI] [PubMed] [Google Scholar]
- 10.Chaulk SG, MacMillan AM. Nucleic Acids Res. 1998;26:3173. doi: 10.1093/nar/26.13.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young DD, Deiters A. Bioorg Med Chem Lett. 2006;16:2658. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Lusic H, Young DD, Lively MO, Deiters A. Org Lett. 2007;9:1903. doi: 10.1021/ol070455u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting R, Lermer L, Perrin DM. J Am Chem Soc. 2004;126:12720. doi: 10.1021/ja046964y. [DOI] [PubMed] [Google Scholar]
- 14.Heckel A, Mayer G. J Am Chem Soc. 2005;127:822. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 15.Matsunaga D, Asanuma H, Komiyama M. J Am Chem Soc. 2004;126:11452. doi: 10.1021/ja0471976. [DOI] [PubMed] [Google Scholar]
- 16.Hobartner C, Silverman SK. Angew Chem Int Ed. 2005;44:7305. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- 17.Shah S, Rangarajan S, Friedman SH. Angew Chem Int Ed. 2005;44:1328. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 18.Mikat V, Heckel A. RNA. 2007;13:2341. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shestopalov IA, Sinha S, Chen JK. Nat Chem Biol. 2007;3:650. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 20.Tang XJ, Maegawa S, Weinberg ES, Dmochowski IJ. J Am Chem Soc. 2007;129:11000. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 21.Tang X, Swaminathan J, Gewirtz A, Dmochowski I. Nucleic Acids Res. 2008;36:559. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ando H, Furuta T, Tsien RY, Okamoto H. Nat Genet. 2001;28:317. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 23.Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. J Biol Chem. 1999;274:20895. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 24.Braasch DA, Corey DR. Biochemistry. 2002;41:4503. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 25.Johansson HE, Belsham GJ, Sproat BS, Hentze MW. Nucleic Acids Res. 1994;22:4591. doi: 10.1093/nar/22.22.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Gene. 2005;361:13. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 27.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Proc Natl Acad Sci USA. 2004;101:7287. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry AR, Rahman A, Dritschilo A. Biochem Biophys Res Commun. 1993;190:952. doi: 10.1006/bbrc.1993.1142. [DOI] [PubMed] [Google Scholar]
- 29.Zhang KJ, Taylor JS. Biochemistry. 2001;40:153. doi: 10.1021/bi001781j. [DOI] [PubMed] [Google Scholar]
- 30.Dussy A, Meyer C, Quennet E, Bickle TA, Giese B, Marx A. Chem Biochem. 2002;3:54. doi: 10.1002/1439-7633(20020104)3:1<54::AID-CBIC54>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Pirrung MC, Zhao XD, Harris SV. J Org Chem. 2001;66:2067. doi: 10.1021/jo001594r. [DOI] [PubMed] [Google Scholar]
- 32.Crey-Desbiolles C, Lhomme J, Dumy P, Kotera M. J Am Chem Soc. 2004;126:9532. doi: 10.1021/ja047976m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details about experimental procedures can be found in the online version.