Abstract
Cancer antigen 125 (CA125) is an antigen that is elevated in the serum of women with ovarian carcinoma, but can also be detected in serum from healthy women. CA125 is expressed in 80% of human ovarian cancers, as well as in normal adult endometrium, lung, and amnion. The gene encoding human CA125 was identified as MUCIN16 (MUC16). A database search identified the orthologous mouse gene, Muc16. Reverse transcription-polymerase chain reaction and RNA in situ hybridization detected Muc16 transcripts in the surface epithelia of the upper respiratory tract, the mesothelia lining body cavities and the internal organs, as well as male and female reproductive organs, and the amnion. Antibodies raised against human MUC16 do not recognize mouse MUC16. Therefore, a rabbit anti-mouse polyclonal antibody against recombinant mouse MUC16 was generated. Immunohistochemistry using this anti-mouse MUC16 antibody revealed expression in the luminal epithelia of the trachea, the epithelia of the secretory glands in the oral cavity, the surface of the olfactory epithelia, as well as mesothelial cells lining body cavities (i.e., pleural, peritoneal, and pelvic cavities), and male and female reproductive organs. In addition, MUC16 protein was detected in other cell types, such as the surface epithelia of the cochlear duct and chief cells of the stomach, suggesting multiple roles for MUC16. In mouse serous epithelial ovarian cancer, MUC16 protein was detected at the apical surface of well-differentiated tumors, but not poorly differentiated tumors. These findings document the presence of MUC16 in murine ovarian cancer and in normal tissues and provide a foundation for future functional studies.
Keywords: CA125, Muc16, mucin, ovarian cancer
Introduction
Cancer antigen 125 (CA125) is a tumor-associated antigen that is cleaved from the surface of ovarian cancer cells and shed into blood providing a useful biomarker for monitoring growth of ovarian cancer (for review see Bast et al., 1998). The monoclonal antibody OC125 recognizes this tumor antigen in epithelial ovarian cancer cells, but not in normal fetal and adult ovaries (Bast et al., 1981). An immunoassay using OC125 detects elevation of CA125 in >80% of ovarian cancer patients’ sera but in <1% of control group sera (Bast et al., 1983). Changes in CA125 antigen levels accurately track response to treatment in >80% of patients and predict recurrence of ovarian cancer with at least 3 months lead time in 70% of cases. CA125 is not, however, an ideal biomarker for early detection of epithelial ovarian carcinomas as serum levels can be elevated by some benign conditions and are increased in only 50%-60% of cases of cancer limited to the ovary (Bast et al., 1998). Still, CA125 is the best serum marker for monitoring the progress of patients with epithelial ovarian cancer and is widely used as a surrogate marker for monitoring the outcome of treatment in ovarian cancer patients.
Studies of CA125 have focused primarily on clinical applications whereas the biological significance of CA125 expression is still poorly understood. Most biochemical evidence indicates that CA125 is a high molecular weight (>1 MDa) glycoprotein with abundant carbohydrate bound by both N- and O-linkage (Davis et al., 1986; Nagata et al., 1991; Yu et al., 1991; de los Frailes et al., 1993; Kobayashi et al., 1993; Lloyd et al., 1997; Lloyd and Yin, 2001). A candidate gene encoding the CA125 antigen was cloned from an expression cDNA library using a rabbit antiserum against purified CA125 (Yin and Lloyd, 2001; Yin et al., 2002). Sequence analysis suggested that the cDNA encoded a novel mucin, designated as MUC16 with a short intracellular domain, a transmembrane domain and a large extracellular domain. The extracellular domain included multiple 154 amino acid tandem repeats and a distinctive N-terminus. Simultaneously, another group used biochemical purification and peptide sequencing followed by database searches to identify the same gene encoding CA125 (O’Brien et al., 2001, 2002). While there is some disagreement regarding the precise number of tandem repeats and its chromosomal location, these findings provide an entry point for the molecular genetic and biological analysis of MUC16 function.
Recent studies suggest that MUC16 may have a biological role in the characteristic pattern of metastasis of ovarian cancer cells, forming implants over the surface of the peritoneal cavity. As a large heavily glycosylated molecule, MUC16 extends from the surface of ovarian cancer cells. MUC16 has been shown to bind mesothelin (Rump et al., 2004; Gubbels et al., 2006; Scholler et al., 2007), a protein that is found on the surface of the mesothelial cells that line the peritoneum. Thus, MUC16 may provide one of the first points of contact and adhesion for metastasizing epithelial ovarian cancer cells.
Although antibodies raised against the CA125 tumor antigen have been used extensively to assess circulating levels of CA125/MUC16 and expression in different cancers, relatively few studies have examined the expression of MUC16 in normal tissues. Human MUC16 distribution in fetal and adult tissues was described by a few groups. MUC16 immunohistochemistry of human tissues using the OC125 antibody detected MUC16 expression in the apical surface of various organs such as the fetal coelomic epithelia and its derivatives—Müllerian duct, fallopian tube, endometrium, endocervix, peritoneum, pleura and pericardium, amnion (Kabawat et al., 1983), trachea, lung, respiratory glands (Nouwen et al., 1986), periderm (Nanbu et al., 1989), breast, prostate (Nap, 1998), the corneal and conjunctival epithelium of the eye (Nap, 1998; Argüeso et al., 2003), and the lacrimal apparatus (Jäger et al., 2007). The OC125 antibody also showed cross-reactivity with other species including rabbit, dog, monkey, and cow (Nouwen et al., 1990; McDonnel et al., 2003). The MUC16 expression pattern in these mammals was conserved as human MUC16. However, the OC125 antibody was not immunoreactive in both mouse and rat tissues (Nouwen et al., 1990).
In addition to the OC125 antibody, other MUC16 antibodies were generated against different epitopes on the MUC16 molecule. Among them, M2 and M11 antibodies were used for immunohistochemical analysis in a few studies. These antibodies detected MUC16 in most mesothelia, consistently with OC125 staining (Hardardottir et al., 1990; Nap, 1998). However, these antibodies also revealed a new distribution of MUC16 in the periderm and its adult derivatives (i.e., lactating mammary glands and secreting apocrine sweat glands), enteric duct remnants in the umbilical cord, notochord, trophoblast (Hardardottir et al., 1990), and esophagus (Nap, 1998).
While numerous studies have examined the expression pattern of MUC16 in human and a few non-human tissues, a complete understanding of the expression pattern of MUC16 during embryogenesis and in adult tissues still has been hampered by difficulties in obtaining appropriate tissue from humans and other animal species. Studies of MUC16 in the mouse should not only permit complete characterization of antigen expression during development, but can also permit more critical experiments regarding the role of MUC16 in ovarian oncogenesis, as genetically defined syngenic epithelial ovarian cancers have been developed that mimic the human disease (Orsulic et al., 2002). We have used DNA sequence databases to identify and characterize the mouse ortholog of the human MUC16 gene. Reverse transcription-polymerase chain reaction (RT-PCR) and in situ hybridization were used to examine Muc16 RNA expression in embryonic and adult mouse tissues. We have also raised a polyclonal antibody to mouse MUC16 in order to localize MUC16 expression in mouse tissues and ovarian carcinomas. These studies provide a foundation for future experiments to understand the biological role of MUC16.
Methods
Mice
Swiss outbred mice were purchased from Taconic Farms (New York). Timed matings were established and noon of the day on which a copulation plug was observed was designated as embryonic day 0.5 (E0.5). All experimental animals were maintained in accordance with the Institutional Animal Care and Use Committee (M.D. Anderson Cancer Center) and the NIH Guide for the Care and Use of Laboratory animals.
Mouse ovarian cancers
Mouse ovarian tumor tissues were obtained from established mouse models of ovarian cancer (Orsulic et al., 2002; Xing and Orsulic 2006). Briefly, ovarian explants from BK5-TVA tg/+; p53-/- mice were infected with different combinations of RCAS (Replication-Competent ASLV long-terminal repeat with a Splice acceptor) viruses carrying human Myc, mouse K-ras G12D, myristoylated mouse Akt1, and truncated rat Her2/neu oncogenes. Ovarian explants from BK5-TVA tg/+; p53 fx/fx; Brca1 fx/fx mice were infected with RCAS-Cre and RCAS-Myc. The ovary explant cultures were passaged three times in the presence of viruses in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% glutamine, and 1% penicillin/streptomycin (Mediatech, Manassas, VA). Because ovarian surface epithelial cells in these mice are the only ovarian cell type that expresses the chicken TVA receptor, which is necessary for transformation, these are the only cells that survive in culture after three rounds of passaging. The transformed ovarian surface epithelial cells were then injected intraperitoneally into nude mice. After 3-8 weeks, the mice developed abundant ascites and intraperitoneal carcinomatosis. Tumor nodules found in the peritoneal cavity were removed and either snap-frozen for RNA isolation or fixed in Carnoy’s fixative (absolute ethanol:chloroform:acetic acid; 6:3:1), transferred to 70% ethanol and embedded in paraffin.
Determination of gene structure of mouse Muc16
Genomic sequence information for mouse chromosome 9 was obtained from the Ensembl genome browser (http://www.ensembl.org, Ensembl gene ID: ENSMUSG00000032134, Location 18,337,838-18,617,880). The predicted cDNA sequence of mouse Muc16 was obtained from the NCBI database (1110008I14 gene, GenBank Accession #: XM_911929). The exon-intron structure of mouse Muc16 was determined by comparison of the predicted cDNA sequence of mouse Muc16 with the genomic sequence of mouse chromosome 9 using VISTA tool (http://genome.lbl.gov/vista/index.shtml). The sequences occurring in both cDNA sequence and genomic sequence were regarded as exons. 7.1 kb of mouse Muc16 cDNA was verified by RT-PCR (Invitrogen, Carlsbad, CA) using E10.5 amnion cDNA and sequencing of the RT-PCR products.
RNA extraction and RT-PCR
Total RNA was extracted from embryonic organs and amnions using RNeasy midipreps (QIAGEN, Valencia, CA), following the manufacturer’s recommendations. Total RNA was extracted from adult tissues using Trizol® reagent (Life Technologies, Carlsbad, CA), according to the manufacturer’s instructions.
cDNA was synthesized by reverse transcription (RT) from 1 μg of total RNA using random hexamers and the SuperScript II first-strand synthesis system for RT-PCR. One microliter of the first-strand cDNA was used as the template for the PCR reactions. RT-PCR of ovary cDNA in the absence of RT served as a negative control. Primers were designed to amplify various subregions of mouse Muc16, as shown in Figure 1. For 5′ Muc16 PCR, PCR conditions were 10 cycles at 94°C for 30 sec, 58°C for 45 sec, and 68°C for 2 min and 20 cycles at 94°C for 30 sec, 58°C for 45 sec, and 68°C for 2 min with an increase of 5 sec per each cycle. For the other PCR reactions (6F, 10R; 22F, 27R; and 38F, 43R), PCR conditions were 30 cycles at 94°C for 30 sec, 55°C for 45 sec, and 72°C for 1 min. A 377 bp β-actin PCR product served as a positive control for cDNA quality. For β-actin, PCR conditions were 30 cycles of 94°C for 30 sec, 58°C for 45 sec, and 72°C for 1 min. Following PCR, 10 μl of each reaction were analyzed on a 1.5% agarose gel and visualized using ethidium bromide staining.
Fig. 1.
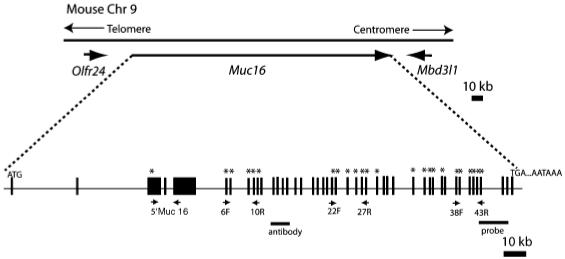
Predicted gene structure of mouse Muc16 locus. Comparison of a predicted cDNA sequence (GenBank Accession #: XM_911929) to genomic sequence on chromosome 9 (Chr 9) suggests a gene structure for mouse Muc16. Exons are indicated by black boxes. Primers used for reverse transcription-polymerase chain reaction analysis are indicated by small arrows under the exons. The probe (probe) used for section in situ hybridization and the peptide used to raise mouse MUC16 antibody (antibody) are indicated as bars under the exons. Exons of which sequences were confirmed by sequencing are indicated by asterisk. Olfr24, olfactory receptor 24; Mbd3l1, methyl-CpG-binding domain protein 3-like 1; ATG, initiation codon; TGA, stop codon; AATAAA, polyadenylation signal.
RNA in situ hybridization
A 700 bp mouse Muc16 cDNA fragment from a mouse EST clone (NCBI Accession #: AK003577, location: 253-973, that corresponds to XM_911929:25539-26259) was subcloned into the pBluescript vector (Fermentas Inc., Glen Burnie, MA) and used for RNA probe synthesis. Antisense mouse Muc16 riboprobe was transcribed with T7 RNA polymerase from BamHI-digested template and sense-strand control riboprobe was transcribed with T3 RNA polymerase from NotI-digested template. Both antisense and sense riboprobes were labeled directly during the RNA polymerase reaction using [35S] uridine 50-(α-thio)triphosphate.35S-labeled RNA probes were synthesized using an in vitro transcription kit (Promega, Madison, WI) according to the manufacturer’s instructions.
The hybridization protocol was modified from a previous report (Simmons et al., 1989). Briefly, individual 7-μM-thick sections were hybridized with a solution containing 5 × SSC, 20% dextran sulfate, 0.25 mg/ml denatured salmon sperm DNA, 50% deionized formamide, 0.1 M dithiothreitol, 0.5× Denhard’s solution, and 35S-labeled probe (1 × 107cpm). Sections were covered with cover slips and incubated in a moist chamber at 60°C overnight (18-20 hr). After removal of the coverslips, the sections were washed twice in a solution containing 2 × SSC, 50% formamide, and 40 mM 2-mercaptoethanol for 30 min at 65°C, then in 1 × NTE (0.5 M NaCl, 10 mM Tris-HCl, pH 8, and 5 mM EDTA) three times for 10 min at 37°C. After digestion with 20 mg/mL RNase A for 30 min at 37°C and washing with 2× SSC and 0.1 × SSC, were dehydrated in ethanol and dried in air. The slides were then exposed to autoradiographic film for 1 day to determine signal strength, and then processed for liquid emulsion autoradiography using NTB-2 emulsion (Eastman Kodak, Rochester, NY). Slides were developed using Kodak D-19 developer and fixer and counterstained with Hoechst. The slides were observed using darkfield/fluorescence microscopy.
Generation of rabbit anti-mouse MUC16 polyclonal antibody
DNA sequence coding for 236 amino acids of the mouse MUC16 fragment (YEEGMNHPGSWKFNATERILQRLLWPLFNKTS ISLRYSNCRLALLRPEKDGAATGVDAICTHHAGPIGPGLDS ERVFWELCNLTNNVTQLGPYTLDKNSLYINGYTHQIQSIT SPTGPVLEHFTINFTIINLKFEEDMSHPGSRKFNITERTLQR LLRILFKKSSVGALYSGCKLSLLRPENDGAGTGVDAVCTH YPDPTGLGLDRKMLYGEISRLTYGVKRLGPYTLNDKSLYVNGY; amino acid location, XM_911929: 6668-6896) was cloned into the VariFLEX protein expression vector pBEn-SBP-SET (Stratagene, La Jolla, CA). The Solubility Expression Tag (SET)-Muc16 fusion protein was purified from bacteria using ion-exchange chromatographic columns by AKTA-FPLC (GE Healthcare, Piscataway, NJ). The purified fusion protein was used to immunize rabbits to obtain antisera against mouse MUC16 (UT MDACC Science Park, Department of Veterinary Science). The antibody was affinity purified by chromatography on a GST-mMUC16-sepharose column and the antibody titer was determined by ELISA, according to the standard protocol (Perlmann and Perlmann, 1994).
Immunofluorescence
Whole mouse embryos or tissues were fixed in 4% paraformaldyhyde (PFA) in phosphate-buffered saline (PBS) at 4°C overnight, then dehydrated and embedded in paraffin and sectioned at 5 μm. Antigen unmasking was performed with 50 mM Tris buffer (pH 9.0) for 20 min in a microwave oven. Nonspecific binding was blocked with PBS and 0.05% Tween-20 (PBS-T) containing 10% normal goat serum for at least 1 hr at room temperature. The slides were incubated with the polyclonal rabbit anti-mouse MUC16 antibody, diluted 1:50 in 10% goat serum overnight at 4°C and then incubated with the secondary biotinylated antibody (Vector Laboratories Inc., Burlingame, CA, diluted 1:250) for 30 min. For the negative control, IgG from rabbit serum (Sigma, St. Louis, MO) was incubated instead of the primary antibody. The slides were next incubated with fluorescein avidin D (Vector Laboratories Inc., diluted 1:300) for another 30 min, were washed three times in PBS, counterstained with VECTASHIELD mounting media with DAPI (Vector Laboratories Inc.), and coverslipped. The slides were examined using fluorescence microscopy.
Results
Characterization of the mouse Muc16 locus
To determine the putative exon/intron organization of mouse Muc16, we obtained the mouse genomic sequence covering 17.5-18 cM on chromosome 9 from the Ensembl database and compared this sequence with the predicted cDNA sequence of mouse Muc16 (GenBank Accession #: XM_357986, currently replaced by XM_911929) using a VISTA tool. This comparison yielded a predicted gene structure of mouse Muc16 (Fig. 1). Mouse Muc16 was located on chromosome 9, spanning about 230 kb. The genomic organization around mouse Muc16 was conserved with the human MUC16 region on chromosome 19. The neighboring genes Olfr24 (Olfactory receptor 24) and Mbd3l1 (methyl-CpG-binding domain protein 3-like 1) are the predicted orthologs of human OR1M1 (olfactory receptor 1M1) and MBD3-like1 (methyl-CpG-binding domain protein 3-like 1), respectively. To verify cDNA sequence information in the public database, we designed primers at different 5′ and 3′ regions of mouse Muc16 and performed RT-PCR using cDNA synthesized from E10.5 amnion. Each RT-PCR product was cloned and sequenced. We have confirmed 7.1 kb of the 22 kb predicted mouse Muc16 cDNA (Fig. 1).
Localization of mouse Muc16/MUC16 during embryonic development
mRNA
To determine the expression of Muc16 during embryonic development, RT-PCR analysis was performed using various embryonic tissues. In embryos, Muc16 mRNA was detected in amnion and gonads from both male and female embryos (Fig. 2A). At E11.5, Muc16 mRNA was barely detected in the embryo whereas Muc16 mRNA expression in the amnion was detected from E10.5 throughout development (Fig. 2A). Muc16 transcripts were also detected in other embryonic tissues including lung and liver (data not shown).
Fig. 2.
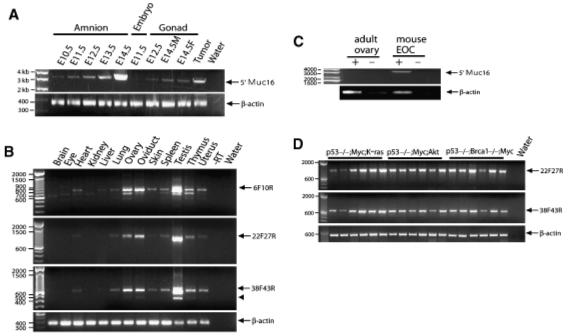
Reverse transcription-polymerase chain reaction (RT-PCR) analysis of mouse Muc16 mRNA expression. Primers used for RT-PCR analysis are shown in Table 1. (A) Muc16 transcripts in extra-embryonic tissues, whole embryo, and gonads. E14.5M, male embryo at embryonic day 14.5; E14.5F, female embryo at embryonic day 14.5; Tumor, mouse epithelial ovarian cancer (Orsulic et al., 2002); Water, reaction without cDNA template. (B) RT-PCR analysis of mouse adult tissues. Muc16 transcripts generated using different primer sets are indicated by an arrow. The reaction without reverse transcriptase (RT) and without cDNA template (Water) serve as negative controls. An arrowhead indicates a splice variant. (C) Mouse Muc16 mRNA expression in mouse epithelial ovarian cancer (EOC). Tumor tissues derived from a mouse model of EOC (Orsulic et al., 2002) were analyzed for Muc16 transcripts and compared with a wild-type ovary. +, reaction with RT; -, reaction without RT. (D) Mouse Muc16 mRNA expression in EOC cell lines. Various cancer cell lines derived from both primary and disseminated tumors derived from the mouse model of EOC (Orsulic et al., 2002; Xing and Orsulic, 2006) were analyzed for Muc16 transcripts. Water, reaction without cDNA template.
To determine the localization of mouse Muc16 mRNA, section in situ hybridization was performed. In E10.5 mice, Muc16 mRNA signal was observed only in the amnion, not in the embryo proper (Fig. 3A). In E13.5 mice, Muc16 mRNA signal was observed in the epithelium covering the gonads (Fig. 3B), peritoneal mesothelium of surrounding paramesonephric duct/mesonephric duct and other peritoneal mesothelia (Fig. 3C). In E16.5 mice, a Muc16 mRNA signal was broadly detected in the mesothelia covering body cavities (e.g., pleura, peritoneum), which include the visceral pleura lining around the lung and heart (Fig. 3D), the visceral peritoneum lining the internal organs (Fig. 3E) as well as the parietal peritoneum lining body wall and the diaphragm. Muc16 mRNA was not detected in the kidney (Fig. 3E). In the female, Muc16 mRNA was detected in the mesothelia covering the ovary (Figs. 3B,3F,3J,3M), oviduct (Figs. 3F,3J,3N), and uterus (Figs. 3F,3O) throughout development. In the male, Muc16 mRNA was initially detected in the surface of the testis and mesonephros (Fig. 3I) and then in the mesothelia covering the testis and other male reproductive organs (Figs. 3K,3L). Muc16 mRNA was also detected in the surface epithelia of the upper respiratory tract of embryos, such as the epithelia lining the trachea lumen (Fig. 3G), the laryngeal aditus (Fig. 3G), and olfactory epithelia (Fig. 3H).
Fig. 3.
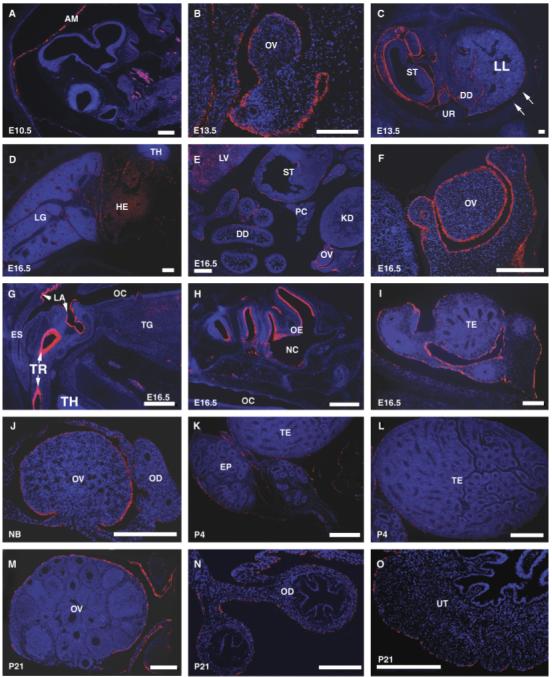
Localization of mouse Muc16 mRNA detected by in situ hybridization. Various mouse tissues at different embryonic and adult stages were used to determine the mouse Muc16 mRNA expression pattern. Red signal indicates mouse Muc16 mRNA. Embryonic (E) or postnatal (P) ages are indicated in each panel. Muc16 expression in the amnion at E10.5 (A), fetal ovary at E13.5 (B), peritoneal cavity at E13.5 (C), pleural cavity at 16.5 (D), female peritoneal cavity at E16.5 (E), ovary at E16.5 (F), oral cavity at E16.5 (G), head region at E16.5 (H), testis at E16.5 (I), newborn (NB) ovary (J), male reproductive organs at P4 (K, L), female reproductive organs at P21 (M, N, O). AM, amnion; DD, duodenum; EP, epididymis; ES, esophagus; HE, heart; KD, kidney; LA, laryngeal aditus; LG, lung; LL, left lobe of lung; LV, liver; NC, nasal cavity; OC, oral cavity; OD, oviduct; OE, olfactory epithelium; OV, ovary; PC, pancreas; ST, stomach; TE, testis; TG, tongue; TH, thymus; TR, lumen of trachea; UR, urogenital ridge; UT, uterus. Scale bars = 200 μm.
Protein
MUC16 protein expression assessed by immunostaining was delayed compared with mRNA expression. At E15.5, MUC16 protein was detected only in the mast cells in the skin (Fig. 4A). At E18.5, MUC16 protein was detected only in the amnion and few mesothelial cells (Fig. 4B). Peritoneal expression of MUC16 protein was not observed until the newborn stage. In postnatal day 4 (P4) mice, MUC16 protein was detected in the surface of oral, nasal cavities, and the inner ear (Figs. 4C-4E). MUC16 signal was observed in ovary surface epithelium that showed some scattered single-positive cells (data not shown). Simultaneously, positive signal was detected in peritoneal mesothelial cells (data not shown). In 2-week-old mice, MUC16 protein expression was observed in the entire surface epithelium covering the ovary, and peritoneal mesothelial cells surrounding the oviduct and uterus (data not shown). The delayed expression of MUC16 protein implies that the epitope region recognized by the MUC16 antibody might not be present during embryonic stages.
Fig. 4.
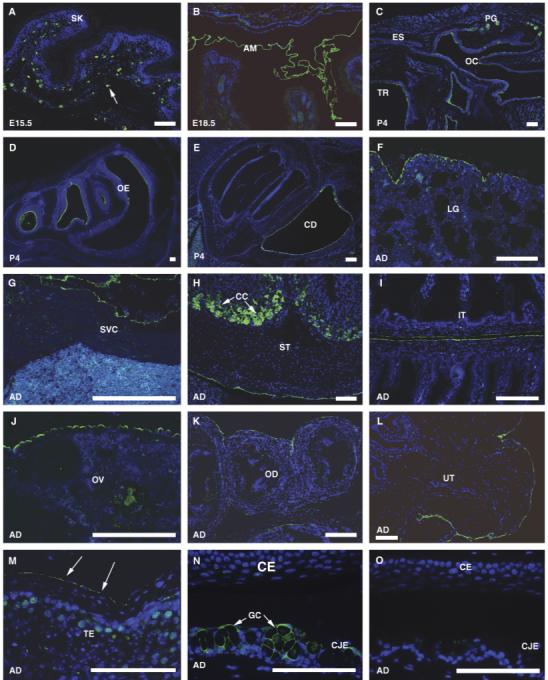
Immunohistochemical localization of mouse MUC16 protein. Green signal indicates mouse MUC16 protein. Negative control showed no signal (O). Embryonic (E), postnatal (P), or adult (AD) stages are indicated in each panel. MUC16 expression in the skin at E15.5 (A). Arrow indicates MUC16 expression in mast cells. MUC16 expression in the amnion at E18.5 (B), oral cavity at P4 (C), nasal cavity at P4 (D), and inner ear at P4 (E). MUC16 expression in adult lung (F), heart (G), stomach (H), intestine (I), female reproductive organs (J, K, L), testis (M), and eye (N). Arrows in M indicate MUC16 expression in the mesothleia. (O), The negative control (no primary antibody) shows no MUC 16 signal in the eye. AM, amnion; CC, chief cell; CD, cochlear duct; CE, corneal epithelia; CJE, conjunctival epithehlia; ES, esophagus; GC, goblet cells; IT, intestine; LG, lung; OC, oral cavity; OD, oviduct; OE, olfactory epithelium; OV, ovary; PG, mucous palatine glands; SK, skin; SP, spleen; ST, stomach; SVC, superior vena cava; TE, testis; TR, lumen of trachea; UT, uterus. Scale bars = 100 μm.
Localization of mouse Muc16/MUC16 in adult tissues
mRNA
In adult, Muc16 transcripts were detected in heart, lung, ovary, oviduct, skin, spleen, testis, thymus, and uterus by RT-PCR (Fig. 2B). This expression pattern was consistently observed using two different primer sets (22F, 27R; 38F, 43R). The products generated by PCR were larger than the expected size. Further sequencing analysis revealed additional sequences, suggesting that current cDNA sequences in the NCBI database are not complete. Occasionally we detected smaller bands; nonspecific bands (smaller bands in the 6F, 10R panel) or a splice variant (indicated by an arrowhead in the 38F, 43R band). To further demonstrate that mouse Muc16 is the ortholog of human MUC16, mouse Muc16 expression were determined in mouse epithelial ovarian tumors (Orsulic et al., 2002). Mouse Muc16 transcripts were detected in mouse epithelial ovarian tumors by RT-PCR (Fig. 2C). However, splice variants were not observed in mouse epithelial ovarian tumors (Fig. 2D). Our RT-PCR results were further confirmed by section in situ hybridization. Consistent with mouse Muc16 expression during embryonic development, mouse Muc16 mRNA was broadly detected in the mesothelial cells lining pleural, peritoneal, and pelvic cavity at the adult stage (data not shown).
Protein
To determine the localization of MUC16 protein, we performed immunofluorescence analysis of various adult tissues using the polyclonal antibody we generated. For the negative control, a rabbit IgG was used instead of the primary antibody. No signal was observed in the negative control (Fig. 4O), showing that MUC16 antibody gives a specific signal. The expression pattern of MUC16 protein was consistent with our RNA in situ hybridization results. MUC16 protein was located in the luminal surface of the trachea (Fig. 4C), the epithelia of the secretory glands of oral cavity such as ducts of mucous palatine glands (Fig. 4C), and the surface of the olfactory epithelia (Fig. 4D). Further, MUC16 expression was broadly detected in all the mesothelial cells in the pleural cavity lining on the lung (Fig. 4F), the heart and pulmonary artery (data not shown), superior vena cava (Fig. 4G), in the peritoneal cavity lining on the internal organs such as stomach (Fig. 4H), spleen (data not shown), and intestine (Fig. 4I), in the pelvic cavity lining on male and female reproductive organs (Figs. 4J-4M), and in the amnion (Fig. 4B). However, there was no MUC16 protein expression observed in oviduct epithelium (Fig. 4K) nor in the endometrium of the uterus (Fig. 4L). We further detected MUC16 protein in other cell types, such as the surface epithelia of the cochlear duct that is the part of the inner ear (Fig. 4E), the chief cells of the stomach (Fig. 4H), the goblet cells of conjunctival epithelium (Fig. 4N), and the mast cells in the skin (Fig. 4A), implying multiple functions for MUC16.
MUC16-positive cells were also pan-keratin positive in the cytoplasm, which is a specific marker of ovarian surface epithelial cells (Fig. 5). Double staining of MUC16 and pan-keratin demonstrated partial overlap of these two proteins in the same cells (Fig. 5), suggesting that MUC16 is localized to the apical surfaces of epithelial cells.
Fig. 5.
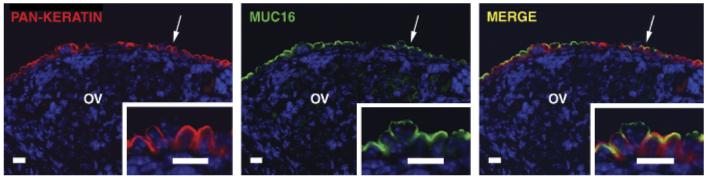
Apical localization of mouse MUC16 protein. Double immunostaining for pan-keratin (red) and mouse MUC16 (green) show overlapping expression (yellow) in adult ovarian surface epithelium. The subcellular distribution of pan-keratin and mouse MUC16 (indicated by arrows) is shown at higher magnification in the insets. OV, ovary. Scale bars = 10 μm.
Immunolocalization of MUC16 in mouse ovarian carcinoma tissue
MUC16 was detected in a genetic mouse model of ovarian cancer (Orsulic et al., 2002; Xing and Orsulic, 2006), which shows morphologies similar to human serous papillary adenocarcinoma of the ovary. MUC16 localization was examined in tumors with different genetic lesions [p53-/-; Myc; Akt (Figs. 6A,6B); p53-/-; Myc; Akt; Neu (Figs. 6C-6F); p53-/-; Brca1-/-; Myc (Figs. 6G,6H)]. MUC16-positive signal was pre-dominantly present at the plasma membrane of cells (Figs. 6B,6D) or in the apical membrane of the outer layer of cells of cancer papillae (Figs. 6B,6F). In addition, MUC16 signal was detected at the apical surface of well-differentiated tumors (Figs. 6B,6D,6F), but not in poorly differentiated tumors (Fig. 6H). MUC16 protein was commonly found in either individual tumor cells (Figs. 6B,6D), or at the apical surface of the papillary structures of tumors (Figs. 6B,6F).
Fig. 6.
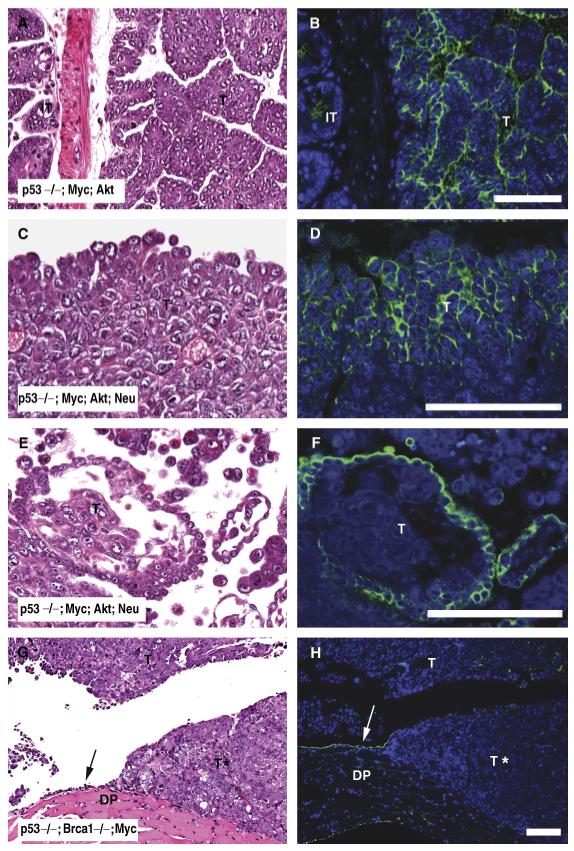
Immunohistochemical localization of mouse MUC16 protein in mouse epithelial ovarian cancer tissues. Tumor tissues derived from mouse model of epithelial ovarian cancer (Orsulic et al., 2002; Xing and Orsulic, 2006) with different genetic lesions [p53-/-; Myc; Akt (A, B,); p53-/-; Myc; Akt; Neu (C, D, E, F); p53-/-; Brca1-/-; Myc (G, H)] were analyzed by hematoxylin and eosin staining (left panels) and MUC16 immunofluorescence (right panels). Green signal indicates MUC16 protein expression in tumors (T) as well as in normal mesothelial cells (arrow) of the diaphragm (DP). Asterisk indicates absence of MUC16 signal in poorly differentiated tumors. Scale bars = 100 μm.
Discussion
MUC16 was originally described as a CA125 in which serum levels were increased in >80 % of ovarian cancers (Bast et al., 1981). However, increased MUC16 serum levels were also detected in other types of cancers and in some benign conditions (reviewed in Bast et al., 1998), suggesting that MUC16 is not a specific marker for ovarian cancer. Further, MUC16 is expressed in a number of different tissues in human and some mammals, implying that there may be multiple biological functions for MUC16. However, functional studies of MUC16 have been hampered due to the absence of MUC16 studies in small laboratory animals such as the mouse and the rat.
We have identified the mouse ortholog of the human MUC16 gene. The general organization of the mouse Muc16 gene is similar to that of human MUC16 gene (O’Brien et al., 2001, 2002). Our sequencing analysis confirmed 7.1 kb of sequence of the 22 kb predicted mouse Muc16 cDNA in current public database (Gen-Bank Accession #: XM_911929), covering half of exon 3, some tandem repeat regions and a short cytosolic tail (exons 6-10, 22-28, 32-36, and 38-46).
Mouse Muc16 has two large exons in the N-terminal region that have sequence homology to exons 1 and 3 of human MUC16 (O’ Brien et al., 2002). Each tandem repeat of mouse Muc16 also has two conserved cysteines like human MUC16 (O’ Brien et al., 2001). We have not found any new protein domains in this region except for one SEA domain characterized by Maeda et al. (2004). The C-terminal domain of mouse Muc16 shows 66% identity with human MUC16. Furthermore, the genomic organization around the mouse Muc16 gene (Olfr24-CA125/Muc16-Mbd3l1) was conserved on human Chr19. However, the size of mouse Muc16 cDNA (22 kb) was much smaller than that of human MUC16 cDNA (66 kb) and the overall sequence similarity of mouse Muc16 cDNA to human MUC16 cDNA was low. These differences might be due to incomplete information on both cDNA and genomic sequences. Indeed, we identified additional cDNA sequences in the tandem repeat region from our RT-PCR analysis, suggesting that sequence information especially in the tandem repeat region is missing. We expect that the size of mouse Muc16 cDNA will be larger once more sequence information is determined.
Our current analysis shows the conserved gene structure and genomic organization of mouse Muc16, indicating that mouse Muc16 is likely the ortholog of human MUC16. Our expression studies further demonstrate that mouse Muc16 shows similar expression patterns as human MUC16, confirming that mouse Muc16 is the ortholog of human MUC16.
Our findings are in general agreement with previous studies. Mouse MUC16 was broadly detected in the pleural and peritoneal mesothelia, the mesothelia lining heart, lung, internal organs, male and female reproductive organs, the surface epithelia of the upper respiratory tract, as well as in the amnion. MUC16 expression in most mesothelial lining including pleural and peritoneal mesothelia is consistent with the previous findings that human MUC16 was detected in the coelomic epithelium and its derivatives (Kawabat et al., 1983; Nouwen et al., 1986, 1987; Nap, 1998). In addition, peritoneal mesothelium is regarded as an endogenous source of MUC16, particularly when inflamed (Zeimet et al., 1998). The mesothelium, derived from fetal co-elomic epithelium, is composed of an extensive monolayer of specialized cells that line the body’s serous cavities (pleura, peritoneum) and internal organs. Initially the mesothelium was regarded as a simple epithelia with a straightforward biological function. However, increasing evidence suggests that the mesothelium performs complex biological functions such as fluid and cell transport, initiation and resolution of inflammation, tissue repair, and tumor dissemination (reviewed in Mutsaers, 2002). The exact biological function of MUC16 in the mesothelia is currently unknown. Given the apical localization in the cell membrane and anti-adhesive properties of MUC16 (Gaetje et al., 2002), it has been speculated that MUC16 might serve as a lubricant, protecting surface epithelia from physical stress. Another interesting finding is that MUC16 can bind to the mesothelial cell surface glycoprotein, mesothelin (Rump et al., 2004). Co-localization of MUC16 and mesothelin in both peritoneal mesothelia and ovarian cancer raises the possibility that ovarian cancer cells might implant on peritoneal mesothelia through MUC16-mesothelin interaction.
Mouse MUC16 is also expressed in the pleural mesothelium and the tracheal epithelium, consistent with Nouwen et al. (1986, 1990). A recent study showed that MUC16 is one of the major mucins secreted by human bronchial epithelial cells (Davies et al., 2007), suggesting a possible role for MUC16 in the upper respiratory tract.
In addition to the mesothelia, mouse MUC16 was expressed by some secretory epithelia such as the epithelia of trachea lumen, the laryngeal aditus, olfactory epithelia, and surface epithelia of the cochlear duct. Furthermore, mouse MUC16 expression was detected in some secretory cells such as the ducts in palatine glands and chief cells in the stomach. Human MUC16 was also detected in some secretory glands such as apocrine sweat glands, submucous glands of the trachea, and lacrimal gland (Hardardottir et al.,1990; Davies et al., 2007; Jäger et al., 2007). Likewise, mouse MUC16 was also detected in some secretory epithelia and secretory cells, suggesting a secretory role for MUC16.
Although the expression of mouse MUC16 in the mesothelia is in general agreement with previous studies, a few differences were noted. The most striking difference is MUC16 expression in female reproductive organs. In human, MUC16 was detected in the luminal epithelia of the fallopian tube and the uterine endometrium, regardless of the type of antibody used (Kawabat et al., 1983; Nanbu et al., 1989; Nap 1998). MUC16 expression in the uterine endometrium was commonly found in other mammals (Nouwen et al., 1990; McDonnel et al., 2003). However, mouse MUC16 protein was neither detected in the luminal surface of oviduct nor in the endometrium of the uterus. We detected mouse MUC16 protein only in the mesothelial cells lining the surface of the oviduct and uterus. One possible explanation is that mouse MUC16 expression in the endometrium is dependent upon the estrous cycle and/or pregnancy. Indeed, bovine MUC16 was expressed in the endometrium in the luteal phase and early pregnancy, but not during estrus nor the postpartum period (McDonnel et al., 2003). In contrast, mouse MUC16 was always detected in the ovarian surface epithelium throughout development. This finding contrasts with the current notion that human MUC16 is not expressed by normal ovarian surface epithelium. While the majority of the findings report no human MUC16 expression by normal ovarian surface epithelium (Kawabat et al., 1983; Bast et al., 1998), there are a few reports of weak MUC16 expression by normal ovarian surface epithelium (Nouwen et al., 1987). Normal ovarian surface epithelium, also called germinal epithelium, is a layer of epithelial cells on the surface of the ovary that is continuous with the mesothelium.
In addition to female reproductive organs, mouse MUC16 shows distinct expression patterns in embryonic skin, lung, and eye. Unlike Nanbu et al. (1989), we did not detect MUC16 in the surface of the embryonic skin except in the mast cells. Also mouse MUC16 was not detected in the corneal epithelia in contrast to human MUC16 (Nap, 1998; Argüeso et al., 2003). We did detect MUC16 in the goblet cells in the conjunctival epithelia, but not in the apical surface (Argüeso et al., 2003).
The differences in the expression pattern of mouse MUC16 from human MUC16 might be explained in several ways. First, there may be different epitopes present in mouse MUC16. Indeed, mouse MUC16 did not show cross-reactivity with the OC125 monoclonal antibody, that showed inter-specific cross-reactivity to rabbit, monkey, dog, and cow (Nouwen et al., 1990; McDonnel et al., 2003), implying that mouse MUC16 may have different epitopes that cannot be recognized by the OC125 antibody. This problem might be resolved by the development of another anti-mouse MUC16 antibody targeting a different epitope region. Another possibility is simply that mouse MUC16 is expressed differently from humans and other mammals. Regardless of differences in expression pattern in normal tissues, mouse MUC16 was expressed in the apical cell membrane of mouse serous adenocarcinoma of the ovary, which is consistent with human MUC16 expression in ovarian carcinoma (Nouwen et al., 1986). Therefore, mouse MUC16 is still a useful target to study the role of MUC16 in normal organs as well as in ovarian cancer.
Detection of mouse MUC16 in mouse ovarian carcinomas provides further confirmation that genetically transformed serous cystadenocarcinomas faithfully mimic the human disease. Mouse MUC16 might be used as a serum marker for tracking tumor growth in this experimental system. Conversely, the mouse model may be used to study the regulation of MUC16 during neoplastic transformation as well as the role of MUC16 in peritoneal metastasis.
In conclusion, we found that mouse Muc16 has a conserved gene structure, genomic organization, and expression pattern as human MUC16, suggesting that mouse Muc16 is the ortholog of human MUC16. Our study established the mouse as a promising model system to study the function of MUC16. Mice are a good model system for functional studies, as timed matings allow for ease of tissue collection at designated time points and disruption of genes is routine. Given its expression pattern, MUC16 has been implicated in tissue protection, immune suppression, and reproduction. Future functional studies of mouse MUC16 may unveil new biological functions for MUC16.
Table 1.
Summary of PCR primer sequences, annealing temperatures, and expected product sizes
| Primers | Sequence of forward (F) and reverse (R) primers | Annealing temperature (°C) | Product size (bp) | Accession # (location) |
|---|---|---|---|---|
| 5′Muc16_F | 5′-CATCATATGAAAATACAAGTAGTGG-3′ | 49 | 3500 | XM_911929 |
| 5′Muc 16_R | 5′-CCTTCAGAGCTCTGGTAGTAG-3′ | 53 | (3830-54; 7262-82) | |
| 6F10R_F | 5′-GCCCATGTTCAAGAATAGCAGTATTGG-3′ | 56 | 698 | XM_911929 |
| 6F10R_R | 5′- GTAGCAGAGAGGGGCTTGTGGTTG-3′ | 58 | (18903-29; 19577-600) | |
| 22F27R_F | 5′-CACAACATTACTCAACTGGGTCCC-3′ | 53 | 855 | XM_911929 |
| 22F27R_R | 5′-GTTCACTGTGAACAGCTCCTC-3′ | 52 | (22111-32; 22945-65) | |
| 38F43R_F | 5′-CCCTTGGTCCAGAATGAATCCC-3′ | 53 | 574 | XM_911929 |
| 38F43R_R | 5′-CACGTGCAGATCTTTCAACTGGTAGG-3′ | 58 | (25066-87; 25610-35) |
Acknowledgements
This work was supported by a United States Department of the Army grant DAMD 17-03-1-0449 to R. R. B. and R. C. B, Jr., the Ben F. Love Endowment to R. R. B. and NIH R01CA103924 to S. O. D.-J. Cheon was supported by a Bettyann Asche-Murray Ovarian Cancer Fellowship and a Schissler Foundation Fellowship in Genetics of Human Disease. DNA sequencing and veterinary resources were supported by the NIH Cancer Center Support (Core) Grant, CA16672.
Contributor Information
Ying Wang, Department of Molecular Genetics, M. D. Anderson Cancer Center, University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Dong-Joo Cheon, Department of Molecular Genetics, M. D. Anderson Cancer Center, University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Zhen Lu, Department of Experimental Therapeutics, M. D. Anderson Cancer Center, University of Texas, Houston, TX 77030, USA.
Sheena L. Cunningham, Des Moines University of College of Osteopathic Medicine, Des Moines, IA 50312, USA
Chun-Ming Chen, Department of Molecular Genetics, M. D. Anderson Cancer Center, University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
Robert Z. Luo, Department of Experimental Therapeutics, M. D. Anderson Cancer Center, University of Texas, Houston, TX 77030, USA
Deyin Xing, Center for Cancer Research, Massachusetts General Hospital, Boston, MA 02114, USA.
Sandra Orsulic, Center for Cancer Research, Massachusetts General Hospital, Boston, MA 02114, USA.
Robert C. Bast, Jr., Department of Experimental Therapeutics, M. D. Anderson Cancer Center, University of Texas, Houston, TX 77030, USA
Richard R. Behringer, Department of Molecular Genetics, M. D. Anderson Cancer Center, University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
References
- Argüeso P, Spurr-Michaud S, Russo CL, Tisdale A, Gipson IK. MUC16 mucin is expressed by the human ocular surface epithelia and carries the H185 carbohydrate epitope. Invest Ophthalmol Vis Sci. 2003;44:2487–2495. doi: 10.1167/iovs.02-0862. [DOI] [PubMed] [Google Scholar]
- Bast RC, Jr., Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC, Jr., Klug TL, St.John E, Jenison E, Niloff JM, Lazarus H, Berkowitz RS, Leavitt T, Griffiths CT, Parker L, Zurawski VR, Jr., Knapp RC. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N Engl J Med. 1983;309:883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- Bast RC, Jr., Xu FJ, Yu YH, Barnhill S, Zhang Z, Mills GB. CA125: the past and the future. Int J Biol Markers. 1998;13:179–187. doi: 10.1177/172460089801300402. [DOI] [PubMed] [Google Scholar]
- Davies JR, Kirkham S, Svitacheva N, Thornton DJ, Carlstedt I. MUC16 is produced in tracheal surface epithelium and submucosal glands and is present in secretions from normal human airway and cultured bronchial epithelial cells. Int J Biochem Cell Biol. 2007;39:1943–1954. doi: 10.1016/j.biocel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Davis HM, Zurawski VR, Jr., Bast RC, Jr., Klug TL. Characterization of the CA125 antigen associated with human epithelial ovarian carcinomas. Cancer Res. 1986;46:6143–6148. [PubMed] [Google Scholar]
- De los Frailes MT, Stark S, Jaeger W, Hoerauf A, Wildt L. Purification and characterization of the CA125 tumor-associated antigen from human ascites. Tumour Biol. 1993;14:18–29. doi: 10.1159/000217821. [DOI] [PubMed] [Google Scholar]
- Gaetje R, Winnekendonk DW, Ahr A, Kaufmann M. Ovarian cancer antigen CA125 influences adhesion of human and mammalian cell lines in vitro. Clin Exp Obstet Gynecol. 2002;29:34–36. [PubMed] [Google Scholar]
- Gubbels JA, Belisle J, Onda M, Rancourt C, Migneault M, Ho M, Bera TK, Connor J, Sathyanarayana BK, Lee B, Pastan I, Patankar MS. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol Cancer. 2006;5:50–65. doi: 10.1186/1476-4598-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardardottir H, Parmley TH, II, Quirk JG, Jr., Sanders MM, Miller FC, O’Brien TJ. Distribution of CA125 in embryonic tissues and adult derivatives of the fetal periderm. Am J Obstet Gynecol. 1990;163:1925–1931. doi: 10.1016/0002-9378(90)90775-3. [DOI] [PubMed] [Google Scholar]
- Jäger K, Wu G, Sel S, Garreis F, Bräuer L, Paulsen FP. MUC16 in the lacrimal apparatus. Histochem Cell Biol. 2007;127:433–438. doi: 10.1007/s00418-006-0246-6. [DOI] [PubMed] [Google Scholar]
- Kabawat SE, Bast RC, Jr., Bhan AK, Welch WR, Knapp RC, Colvin RB. Tissue distribution of a coelomic-epithelium-related antigen recognized by the monoclonal antibody OC125. Int J Gynecol Pathol. 1983;2:275–285. doi: 10.1097/00004347-198303000-00005. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Ida W, Terao T, Kaeashima Y. Molecular characteristics of the CA125 antigen produced by human endometrial epithelial cells: comparison between eutopic and heterotopic epithelial cells. Am J Obestet Gynecol. 1993;169:725–730. doi: 10.1016/0002-9378(93)90652-y. [DOI] [PubMed] [Google Scholar]
- Lloyd KO, Yin BWT, Kudryashov V. Isolation and characterization of ovarian cancer antigen CA125 using a new monoclonal antibody (VK-8): identification as a mucin-type molecule. Int J Cancer. 1997;71:842–850. doi: 10.1002/(sici)1097-0215(19970529)71:5<842::aid-ijc24>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Lloyd KO, Yin BWT. Synthesis and secretion of the ovarian cancer antigen CA125 by the human cancer cell line NIH:OVCAR-3. Tumor Biol. 2001;22:77–82. doi: 10.1159/000050600. [DOI] [PubMed] [Google Scholar]
- Maeda T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Motoda Y, Kobayashi A, Hiroyasu F, Shirouzu M, Terada T, Hayami N, Ishizuka Y, Shinya N, Tatsuguchi A, Yoshida M, Hirota H, Matsuo Y, Tani K, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kigawa T, Yokoyama S. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16) J Biol Chem. 2004;279:13174–13182. doi: 10.1074/jbc.M309417200. [DOI] [PubMed] [Google Scholar]
- McDonnel AC, Van Kirk EA, Austin KJ, Hansen TR, Belden EL, Murdoch WJ. Expression of CA-125 by progestational bovine endometrium: prospective regulation and function. Reproduction. 2003;126:615–620. [PubMed] [Google Scholar]
- Mutsaers SE. Mesothelial cells: their structure, function and role in serosal repair. Respirology. 2002;7:171–191. doi: 10.1046/j.1440-1843.2002.00404.x. [DOI] [PubMed] [Google Scholar]
- Nagata A, Hirota N, Sakai T, Fujimoto M, Komoda T. Molecular nature and possible presence of a membranous glycan-phosphatidylinositol anchor of CA125 antigen. Tumour Biol. 1991;12:279–286. doi: 10.1159/000217716. [DOI] [PubMed] [Google Scholar]
- Nanbu Y, Fujii S, Konishi I, Nonogaki H, Mori T. CA125 in the epithelium closely related to the embryonic ectoderm: the periderm and the amnion. Am J Obstet Gynecol. 1989;161:462–467. doi: 10.1016/0002-9378(89)90542-5. [DOI] [PubMed] [Google Scholar]
- Nap M. Immunohistochemistry of CA125. Unusual expression in normal tissues, distribution in the human fetus and questions around its application in diagnostic pathology. Int J Biol Markers. 1998;13:210–215. [PubMed] [Google Scholar]
- Nouwen EJ, Pollet DE, Eerdekens MW, Hendrix PG, Briers TW, De Broe ME. Immunohistochemical localization of placental alkaline phosphatase, carcinoembryonic antigen, and cancer antigen 125 in normal and neoplastic human lung. Cancer Res. 1986;46:866–876. [PubMed] [Google Scholar]
- Nouwen EJ, Hendrix PG, Dauwe S, Eerdekens MW, De Broe ME. Tumor markers in the human ovary and its neoplasms. A comparative immunohistochemical study. Am J Pathol. 1987;126:230–242. [PMC free article] [PubMed] [Google Scholar]
- Nouwen EJ, Dauwe S, De Broe ME. Occurrence of the mucinous differentiation antigen CA125 in genital tract and conductive airway epithelia of diverse mammalian species (rabbit, dog, monkey) Differentiation. 1990;45:192–198. doi: 10.1111/j.1432-0436.1990.tb00473.x. [DOI] [PubMed] [Google Scholar]
- O’Brien TJ, Beard JB, Underwood LJ, Dennis RA, Santin AD, York L. The CA125 gene: an extracellular superstructure dominated by repeat sequences. Tumour Biol. 2001;22:348–366. doi: 10.1159/000050638. [DOI] [PubMed] [Google Scholar]
- O’Brien TJ, Beard JB, Underwood LJ, Shigemasa K. The CA 125 gene: a newly discovered extension of the glycosylated N-terminal domain doubles the size of this extracellular superstructure. Tumour Biol. 2002;23:154–169. doi: 10.1159/000064032. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus H. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann H, Perlmann P. Cell biology: a laboratory handbook. Academic Press Inc.; San Diego: 1994. [Google Scholar]
- Rump A, Morikawa Y, Tanaka M, Minami S, Umesaki N, Takeuchi M, Miyajima A. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–9198. doi: 10.1074/jbc.M312372200. [DOI] [PubMed] [Google Scholar]
- Scholler N, Garvik B, Hayden-Ledbetter M, Kline T, Urban N. Development of a CA125-mesothelin cell adhesion assay as a screening tool for biologics discovery. Cancer Lett. 2007;247:130–136. doi: 10.1016/j.canlet.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radio-labeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- Xing D, Orsulic S. A mouse model for the molecular characterization of brca1-associated ovarian carcinoma. Cancer Res. 2006;66:8949–8953. doi: 10.1158/0008-5472.CAN-06-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin BWT, Lloyd KO. Molecular cloning of the CA125 ovarian cancer antigen: identification as a new mucin, MUC16. J Biol Chem. 2001;276:27371–27375. doi: 10.1074/jbc.M103554200. [DOI] [PubMed] [Google Scholar]
- Yin BWT, Dnistrian A, Lloyd KO. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int J Cancer. 2002;98:737–740. doi: 10.1002/ijc.10250. [DOI] [PubMed] [Google Scholar]
- Yu YH, Schlossman DM, Harrison CL, Rhinehardt-Clark A, Soper JT, Klug TL, Zurawski VR, Jr., Bast RC., Jr. Coexpression of different antigenic markers on moieties that bear CA125 determinants. Cancer Res. 1991;51:468–475. [PubMed] [Google Scholar]
- Zeimet AG, Offner FA, Muller-Holzner E, Widschwendter M, Abendstein B, Fuith LC, Daxenbichler G, Marth C. Peritoneum and tissues of the female reproductive tract as physiological sources of CA-125. Tumour Biol. 1998;19:275–282. doi: 10.1159/000030018. [DOI] [PubMed] [Google Scholar]


