Introduction
Genetic approaches have yielded progress toward characterizing the relationship of molecularly defined neuronal cell types to behavior. However, relatively little is known about the functional properties of the connections between these molecularly defined cell types. This limitation arises because multiple cell types and their projections are typically intermingled, and methods for selective activation have been lacking. Traditional methodology for probing synaptic connections, such as electrical stimulation, is not specific for axons arising from separate cell types, and recording connections between identified neuron pairs typically becomes prohibitively inefficient as the distance between neurons increases. Recently, however, channelrhodopsin-2 (ChR2) has been used as an alternative approach to activate neurons and their axons (Boyden et al., 2005; Petreanu et al., 2007; Wang et al., 2007), in this case with light. Because ChR2 can be genetically targeted, it is well suited for mapping the connections of molecularly defined cell populations using photostimulation instead of electrical stimulation. Nevertheless, this approach is not yet routine because many cell type-selective promoters cannot achieve levels of ChR2 expression sufficient for photostimulation, especially in axons far from the cell body.
To overcome these problems, we developed a Cre recombinase-dependent viral vector for targeting ChR2 to spatially restricted subsets of molecularly defined neurons with expression levels sufficient to permit reliable somatic and axonal photostimulation. By tagging ChR2 with the fluorescent protein mCherry (ChR2mCherry), this tool can also be used for characterizing cell morphology and the anatomy of axon projections. In this study, we set out to demonstrate the utility of this approach by targeting ChR2mCherry to two separate molecularly defined neuron populations in the hypothalamus. Furthermore, we used this tool to record, for the first time, long-range synaptic connections of molecularly defined cell types in neural circuits that regulate feeding behavior.
Materials and Methods
All experimental protocols were conducted according to United States National Institutes of Health guidelines for animal research and were approved by the Institutional Animal Care and Use Committee at Janelia Farm Research Campus.
Viral vector
The flip-excision (FLEX) switch was synthesized de novo (DNA 2.0) with flanking BamH1 and EcoRI restriction sites and an internal SpeI site (GenBank accession number EU784738) (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material). The vector was digested with SpeI and blunt-ended. Codon-optimized ChR2mCherry with the point mutation H134R was used for these studies. Blunt-end ChR2mCherry was ligated into the FLEX switch. Forward and reverse orientations of the corresponding construct were digested with BamH1/EcoRI and ligated into a vector (supplied by A. Karpova, Janelia Farm Research Campus, Ashburn, VA) containing the CAG promoter, a WPRE sequence, SV40 polyA sequence, and two inverted terminal repeats required for recombinant adeno-associated virus (rAAV) production. rAAV-FLEX-rev-ChR2mCherry was produced by the University of Pennsylvania Gene Therapy Program Vector Core.
HEK 293 cells were transfected using Fugene HD (Roche Diagnostics) according to the manufacturer's instructions. FLEX-ChR2mCherry vectors were cotransfected with Cre-IRES-EGFP in the pcDNA3.1 vector (Invitrogen) or with empty pcDNA3.1. After 48 h, cells were imaged for mCherry and EGFP fluorescence using mRFP and EGFP filter cubes (Chroma).
Stereotaxic rAAV injections
Mice were anesthetized with isoflurane and were placed into a stereotaxic apparatus (David Kopf Instruments). The skull was exposed via a small incision, and a small hole was drilled for injection. For injection into the arcuate nucleus of the hypothalamus (ARC), a pulled-glass pipette with 20–40 μm tip diameter was inserted into the brain, and three 50 nl injections were made (coordinates, bregma: −1.5 mm; midline: +0.2 mm; skull surface: −5.7 mm, −5.5 mm, −5.3 mm). A micromanipulator (Narishige) was used to control injection speed at 30 nl/min, and the pipette was withdrawn 15 min after the final injection. For injection, rAAV-FLEX-rev-ChR2mCherry titer was 3.4 × 1013 genomic copies (GC)/ml; rAAV-ChR2EGFP titer was 1.8 × 1012 GC/ml.
Imaging
Mice were deeply anesthetized and then perfused with cold saline followed by 4% paraformaldehyde (PFA) in PBS and fixed overnight in PFA. Brain sections (60 μm) were cut, mounted on glass slides, and coverslipped for imaging. Neuron images were collected by confocal microscopy (Zeiss).
Electrophysiology and photostimulation
Experimental techniques were similar to those reported previously (Petreanu et al., 2007), and only the differences are described here. Mice [postnatal day 21 (P21)–P25] were infected as above, and 5–8 d later they were deeply anesthetized with isoflurane and decapitated. Coronal brain slices were prepared in chilled cutting solution containing the following (in mm): 119 NaCl, 25 NaHCO3, 11 d-glucose, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, and 1.25 mm NaH2PO4, aerated with 95:5 O2/CO2. Slices were transferred to artificial CSF (ACSF) containing the following (in mm): 119 NaCl, 25 NaHCO3, 11 d-glucose, 2.5 KCl, 1.25 MgCl2, 2 CaCl2, and 1.25 NaH2PO4, aerated with 95% O2/5% CO2. For mapping synaptic connections from proopiomelanocortin (POMC) neurons→paraventricular hypothalamic nucleus (PVN) neurons, divalent concentrations in ACSF were modified (0.1 mm MgCl2 and 4.0 mm CaCl2) to increase release probability. Slices were incubated at 34°C for 30 min and then transferred to a recording chamber with ACSF at room temperature (20–24°C). Neurons were identified, in some cases by mCherry fluorescence emission (excitation, 575 nm; emission, 640 nm; dichroic, 610 nm longpass; Chroma), and then visually targeted with infrared gradient optics. Neurons were patched using electrodes with tip resistances 4–5 MΩ. The intracellular solution for current clamp recordings contained the following (in mm): 125 potassium gluconate, 10 KCl, 10 HEPES, 1 EGTA, 4 Mg-ATP, 0.3 Na2GTP, and 10 sodium phosphocreatine, pH 7.25 and 290 mOsm. The intracellular solution for voltage-clamp recordings contained the following (in mm): 125 CsCl, 5 NaCl, 10 HEPES, 0.6 EGTA, 4 Mg-ATP, 0.3 Na2GTP, and 10 lidocaine N-ethyl bromide (QX-314), pH 7.35 and 290 mOsm. The holding potential for voltage-clamp recordings was −60 mV. Responses were digitized at 10 kHz and filtered at 1 kHz.
A blue laser (473 nm; CrystaLaser) was used to deliver photostimuli ranging from 0.01 to 150 μW. Laser power was measured with a photodiode (Edmund Optics). The response of the photodiode was calibrated by measuring laser power at the specimen using a light power meter (Coherent). Neutral density filters were used to control the laser power at the specimen. Pulse duration was controlled by a Pockels cell (ConOptics) and a Uniblitz shutter (Vincent Associates). For measuring photocurrents, pulse duration was 500 ms. For photostimulation and for channelrhodopsin-assisted circuit mapping, laser pulse duration was 1 ms, and laser power was 50–150 μW at the specimen. For laser light delivery by optical fiber, laser power was 1.6 mW/mm2. Photostimulation was performed with glutamate receptor blockade (50 μm AP-5 and 10 μm CNQX). Light pulses were delivered every 400 ms on a grid with 75 μm spacing between stimulation sites and were given to maximize time between stimuli to neighboring spots.
Results
For the viral vector, we selected rAAV, which is widely used for gene delivery in the CNS. rAAV serotype 2/1 exhibits neuronal tropism, and it can be readily produced at high titer. This facilitates high expression levels, because multiple viral particles are taken up by each cell (Bartlett et al., 2000). Moreover, use of rAAV2/1 is risk group 1 and does not require special facilities for delivering the virus or housing infected animals.
FLEX switch in rAAV for ChR2mCherry expression
Because rAAV2/1 will infect neurons throughout an injection site, we sought to eliminate the possibility of background ChR2mCherry expression in cells not expressing Cre. For this, we investigated Cre-mediated inversion of the ChR2mCherry coding sequence. In this approach, the coding sequence is inverted and is therefore in the wrong orientation for transcription until Cre inverts the sequence, turning on transcription of ChR2mCherry. However, although DNA inversion can be achieved with site-specific recombination between loxP sites placed in an antiparallel orientation, inversion is not stable and leads to a mixture of reverse and forward configurations, which would reduce ChR2mCherry expression levels.
To achieve stable transgene inversion, we tested a FLEX switch (Schnütgen et al., 2003). The FLEX switch uses two pairs of heterotypic, antiparallel loxP-type recombination sites, which first undergo an inversion of the coding sequence, followed by excision of two sites, leading to one of each orthogonal recombination site oppositely oriented and incapable of further recombination (Fig. 1A). For this, we used loxP and lox2272 sequences, which show efficient homotypic but not heterotypic recombination (Lee and Saito, 1998).
Figure 1.
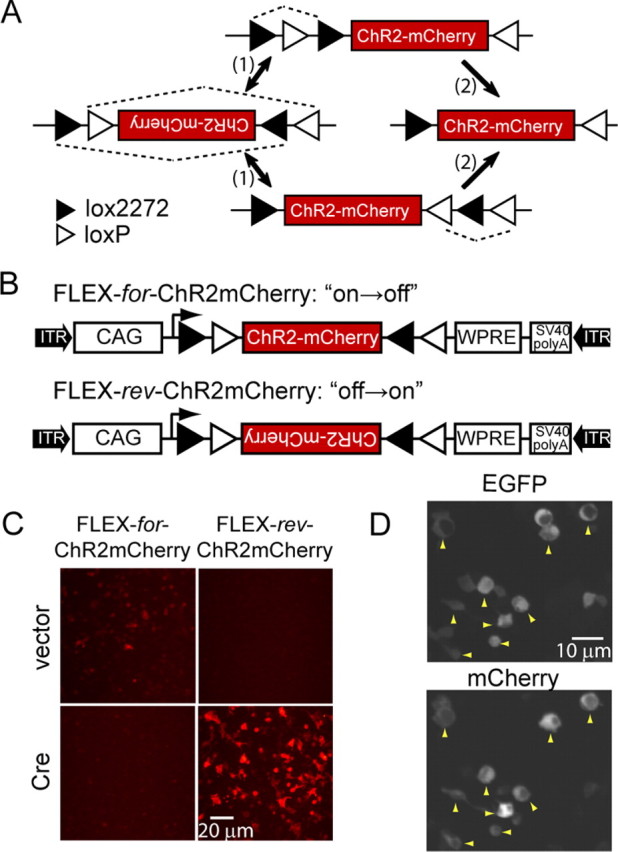
Design and characterization of a FLEX switch for ChR2mCherry. A, FLEX switch recombination sequence for stable inversion proceeds in two steps: (1) inversion followed by (2) excision. loxP and lox2272 are orthogonal recombination sites. B, Construct design for FLEX-for-ChR2mCherry and FLEX-rev-ChR2mCherry. CAG, CMV enhancer/β-globin chimeric promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; ITR, inverted terminal repeat. C, Images showing mCherry fluorescence in HEK 293 cells for FLEX-for-ChR2mCherry and FLEX-rev-ChR2mCherry in the presence and absence of Cre. D, Colocalization of EGFP and mCherry fluorescence (yellow arrowheads) in HEK 293 cells cotransfected with FLEX-rev-ChR2mCherry and Cre-IRES-EGFP.
We characterized the efficiency, stability, and background expression of this stable inversion approach with comple-mentary gain- and loss-of-function experiments in transfected HEK 293 cells, using mCherry fluorescence as the readout. First, we considered inversion efficiency and irreversibility by switching off expression in a construct in which ChR2mCherry coding sequence was oriented in the forward direction with respect to the promoter (FLEX-for-ChR2mCherry) (Fig. 1B). In this “on→off” construct, 0/73 Cre-expressing cells showed mCherry fluorescence, which demonstrates both the high efficiency and the desired irreversibility of this FLEX switch (Fig. 1C). Next, we tested the “off→on” construct, in which the ChR2mCherry coding sequence is in the reverse orientation (FLEX-rev-ChR2mCherry) (Fig. 1B). For this configuration, ChR2mCherry was strongly expressed in the presence but not the absence of Cre (Fig. 1C). Furthermore, mCherry fluorescence was only found in Cre-expressing cells when FLEX-rev-ChR2mCherry was cotransfected with Cre-IRES-EGFP as revealed by colocalization with EGFP fluorescence (Fig. 1D). Thus, these data show that FLEX-rev-ChR2mCherry provides stable, Cre-dependent transgene expression in HEK 293 cells.
For targeting ChR2mCherry to the CNS, FLEX-rev-ChR2mCherry was packaged into rAAV2/1 (rAAV-FLEX-rev-ChR2mCherry). This virus was first tested to confirm the lack of background expression in vivo by injection of a large volume (150 nl) of rAAV-FLEX-rev-ChR2mCherry into the hypothalamus of wild-type mice. To mark the extent of the injection site, we coinfected with rAAV-ChR2-EGFP (not requiring Cre for expression). Although the fluorescence resulting from EGFP expression extended over much of the hypothalamus (Fig. 2A, left), no mCherry fluorescence was seen (Fig. 2A, right), verifying that rAAV-FLEX-rev-ChR2mCherry has no detectable background expression in vivo in the absence of Cre.
Figure 2.
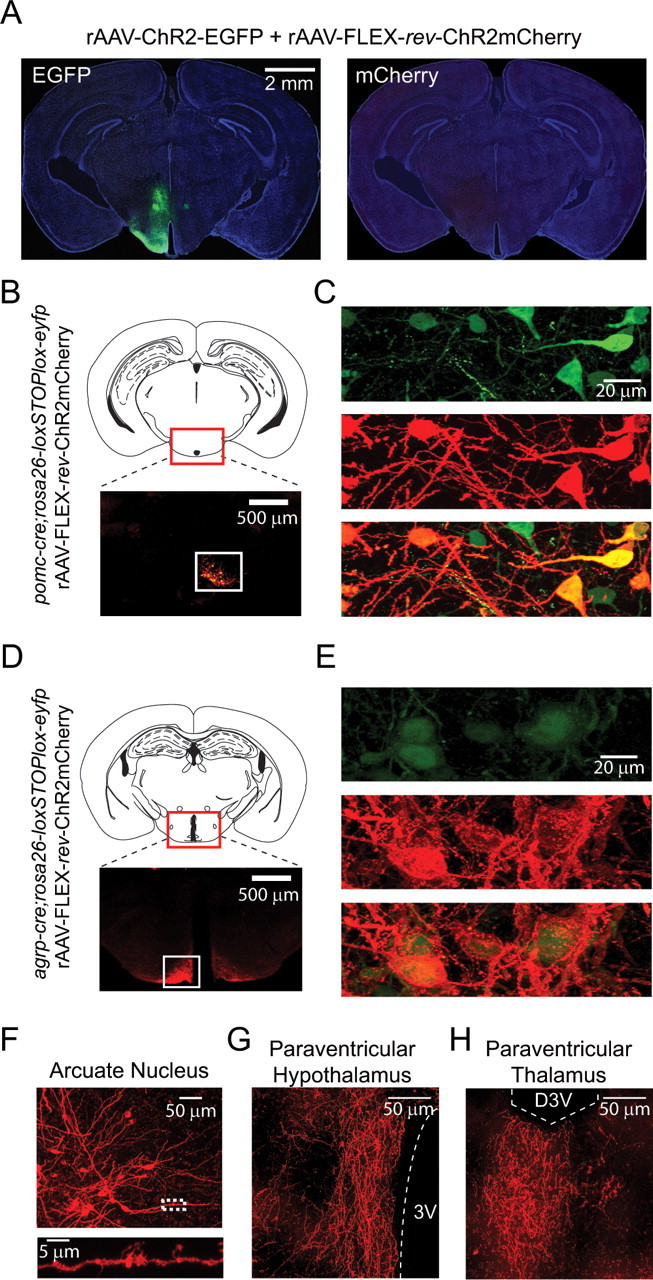
Cre-dependent ChR2mCherry expression in transgenic mice using rAAV-FLEX-rev-ChR2mCherry. A, Distribution of fluorescent neurons resulting from a large coinjection (150 nl) of rAAV-ChR2-EGFP and rAAV-FLEX-rev-ChR2mCherry into the hypothalamus of wild-type mice. Extensive fluorescence from EGFP (left) but no fluorescence from mCherry in brain slices (right) shows the absence of background expression with rAAV-FLEX-rev-ChR2mCherry. The background image of the slice was obtained from 4′,6′-diamidino-2-phenylindole fluorescence. B, Top, Schematic showing location of the imaged area in caudal arcuate nucleus. Bottom, mCherry fluorescence only in the arcuate nucleus after a large injection of rAAV-FLEX-rev-ChR2mCherry into the hypothalamus of pomc-cre;rosa26-loxSTOPlox-eyfp mice. Compare distribution of fluorescence with A. C, Colocalization of mCherry and EYFP fluorescence in arcuate nucleus. D, E, Similar to B and C; in this case, agrp-cre;rosa26-loxSTOPlox-eyfp mice were used with rAAV-FLEX-rev-ChR2mCherry virus injections. F, Top, Image showing neuron morphology from the arcuate nucleus of labeled POMC neurons. Bottom, Higher-magnification image of boxed area. G, H, Axonal projections of AGRP neurons infected with rAAV-FLEX-rev-ChR2mCherry. Strong axonal labeling was observed in the paraventricular nucleus of the hypothalamus (G) and paraventricular thalamus (H). 3V, Third ventricle; D3V, dorsal third ventricle.
Next, we characterized expression of rAAV-FLEX-rev-ChR2mCherry in pomc-cre (Balthasar et al., 2004) and agrp-cre (Kaelin et al., 2004) transgenic mice. POMC and agouti-related protein (AGRP) gene expression each define separate neuron populations intermingled in the ARC, which, respectively, inhibit and stimulate feeding (Morton et al., 2006). Injection of rAAV-FLEX-rev-ChR2mCherry into the medial hypothalamus of pomc-cre or agrp-cre mice labeled neurons only in the ARC (Fig. 2B,D) (for comparison with Cre-independent infection pattern, see Fig. 2A). We also used a rosa26-loxSTOPlox-eyfp (Srinivas et al., 2001) reporter mouse to further demonstrate the Cre dependence of ChR2mCherry expression. After viral transduction of pomc-cre;rosa26-loxSTOPlox-eyfp or agrp-cre;rosa26-loxSTOPlox-eyfp, mCherry fluorescence colocalized, in all cases, with EYFP-expressing cells (Fig. 2C,E). For example, in six brain slices from agrp-cre;rosa26-loxSTOPlox-eyfp mice infected with rAAV-FLEX-rev-ChR2mCherry, 254/254 mCherry-expressing cells were also GFP positive. Thus, these data show that rAAV-FLEX-rev-ChR2mCherry is well suited for cell type-specific transgene expression in the mouse brain.
Imaging molecularly defined cell types
Because ChR2mCherry distributes efficiently into the membranes of neuronal processes, it is a useful tool for characterizing neuron morphology and anatomy. Using rAAV-FLEX-rev-ChR2mCherry to label POMC neurons allows clear identification of the long dendrites that run along the lateral edge of the ARC (Fig. 2F, top). Moreover, close inspection of ChR2mCherry-labeled POMC neurons reveals numerous dendritic filopodia (Fig. 2F, bottom), which have been previously observed in Golgi-stained ARC neurons (van den Pol and Cassidy, 1982). In contrast, the EYFP fluorescence in pomc-cre;rosa26-loxSTOPlox-eyfp or agrp-cre;rosa26-loxSTOPlox-eyfp was, in most cases, confined to the soma or proximal dendrites (Fig. 2C,E).
Axons from Cre-expressing cell populations are also clearly labeled with ChR2mCherry. We observed strong AGRP neuron projections in the paraventricular hypothalamic nucleus (Fig. 2G) and the paraventricular thalamic nucleus (Fig. 2H). Furthermore, the localization of rAAV delivery permits unambiguous mapping of projections from spatially defined neuron populations in mouse lines that express Cre in multiple brain regions. Thus, rAAV-FLEX-rev-ChR2mCherry is also useful for mapping the projection patterns of molecularly defined neurons.
Photostimulation of molecularly defined cell types
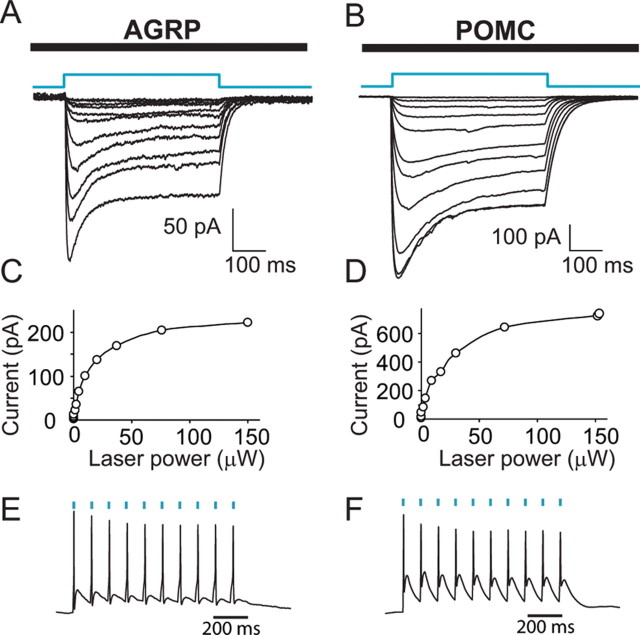
Next, we set out to demonstrate ChR2mCherry-mediated photostimulation in multiple cell types using rAAV-FLEX-rev-ChR2mCherry. Strong photocurrents were evoked with focused laser light (473 nm) during whole-cell voltage-clamp recordings of AGRP (Fig. 3A) and POMC (Fig. 3B) neurons identified by mCherry fluorescence in brain slices. Current amplitudes reached a plateau at ∼100 μW laser power (Fig. 3C,D), and both neuron populations could also be repetitively photostimulated to fire action potentials with brief (1 ms) laser flashes (Fig. 3E,F). Spike latencies were 6.9 ± 0.6 ms for AGRP neurons (n = 5) and 8.9 ± 2.1 ms for POMC neurons (n = 4). Neurons could also be photostimulated using an optical fiber for light delivery to the brain slice (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material).
Figure 3.
Photostimulation of AGRP and POMC neurons in the hypothalamus. A, B, Whole-cell voltage-clamp recordings from mCherry positive neurons in hypothalamic slices from agrp-cre or pomc-cre mice infected with rAAV-FLEX-rev-ChR2mCherry. Light pulses (500 ms) of varying power elicited ChR2mCherry-mediated inward currents. C, D, The peak current is plotted as a function of laser power for AGRP (C) and POMC (D) neurons. E, F, Perisomatic repetitive stimulation with 1 ms light pulses at 10 Hz in AGRP (E) and POMC (F) neurons. Blue dashes mark timing of light flashes.
Because rAAV-FLEX-rev-ChR2m-Cherry labels axons from molecularly defined cell types, we also tested whether photostimulation of these axons could be used for cell type-specific, ChR2-assisted circuit mapping (CRACM) (Petreanu et al., 2007). Anatomic, genetic, and pharmacologic studies have shown that projections from AGRP and POMC neurons converge in the PVN to regulate energy homeostasis (Cowley et al., 1999). Using rAAV-FLEX-rev-ChR2mCherry, we performed CRACM between POMC or AGRP neurons and PVN neurons (Fig. 4A). Because POMC neurons, AGRP neurons, and their projections are intermingled with each other and additional axon projections, it has not previously been possible to selectively record the synaptic connections of these neurons in brain slices.
Figure 4.
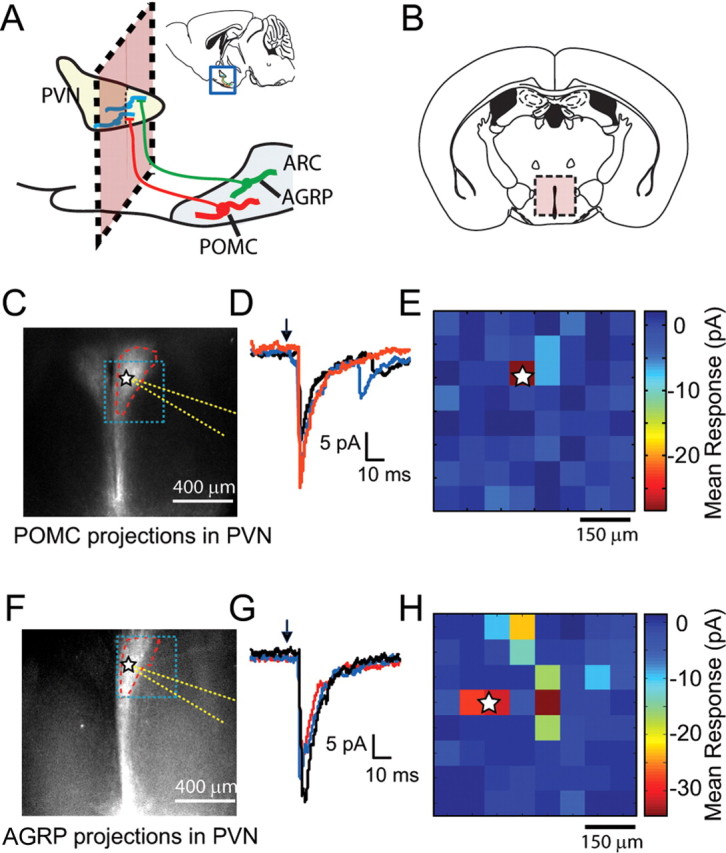
Channelrhodopsin-assisted circuit mapping for hypothalamic neuronal circuits: AGRP→PVN and POMC→PVN. A, Diagram of a sagittal hypothalamic section depicting anatomy of connections between ARC and PVN. The pink box denotes the plane of the coronal slice. B, Coronal slices containing PVN, but not arcuate nucleus, were used for whole-cell voltage-clamp recordings from PVN neurons. C, Fluorescence image showing POMC axonal projections to PVN. Blue box outlines region of laser stimulation in E, PVN boundary is outlined in red, location of recorded cell body is marked by a star, and recording pipette is outlined in yellow. D, Overlay of POMC→PVN IPSCs resulting from three photostimulation trials at a site perisomatic to a voltage-clamped PVN neuron. E, Synaptic input map shows mean current responses over 100 ms time window as a color map in voltage-clamped PVN neuron resulting from LSPS of axons originating from POMC neurons. The position of the soma is marked with a star. F–H, Similar to C–E, but in this case, projections arise from AGRP neurons.
Coronal brain slices containing the PVN (Fig. 4B), which is anterior to the ARC, lack AGRP or POMC neurons. However, in slices from pomc-cre and agrp-cre mice infected with rAAV-FLEX-rev-ChR2mCherry, mCherry fluorescence is strong in axonal afferents, which fill the PVN (Fig. 4C,F; see also Fig. 2G). Using laser-scanning photostimulation (LSPS), we recorded whole-cell inhibitory currents in PVN neurons while targeting a focused laser beam to discrete positions in the field of ChR2-labeled AGRP or POMC axons (Fig. 4E,H). We found that perisomatic as well as more distal stimulation sites evoked synaptic currents arising from AGRP and POMC axons (Fig. 4D,G), and afferents from both cell types released GABA as their neurotransmitter as synaptic responses were blocked by picrotoxin (data not shown). Evoked currents were also observed using light delivery by optical fiber (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). These data demonstrate that rAAV-FLEX-rev-ChR2mCherry can be used for CRACM in molecularly defined circuits.
Discussion
Optogenetic mapping of molecularly defined circuits
We have developed a viral vector, rAAV-FLEX-rev-ChR2mCherry, for labeling and photostimulation of molecularly defined neurons. Because ChR2mCherry distributes efficiently into cell membranes, rAAV-FLEX-rev-ChR2mCherry is well suited for recording from the long-range connections of molecularly defined neurons. Also, because the virus is targeted by injection to a spatially defined subset of neurons, the origin of labeled projections is unambiguous, even if Cre is expressed in multiple brain regions. Transduction of Cre-expressing cell types with rAAV-FLEX-rev-ChR2mCherry also enables efficient photostimulation with only brief exposure (1 ms) to <100 μW focused laser light or to light from an optical fiber placed over the brain slice. These properties are desirable for mapping neural circuits with CRACM. In this regard, we used rAAV-FLEX-rev-ChR2mCherry to selectively photo-activate presynaptic inputs from AGRP→PVN and POMC→PVN. Although electrophysiological characterization of afferent inputs to the PVN has been reported previously (Boudaba et al., 1996), it has not been possible to determine the cell types from which these afferents originated. Using rAAV-FLEX-rev-ChR2mCherry, investigation of these and other circuits involving molecularly defined neurons will be possible. In addition, the high expression levels and low laser power for photostimulation demonstrated here are consistent with the requirements for effective in vivo photostimulation for use in behavioral studies.
Technical considerations for Cre-dependent rAAV
The FLEX strategy is well suited for Cre-conditional gene delivery by rAAV because transgene expression is mechanistically constrained by Cre-dependent inversion. This is preferable to standard approaches involving excision of a transcriptional terminator sequence because these sequences can show “read-through” of transcription, especially when using strong promoters. This difficulty was reported recently for a Cre-conditional rAAV2/9 vector for fluorescent proteins and ChR2mCherry that used a loxP-flanked transcriptional terminator sequence (Kuhlman and Huang, 2008). Furthermore, the FLEX switch sequence is short (∼300 bp), which is ideal for use in the small, 4800 bp rAAV genome (Dong et al., 1996). Conversely, transcriptional terminator sequences are typically 5–7 times longer the FLEX switch (Kuhlman and Huang, 2008). Thus, the rAAV-FLEX-rev delivery approach permits expression of larger transgenes.
In summary, the ongoing systematic efforts to define gene expression in the mouse brain have identified promoters that could be used to deliver transgenes to neuron populations, and mouse genetic techniques now routinely target Cre to these cell types (Gong et al., 2007). The rAAV-FLEX-rev delivery approach could be used for expression of transgenes in a temporally and spatially restricted manner, using any Cre driver line as the template. These approaches are critical to expand the study of neural circuitry to molecularly defined neuron populations. Here, we show that anatomically complex circuits, such as those in the hypothalamus, can be readily dissected using these molecular genetic and optogenetic tools.
Footnotes
This work was supported by the Howard Hughes Medical Institute. We thank K. Svoboda and L. Petreanu for technical discussions, A. Karpova for the rAAV CAG vector backbone, N. Ghitani for histology and imaging assistance, and H. White for cell culture assistance.
Editor's Note: Toolboxes are intended to briefly highlight and evaluate an emerging approach or a resource that is becoming widely used in neuroscience. For more information, see http://www.jneurosci.org/misc/itoa.shtml.
References
- Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC, Jr, Elmquist JK, Lowell BB. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaba C, Szabó K, Tasker JG. Physiological mapping of local inhibitory inputs to the hypothalamic paraventricular nucleus. J Neurosci. 1996;16:7151–7160. doi: 10.1523/JNEUROSCI.16-22-07151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Cowley MA, Pronchuk N, Fan W, Dinulescu DM, Colmers WF, Cone RD. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/s0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin CB, Xu AW, Lu XY, Barsh GS. Transcriptional regulation of agouti-related protein (Agrp) in transgenic mice. Endocrinology. 2004;145:5798–5806. doi: 10.1210/en.2004-0956. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS ONE. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN, Cassidy JR. The hypothalamic arcuate nucleus of rat–a quantitative Golgi analysis. J Comp Neurol. 1982;204:65–98. doi: 10.1002/cne.902040108. [DOI] [PubMed] [Google Scholar]
- Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, Kasai H, Hall WC, Feng G, Augustine GJ. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]