Abstract
Estrogens have cell-specific effects on a variety of physiological endpoints including regulation of mitochondrial biogenesis and activity. Estrogens regulate gene transcription by the classical genomic mechanism of binding to estrogen receptors α and β (ERα and ERβ) as well as the more recently described nongenomic pathways involving plasma membrane-associated ERs that activate intracellular protein kinase-mediated phosphorylation signaling cascades. Here I will review the rapid and longer-term effects of estrogen on mitochondrial function. The identification of ERα and ERβ within mitochondria of various cells and tissues is discussed with a model of estrogen regulation of the transcription of Nuclear Respiratory Factor-1 (NRF-1, NRF1). NRF-1 subsequently promotes transcription of mitochondrial transcription factor Tfam (mtDNA maintenance factor, also called mtTFA) and then Tfam targets mtDNA-encoded genes. The nuclear effects of estrogens on gene expression directly controlling mitochondrial biogenesis, oxygen consumption, mtDNA transcription, and apoptosis are reviewed. Overall, we are just beginning to evaluate the many direct and indirect effects of estrogens on mitochondrial activities.
Keywords: estrogen, estrogen receptor, NRF-1, Tfam
Introduction
Women have longer life expectancies in developed countries than men and this difference has been attributed to protective effects of premenopausal estrogens [Vina et al., 2005]. Mitochondrial reactive oxygen species (ROS) production during ATP generation by the electron transport chain (ETC) damages macromolecules including mitochondrial DNA (mtDNA). Damaged molecules and mitochondrial dysfunction accumulate over time and contribute to aging [Guarente, 2008]. Estrogens exert direct and indirect effects on mitochondrial function in a variety of tissues. While the neuroprotective effects of estrogen are arguably the best-studied, the mechanisms by which estrogens achieve their physiological and pharmacological effects are complex and not completely elucidated (reviewed in [Simpkins et al., 2008]). Indeed, the impact of steroid hormones, including estradiol (E2) on mitochondrial function “is a new and novel area of investigation” [Gavrilova-Jordan and Price, 2007]. The ability of estrogen to regulate nuclear DNA- and mitochondrial DNA (mtDNA)- encoded mitochondrial genes in MCF-7 cells was recently reviewed and the authors concluded that mitochondrial effects of estrogen may play a role in breast cancer, cardiovascular function, and gender-differences in neuroprotection [Gavrilova-Jordan and Price, 2007]. The role of estrogens in mitochondria and the role of mitochondria on nuclear estrogen action has been reviewed [Chen et al., 2008; Felty and Roy, 2005a; Felty and Roy, 2005b]. In breast cancer cells, Felty and Roy speculated that E2 acts via membrane ER (mER) or mitochondrial ER (mtER) in breast cancer cell lines to alter three mitochondrial pathways: 1) direct interaction with the mitochondrial respiratory complexes (MRC) generates ROS leading to protein kinase activation; 2) mER activation of Ca++-dependent proteases and protein kinases; 3) mER activation causes cytoskeletal changes leading to mitochondrial changes and protein kinase activation [Felty and Roy, 2005a]. Whether this model differs in a cell-type-specific manner remains to be evaluated. Further, the role of estrogens in protecting against mitochondrial ROS damage associated with aging has been disputed based on study of C57Bl/6 mice in which males and females have similar longevity [Sanz et al., 2007]. This observation reflects the complexity of estrogen action at the molecular level.
Another topic of debate in understanding how estrogens affect mitochondrial function is whether there is a direct role for estrogen receptors α and β (ERα and ERβ) localized within mitochondria on mtDNA gene transcription and mitochondrial function. A number of steroid/nuclear receptors (NR) in addition to ERα and ERβ, i.e., glucocorticoid receptor (GR), androgen receptor (AR), thyroid receptor (TR), retinoid X receptor (RXR), retinoic acid receptor (RAR), and peroxisome proliferator activated receptor gamma (PPARγ2), have been reported to be present in the mitochondria of various cell types and tissues (reviewed in [Psarra and Sekeris, 2008]). However, there are those who dispute these findings. For example, Gustafsson’s group reported that ERβ could not be positively identified in mouse liver mitochondria by MALDI-TOF mass spectrometry [Schwend and Gustafsson, 2006]. This finding was refuted based on the fact that ERβ expression is low in mouse liver and that the mitochondrial localization of ER may be cell-specific because ERβ was identified in human heart mitochondrial proteins by MALDI-TOF [Yang et al., 2006].
We recently reported that E2 increases the transcription and protein expression of nuclear respiratory factor-1 (NRF-1) and upregulates mitochondrial biogenesis in MCF-7 human breast cancer cells through activation of genomic ERα function [Mattingly et al., 2008]. NRF-1 is a key nuclear-encoded transcription factor that regulates the expression of a number nuclear-encoded genes regulating mitochondrial function [Scarpulla, 2006]. Interestingly, NRF-1 and many MRC genes were recently identified as E2-regulated in opposite directions by ERα and ERβ in mouse aorta [O'Lone et al., 2007]. Many questions regarding the mechanisms by which E2 regulates mitochondrial function remain to be addressed [Burris and Krishnan, 2005]. This review will summarize the mechanisms of estrogen action, discuss transcriptional regulation of the mitochondrial genome, and provide an overview on various aspects of estrogen action in mitochondria. I apologize in advance for the many scientists/references that are not cited directly but whose work is included in reviews that are cited due to limitations of the format for this article.
I. Genomic estrogen action
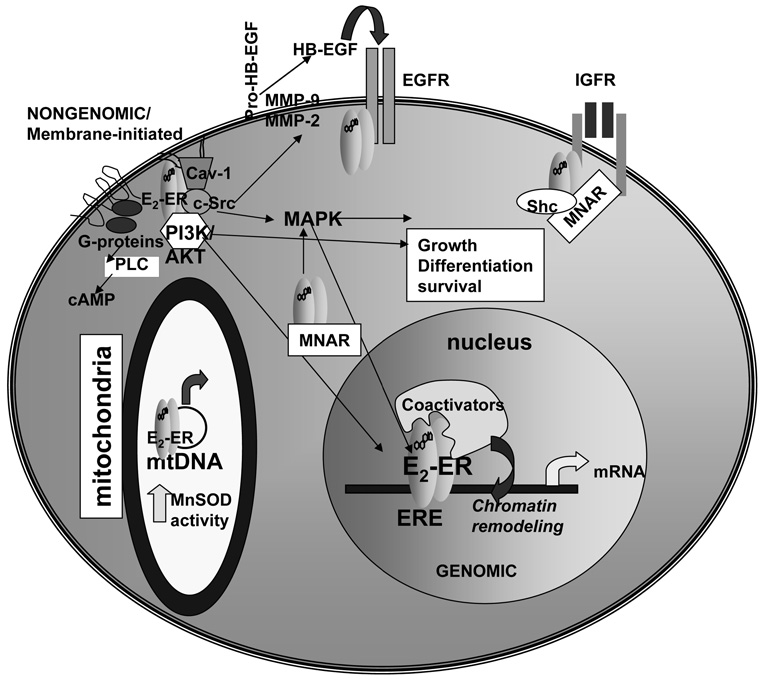
The genomic activity of estrogens is mediated by ERα and ERβ that are members of the steroid/nuclear receptor superfamily of transcription factors. The ER protein is divided into six domains: A-F [Kuiper et al., 1998]. The A/B N-terminal domain has low amino acid (aa) homology between ERα and ERβ and regulates transcription by activating function 1 (AF-1) in a ligand-independent, cell-specific manner. In contrast, the DNA binding domain (DBD), domain C, has high aa homology between the two ER subtypes. The ligand binding domain (LBD) and AF-2 in the E and F domains share 59 and 18% aa homology, respectively [Kuiper et al., 1996]. Once activated by E2, or an estrogen-like compound, ERs form homodimers or heterodimers of ERα/ERβ and bind with high affinity to estrogen response elements (EREs) in the promoters, introns, or 3’ untranslated regions of target genes (reviewed in [Klinge, 2000]). The ERE is composed of a 15 bp palindromic inverted repeat consensus sequence: 5'-AGGTCAnnnTGACCT-3' (n=any nucleotide) [Klinge, 2003]. Figure 1 shows E2-ER bound to an ERE within the nucleus. In addition to direct ERE binding, ERα and ERβ interact with DNA-bound transcription factors, e.g., Sp1 or AP1, at their binding sites and via a ‘tethering mechanism’ activate transcription (reviewed in [Klinge, 2000]). ERα and ERβ interact with NF-κB to inhibit transcription [Kalaitzidis and Gilmore, 2005]. Ligand-activated ERs mediates their genomic effects through interactions with coactivators that recruit chromatin remodeling complexes, alter nucleosomal structure, enhance recruitment of general transcription factors, and increase recruitment of RNA polymerase II to transcribe target genes [McKenna and O'Malley, 2002]. According to my count of the literature, 60 different protein coactivators have been reported to interact with ERα and/or ERβ. The total number of NR coactivators is 200 [Lonard and O'Malley, 2006]. Coactivators exist in large complexes of proteins with many activities in addition to the initiation of transcription, including topoisomerase II [Ju et al., 2006], mRNA translation and splicing, and post-translational protein modifications (reviewed in [Klinge, 2000]). When ERα is occupied by the selective ER modulator (SERM) 4-hydroxytamoxifen (4-OHT), the conformation of a critical α-helix (helix 12) within the LBD, is altered such that the coactivator binding groove is unavailable and corepressors, e.g., silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) and nuclear receptor corepressor (NCoR), interact with the LBD and recruit histone deacetylase (HDAC) complexes to suppress gene transcription in a cell type- and gene- dependent manner [Smith and O'Malley, 2004]. The NURSA website has further information on coactivators, corepressors and chromatin remodeling at http://www.nursa.org.
Figure 1. ERα and ERβ have genomic and nongenomic/membrane initiated activities.
As reviewed in the text, ERα and ERβ are located in the cytoplasm and nucleus, within caveolae in the plasma membrane and within mitochondria. For genomic (nuclear) ER activity, E2 binds and activates ER causing dimerization, ERE binding (or interaction with other transcription factors bound to DNA, not shown), coactivator recruitment, chromatin remodeling, and increased transcription in the nucleus. For nongenomic/membrane initiated estrogen signaling, E2 binds ER in caveolae in the plasma membrane. ER interacts with G-proteins, the p85 subunit of PI3K, with c-Src, and caveolin-1 (Cav-1) to initiate PI3K/AKT and MAPK signaling cascades. In the cytoplasm, ER interacts with MNAR and Shc which play a role in the nongenomic activities of ER. ER interacts with the EGF- and IGF-1 receptors in plasma membranes. ER in mitochondria interact with the D-loop of mtDNA and directly increase MnSOD activity.
In addition to regulation by ligand binding and coregulator protein recruitment, ERs are regulated by phosphorylation in both a ligand-dependent [Lannigan, 2003] and ligand-independent manner [Feng et al., 2001]. Depending on which sites of ERα are phosphorylated, phosphorylation can either increase [Coleman et al., 2003; Denton and Notides, 1992; Tzeng and Klinge, 1996] or inhibit ERα-DNA binding [Al-Dhaheri and Rowan, 2007] and alter nuclear localization [Picard et al., 2008], and thus alter gene transcription. MAPK phosphorylates the N-terminus of ERβ, enhancing ERβ-SRC-1-CBP interaction and gene transcription [Tremblay and Giguere, 2001]. E2 binding promotes ERα ubiquitination and targeting to the 26S proteasome as a regulatory mechanism for ER turnover [Wijayaratne and McDonnell, 2001]. Likewise, ERβ stability is also regulated by phosphorylation and ubiquitination [Picard et al., 2008]. Other post-translational modifications including glycosylation [Cheng and Hart, 2001; Li et al., 2003] and sumoylation also regulate ERα and ERβ activity.
II. Nongenomic estrogen action
E2 has “nongenomic, extra-nuclear, or membrane-initiated” effects, i.e., independent of ER-mediated transcription, that occur within minutes after E2 administration in a G-protein-coupled manner (as reviewed in [Arpino et al., 2008; Pedram et al., 2006a; Watson et al., 2007]). Since ERα lacks a transmembrane domain, how it gets to the plasma membrane (PM) has been controversial but it appears to require palmitoylation [Moriarty et al., 2006]. ERα or its splice variant ERα46 (N-terminally truncated so that the A/B domain and thus AF-1 is missing) also interacts with a number of proteins that are part of a ‘signalsome complex’ including c-Src, the p85 subunit of PI3K, caveolin-1, straitin, MNAR (modulator of nongenomic activity of ER), and eNOS located in caveolae in the PM [Li et al., 2007]. ERα has been shown to interact with the EGF-receptor (EGFR), IGFR1, and HER2 in the plasma membrane of breast cancer cells (reviewed in [Pietras and Marquez-Garban, 2007]). Figure 1 depicts an overview of nongenomic ER interactions. In MCF-7 cells, the adaptor protein Shc shuttles ERα from the nucleus to the PM where ERα interacts with the IGF-1 receptor (IGF-1R) [Song et al., 2004] and arginine methylation of ERαR260 by PRMT1 was recently shown to be essential for complex formation [Le Romancer et al., 2008]. In endothelial cells (EC), E2 rapidly increased intracellular cAMP [Farhat et al., 1996], inhibited of Ca+2 influx [Mueck et al., 1996], stimulated Ca+2 release from internal stores [Improta-Brears et al., 1999], and stimulated nitric oxide (NO) production [Kauser and Rubanyi, 1997]. In MCF-7 human breast cancer cells, E2 rapidly increased PIP2-phospholipase C activity [Graber et al., 1993], mobilized intracellular Ca2+, and activated the MAPK [Improta-Brears et al., 1999] and PI3K/AKT pathways [Stoica et al., 2003].
An ‘orphan’ 7-transmembrane G protein-coupled receptor, GPR30, was reported to bind E2 with high affinity (Kd = 2.7nM) resulting in activation of adenylate cyclase, MAPK, AKT, and calcium signaling (reviewed in [Prossnitz et al., 2008]). Interestingly, and in contrast to ERα or ERβ, tamoxifen and ICI 182,780 (Fulvestrant) also bind GPR30 with high affinity and mimic the effects of E2 [Thomas et al., 2005]. Although the role of GPR30 has been questioned [Otto et al., 2008; Pedram et al., 2006a], it appears likely that differences between cell/tissue types, e.g., thyroid [Vivacqua et al., 2006] or brain [Raz et al., 2008], may be responsible for whether GPR30 is a bone fide novel membrane estrogen receptor.
III. Transcriptional regulation of the mitochondrial genome
Essential metabolic pathways for energy production and homeostasis are located in mitochondria, e.g., the MRC of oxidative phosphorylation for ATP production, heme biosynthesis, β-oxidation, metabolism of certain amino acids, and steroid synthesis. Mitochondrial DNA (mtDNA) is a 16.5 kb circular genome encoding 13 mRNAs, 2 rRNAs, and 22 tRNAs with 103–104 copies of mtDNA/ mammalian cell [Chen et al., 2005]. Thirteen of the 80 proteins of the ETC complexes I, II, III, IV, and V are encoded by mtDNA [Chen et al., 2005]. The remaining subunits of the ETC as well as other proteins involved in mtDNA metabolism and function are nuclear-encoded [Chen et al., 2005]. Once translated, these proteins are targeted to mitochondria via a signal sequence and imported by translocases [Schmidt et al., 2001]. MtDNA transcription is initiated at two promoters (PL and PH) located in the D-loop regulatory region through the binding of mitochondrial RNA (mtRNA) polymerase and the mitochondrial transcription factors Tfam (mtDNA maintenance factor, also called mtTFA) and TFB (mitochondrial transcription factor B, TFB1M and TFB2M) [Scarpulla, 2006]. TFAM, TFB1M, and TFB2M are nuclear-encoded genes whose transcription is regulated by NRF-1 [Scarpulla, 2006]. Although mitochondrial gene expression depends on nuclear genome function and reciprocally by “retrograde communication” mitochondrial activity regulates nuclear gene expression [Poyton and McEwen, 1996], the mechanisms coordinating these events remain to be clarified.
ERα and ERβ have been identified in mitochondria and to bind to the D-loop of mouse and human mtDNA (reviewed in [Chen et al., 2008]). It is not clear whether ERα and ERβ directly regulate mtDNA transcription or whether this effect is mediated through nuclear effects on nuclear-encoded genes that in turn regulate mtDNA transcription. Nonetheless, the presence of ER in both nucleus and mitochondria provide a possible mechanism for E2 coordination of the expression of mtDNA and nuclear-encoded mitochondrial respiratory complex (MRC) genes. Whether ER is involved in retrograde communication between the mitochondria and nucleus is unknown. E2 induces the transcription of nuclear-encoded MRC proteins including mitochondrial ATP synthase subunit E, COVII, and a number of other genes reviewed in [Chen et al., 2008].
IV. NRF-1 is a nuclear transcription factor that regulates transcription of mtDNA transcription factors
NRF-1 is a 68 kDa protein containing an N-terminal nuclear localization signal and a C-terminal transcriptional activation domain that is ubiquitously expressed [Gugneja et al., 1996]. Homozygous deletion of murine NRF-1 results in embryonic death [Huo and Scarpulla, 2001]. NRF-1 homodimerizes and binds to palindromic NRF-1 response elements (RE) in the promoter of nuclear-encoded mitochondrial genes, thus regulating the transcription of genes regulating mitochondrial activity in a cell-type specific manner [Scarpulla, 2002]. Research from Scarpulla’s lab has demonstrated that NRF-1 plays a critical role in integrating nucleo-mitochondrial interactions by initiating transcription of nuclear-encoded mtDNA-specific transcription factors including Tfam, TFB1M, and TFB2M [Scarpulla, 2006]. Additional NRF-1 target genes include subunits of the five ETC complexes, components of the mtDNA transcription and replication machinery, mitochondrial and cytosolic enzymes of heme biosynthesis, components of mitochondrial protein import, and > 400 genes involved in cell cycle regulation, metabolism, DNA replication, and transcriptional regulation [Cam et al., 2004]. Recently, NRF-1 was shown to bind and increase transcription of all 10 nuclear-encoded mouse COX genes [Dhar et al., 2008]. NRF-1 activity was inhibited by phosphorylation by cyclin D1 and NRF-1 interacts directly with cyclin D1 in MCF-7 cells [Wang et al., 2006]. The reciprocal regulation of cyclin D1 and NRF-1 was proposed to reduce mitochondrial activity and shift glucose metabolism toward glycolysis, i.e., the Warburg effect, in cancer cells [Cuezva et al., 2002]. This hypothesis offers an explanation for the lack of inclusion of NRF1 in gene module analysis revealing an association of nuclear-encoded mitochondrial gene expression with reduced breast cancer survival [Wong et al., 2008]. This study reported increased expression of 218 nuclear-encoded mitochondrial genes including a number of NRF-1 targets, e.g., TFAM SDHB, and COX5B, although not NRF-1 itself, as associated with poor prognosis in breast cancer [Wong et al., 2008]. Notably, what regulates NRF-1 expression and activity is less well-characterized [Scarpulla, 2006]. Thyroid hormone (T3) increased NRF-1 transcription in rat liver “probably through a thyroid response element”, although this element is undefined [Weitzel et al., 2003; Weitzel et al., 2001].
V. Estrogen regulates NRF-1 transcription
Studies from Duckles’ lab reported that NRF-1 was upregulated in the cerebral blood vessels of ovariectomized (ovex) rats chronically treated (2 wks) with E2, suggesting that estrogen may regulate NRF-1 transcription [Stirone et al., 2005]. We followed up that observation at the molecular level and recently reported that NRF-1 mRNA expression was increased by E2 in a time-dependent manner in two estrogen-responsive cell lines: MCF-7 breast cancer and H1793 lung adenocarcinoma cells [Mattingly et al., 2008]. The ER antagonist ICI 182,780 blocked the E2-induced increase in NRF-1 mRNA, indicating that ER mediates the E2-stimulated increase in NRF-1. The transcriptional inhibitor actinomycin D (Act D) and protein synthesis inhibitor cycloheximide (CHX) indicated that the E2-ER mediated increase in NRF-1 was a direct effect of ER at the genomic level. Indeed, pretreatment of MCF-7 cells for 1 h with the MAPK (MEK) and PI3K inhibitors PD98059 and Wortmannin, respectively, did not inhibit the E2-induced increase in NRF-1, indicating that the E2-induction of NRF-1 was not mediated by nongenomic/ membrane-initiated activation of the PI3K/Akt and MAPK signaling pathways. We concluded that NRF-1 is a primary E2-responsive gene [Mattingly et al., 2008]. NRF-1 protein expression increased in a time-dependent manner in E2-treated MCF-7 cells, commensurate with the increase seen in NRF-1 mRNA expression.
We reported that E2 increased the interaction of both ERα and ERβ with a 5’ promoter region of the human NRF-1 gene containing a imperfect ERE [Mattingly et al., 2008]. However, siRNA against ERβ did not inhibit E2-induced NRF-1 transcription, whereas siRNA against ERα did, thus implicating ERα as mediating the increase in NRF-1 induced by E2 in MCF-7 cells. Thus, why E2 increases ERβ recruitment to the NRF1 promoter remains to be defined. However, recent microarray profiling of genes regulated by E2 in aortas of wildtype, ERα, and ERβ knockout (αERKO and βERKO) mice revealed opposite regulation of NRF-1 and many mitochondrial respiratory chain (MRC) gene targets by ERα and ERβ [O'Lone et al., 2007]. Therefore, we hypothesize that the direct interaction of ERα and ERβ with the NRF-1 promoter recruits different coregulators, i.e., coactivators and corepressors, in a ligand- and cell- specific manner. Further, the regulation of NRF-1 transcription by estrogen may be cell type specific. Brown adipose tissue from male versus female Wistar rats showed higher transcript levels of nrf1 and tfam, but not pparg, [Rodriguez-Cuenca et al., 2007]. E2 reduced nrf1 and tfam transcription in primary cultured brown adipocytes from male NMRI mice [Rodriguez-Cuenca et al., 2007], the opposite of E2- induced NRF1 in MCF-7 and H1793 cells [Mattingly et al., 2008]. A microarray gene profiling study showed a 7.3-fold increase in NRF-1 expression in Fulvestrant-resistant versus TAM-resistant MCF-7 cells (Table S2 in [Fan et al., 2006]), but no one has examined how TAM or 4-OHT affects NRF1 expression in MCF-7 or other breast cancer cells.
VI. Estrogen-induced NRF-1 increases Tfam and Tfam increases mtDNA gene expression
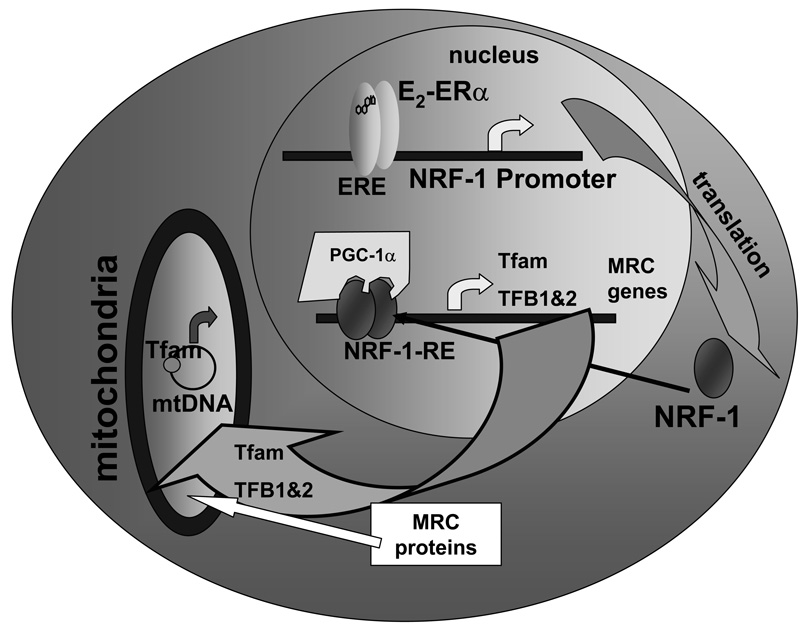
NRF-1 stimulates transcription of the Tfam gene by binding to an NRF-1 response element in the promoter [Scarpulla, 2006]. Subsequently, Tfam increases the transcription of mtDNA-encoded gene targets. We have reported that E2 increased nuclear-encoded Tfam transcription in MCF-7 cells and also two mitochondrial-encoded mRNAs regulated by Tfam: Complex IV, Cytochrome c oxidase subunit I; (MTCO1, COI) and NADH dehydrogenase subunit 1 (MTND1, ND1) [Mattingly et al., 2008]. Tfam mRNA was increased 12–72 h after E2 treatment. The expression of Tfam-regulated, mtDNA-encoded MTCOI and MTND1 mRNAs was increased 48 to 72 h after E2 treatment and was inhibited by ICI 182,780, indicating an ER-dependence. COI and Cytochrome c oxidase (COX IV, nuclear-encoded), both subunits of Complex IV in the MRC, protein expression was similarly increased by E2 treatment [Mattingly et al., 2008]. MTCOI is transcriptionally regulated by Tfam and TFBs while COX IV is a direct target of NRF-1 [Scarpulla, 2006]. Thus, we proposed a model (Fig. 2) in which E2 induced NRF-1 expression results in the increased expression of Tfam, as well as TFB1 and 2, and MRC genes, that subsequently entered mitochondria to increase the expression of mtDNA-encoded genes, mitochondrial biogenesis, and oxidative phosphorylation [Mattingly et al., 2008].
Figure 2. E2-ERα increases NRF-1 gene transcription.
This model is based on our recent report showing that E2-ERα binds a non-consensus ERE in the 5’ promoter of the human NRF-1 gene and increases NRF1 gene transcription [Mattingly et al., 2008]. The subsequent increase in NRF-1 protein increases the transcription of NRF-1 target genes in the nucleus, e.g., increased Tfam transcription. PGC-1α is a coactivator needed for NRF-1 regulated gene transcription. Increased Tfam as well MRC gene products are imported into mitochondria resulting in increased ETC oxidative phosphorylation. Tfam and TFB1 and TFB2 increase transcription of mtDNA and increase mitochondrial biogenesis.
VII. Effect of estrogens on mitochondrial biogenesis
Regulation of mitochondrial biogenesis involves the coordinated actions of both mtDNA and nuclear-encoded gene products including NRF-1, NRF-2, Tfam, and PGC-1α (reviewed in [Kelly and Scarpulla, 2004]). mtDNA copy number is a measure of mitochondrial biogenesis. Increased numbers of mitochondria and increased mitochondrial size were observed in the glandular epithelium and stromal endometrium of pregnant bonnet monkeys, an effect attributed to increased E2 [Rosario et al., 2008]. We reported that E2 significantly increased mtDNA copy number after 24, 48, and 72 h. in MCF-7 cells [Mattingly et al., 2008]. Because Tfam is essential for mtDNA replication [Larsson et al., 1998], these data are consistent with the increase in E2-induced Tfam mRNA that we observed. Notably, siRNA knockdown of NRF-1 inhibited the E2-induced increase in mtDNA, indicating that the E2-induced increase in mitochondrial biogenesis is mediated by NRF-1 and not directly by E2.
We note that the ability to estrogen to increase mitochondrial biogenesis may be cell-specific because E2 treatment of ovariectomized (ovex) rats did not increase mitochondrial biogenesis as measured by the COXII (mitochondrial)/ β-actin (nuclear) ratio in brain [Irwin et al., 2008]. On the other hand, studies in male mouse heart following trauma–hemorrhage revealed that E2 and the ERβ agonist diarylpropionitrile (DPN) increased mitochondrial biogenesis via upregulation of mitochondrial ERβ [Hsieh et al., 2006]. Further, mitochondrial ERβ upregulated mtDNA transcription and MRC activity [Hsieh et al., 2006]. In mouse heart, E2- or DPN- activated mitochondrial ERβ specifically increased mitochondrial respiratory complex-IV (MRC-IV, cytochrome c oxidase) activity [Hsieh et al., 2006]. E2 also increased PGC-1α mRNA and protein levels in the trauma–hemorrhage mouse heart model [Hsieh et al., 2005].
Isoflavones, e.g., daidzen and genistein, are phytoestrogens that bind ERβ and ERα with µM affinities and have additional activities that are independent of ER binding [Kuiper et al., 1997]. Isoflavones were reported to increase mitochondrial biogenesis in primary rabbit renal proximal tubule cells (RPTC) in an ER-independent manner by increasing the expression and activity of sirtuin 1 (SIRT1) which in turn deacetylates and activates PGC-1α [Rasbach and Schnellmann, 2008]. The coactivator PGC-1α is considered a master regulator of mitochondrial biogenesis in mammals (reviewed in [Ventura-Clapier et al., 2008]). On the other hand, genistein (0.5 µM) was shown to decrease H2O2 levels in cells via nongenomic ER-activation of the MAPK pathway leading to NFκB activation and increased MnSOD expression in MCF-7 cells [Borras et al., 2006]. Again, these results point to cell type-specific responses to phytoestrogens in affecting mitochondrial activities.
VIII. Effect of estrogens on O2 consumption
There is surprisingly little information regarding the role of estrogen in regulating oxygen consumption as an index of MRC activity. An early study reported that E2 inhibited oxygen consumption/mitochondrial respiration in isolated rat liver mitochondria with an IC50 > 0.1mM E2 [Vallejos and Stoppani, 1967], a concentration which is not physiologically relevant since serum E2 is 0.28nM in follicular phase and 1.1. nM in luteal phase in premenopausal women [Clarke et al., 2003]. Rats in estrous had enhance mitochondrial respiration compared to pseudopregnant or ovex rats, suggesting that E2 increases mitochondrial respiration [Gigli and Bussmann, 2001]. Another study revealed that brain mitochondria isolated from E2-treated ovex rats displayed significantly greater oxygen consumption following Ca++ challenge in State 3 respiration and a significantly higher respiratory control ratio (RCR) than mitochondria from control rats [Nilsen et al., 2006]. We reported a significant increase in oxygen consumption after 4 and 6 d of E2 treatment in MCF-7 cells [Mattingly et al., 2008]. This delayed time of response was consistent with the upregulation of COI and COIV proteins beginning at 48 h. Interestingly, E2 did not increase oxygen consumption in MDA-MB-231 ERα-negative breast cancer cells [Mattingly et al., 2008].
IX. Direct effects of E2, 4-OHT, and raloxifene within mitochondria
Although µM concentrations of E2 have antioxidant activity in isolated hepatocytes [Leal et al., 1998], such levels are not physiologically relevant as indicated previously in this review. Likewise, although 4-hydroxytamoxifen was shown to specifically inhibit MRC Complexes III and IV (cytochrome c oxidase) in isolated rat liver mitochondria with IC50s ~ 45–80 µM [Tuquet et al., 2000], the serum concentration of 4-OHT in breast cancer patients on oral tamoxifen is ~ 0.2 µM [Clarke et al., 2003]. Numerous reports from Simpkins’ lab have demonstrated the protective effects of E2 against exogenous or endogenous ROS in neuronal and lens epithelial cells as measured by maintenance of ATP levels and inhibition of mitochondrial Ca++ influx (reviewed in [Simpkins et al., 2008]). Studies from Yager’s lab have demonstrated that E2 increases superoxide production, O2 uptake, and intracellular ATP levels through ER-activation because these effects are inhibited by Fulvestrant (ICI 182,780, a pure steroidal ER antagonist) (reviewed in [Chen et al., 2008]). These data argue against any effect of the putative membrane estrogen receptor GPR30 since Fulvestrant activates GPR30 (reviewed in [Filardo et al., 2006]).
However, estrogen activation of membrane-associated ER (here referring to either ERα and/or ERβ, i.e., the ESR1 and ESR2 gene products) thus initiating MAPK and NF-κB signaling resulting in increased transcription of mtDNA-encoded manganese-superoxide dismutase (MnSOD) and glutathione peroxidase (GPx) [Vina et al., 2005] offers a mechanism for estrogen’s mitochondrial-protective activity. These enzymes detoxify ROS and thus protect mtDNA and macromolecules despite E2-activation of the MRC to increase ATP production and thus cell proliferation and movement [Chen et al., 2008]. However, this mechanism may be cell type-specific. In viral-transformed human lens epithelial cells (HLE-B3), E2 rapidly activated MnSOD activity, but had no effect on MnSOD mRNA or protein expression [Gottipati and Cammarata, 2008]. In human brain microvascular endothelial cells (HBMECs), E2 and Raloxifene (RAL) suppressed mitochondrial superoxide production through ERα, not ERβ, activation, but did not affect MnSOD RNA or protein levels in isolated mitochondria [Razmara et al., 2008]. E2 increased the ratio of aconitase/fumerase enzyme activity, indicative of reduced ROS production in HBMECs. E2 increased the transcription of cytochrome c through ERα activation in HBMECs and the authors suggested that the increase in cytochrome c by activating the cytochrome c-mediated electron-leak pathway, in which the electron of ferrocytochrome c migrates directly to H2O2 rather than to cytochrome c oxidase [Zhao and Xu, 2004], offers a mechanism by which E2 suppresses ROS [Razmara et al., 2008]. Interestingly, NRF-1, which, as discussed previously, is directly upregulated by E2-ERα binding to an ERE in the NRF1 promoter [Mattingly et al., 2008], directly regulates cytochrome c gene transcription by interacting with its palindromic response element in the promoter [Scarpulla, 2005], but whether the observed E2-ERα-induced increase in cytochrome c was mediated directly or through NRF-1 in HBMECs was not evaluated [Razmara et al., 2008].
E2 directly increased MnSOD activity in isolated mitochondria from MCF-7 human breast cancer cells and thus reduced ROS [Pedram et al., 2006b]. The investigators in the Levin group also transfected ER-null CHO cells with a targeting vectors that specifically directed the expression of the E domain (ligand binding domain) of ERα to the plasma membrane, nucleus, or mitochondria and examined the effect of UV to increase apoptosis in the E2-treated cells [Pedram et al., 2006b]. Only the cells in which the ERα-E domain was targeted to mitochondria did E2 prevent the UV-stimulated cytochrome c release [Pedram et al., 2006b], thus demonstrating the requirement of E2-E-domain interaction within mitochondria to inhibit apoptosis.
Physiological evidence for a role of ERα in mitochondrial function in vivo comes from studies with αERKO mice and ovex rodents. After subjecting αERKO versus wildtype(wt) mouse hearts to cardiac ischemia-reperfusion, mitochondria in the αERKO hearts showed ultrastructural damage and reduced ETC function [Zhai et al., 2000a]. In a similar study comparing myocardial ischemia-reperfusion injury in the hearts of ovex or E2-treated ovex rats, the E2-treated rats showed less myocardial mitochondrial damage and greater maintenance of ETC function [Zhai et al., 2000b]. Microarray profiling of aortas from ovex versus E2-supplemented ovex αERKO, βERKO, and wt mice identified > 18 nuclear-encoded MRC subunits that are E2-ERα regulated [O'Lone et al., 2007]. Importantly, NRF-1 mRNA was decreased in the αERKO but not βERKO mice, indicating that E2-ERα regulates NRF-1 expression in mouse aorta. Notably, NRF-1 was downregulated by E2-ERα and genes including an NRF-1-RE in the -1kB promoter were also downregulated in the aortas of αERKO mice indicating that ERβ actively represses a subset of MRC genes. A caveat of this study is that the mice were given one week of E2 treatment [O'Lone et al., 2007]; thus, many identified genes are unlikely to be direct E2 targets, but rather secondary or tertiary targets.
In vivo E2 or progesterone treatment of ovex rats increased respiratory activity in isolated brain mitochondria [Irwin et al., 2008]. Both E2 and progesterone also increased the mRNA levels of both nuclear and mtDNA-encoded components of the MRC, i.e., mt-encoded COXI, COXII, and COXIII and nuclear encoded COXIV [Irwin et al., 2008]. Studies on changes in the proteome of mitochondria isolated from the brains of ovex rats treated for 24 h with E2 revealed increased expression of many proteins that couple glucose utilization to the TCA cycle as well as ETC proteins, e.g., complex IV subunits I–IV [Nilsen et al., 2007]. The authors concluded that E2 regulates both mitochondrial and nuclear encoded genes, requiring coordinated control of mitochondrial and nuclear encoded gene transcription, a feat presumably regulated by the localization of ERs in both cellular organelles [Nilsen et al., 2007].
X. ERα and ERβ are inside mitochondria
Techniques including immunoblotting of mitochondrial preparations, immunocytochemistry, and mass spectrometry have revealed that ERβ is localized in mitochondria in a variety of tissues, including rabbit ovaries and uterus, human lens epithelial cells, spermatocytes, primary cerebral cortical and cerebral cortical and hippocampal neurons, primary cardiomyocytes, as well as HepG2, SaOS-2, and MCF-7 cell lines (reviewed in [Psarra and Sekeris, 2008]). ERα has also been reported to be in mitochondria of a few tissues, e.g., rabbit uterus [Monje and Boland, 2001] and in MCF-7 cells (reviewed in [Chen et al., 2008]), but many more cell lines have mitochondrial ERβ (reviewed in [Psarra and Sekeris, 2008]). E2 enhanced ERα and ERβ mitochondrial localization in MCF-7 cells (reviewed in [Chen et al., 2008]). Other nuclear transcription factors, e.g., p53, NF-κB, AP-1, CREB, wnt, c-src, and c-myc, have also been observed in mitochondria (reviewed in [Psarra and Sekeris, 2008]). This raises the possibility that ER interaction with these proteins in the mitochondria may impact mtDNA gene expression or other functions. On the other hand, a recent proteomic/bioinformatic analysis of mitochondrial proteins in 14 different C57Bl/6 mouse tissues did not reveal either ERα or ERβ in any tissue, even with an estimated 85% of proteins identified and a false discovery rate of 10% [Pagliarini et al., 2008], see also http://www.broad.mit.edu/publications/MitoCarta/.
ER is transcriptionally active within mitochondria. An E2-stimulated increase in mRNA levels of mtDNA-encoded genes cytochrome c oxidase subunits I and II (COI and COII) was inhibited by ICI 182,780, indicating ER-dependence [Chen et al., 2004a]. Yager’s group identified a mitochondrial targeting protein sequence (mTPS) in ERβ aa 220–270 and reported that ERα lacks a mTPS [Chen et al., 2004a]. Other studies indicate that ERβ interaction with Tom70 and Hsp70 may play a role in mitochondrial trafficking of ERβ (reviewed in [Simpkins et al., 2008]). The D-loop of mtDNA contains replication and transcriptional control elements. ERβ bound to D-loop EREs in MCF-7 mtDNA in vitro [Chen et al., 2004b]. Recently, ERβ was reported by be predominantly localized in mitochondria of immortalized ‘normal’ and E2-transformed human breast epithelial cell lines (MCF-10F) [Chen et al., 2007]. Further, the Russo group reported that ERβ shifts from mitochondria to nuclei during E2-induced neoplastic transformation and that E2 and the ERβ-selective agonist DPN, but not the ERα-selective agonist PPT, induced expression of mtDNA-encoded MRC genes and proteins COI, COII, and ND1 [Chen et al., 2007]. However, the authors did not state how long the cells were treated with E2 or DPN and thus whether the effect is primary or secondary, i.e., due to increased NRF-1, Tfam, TFB1, etc (as in Fig. 1), is unknown.
XI. Estrogens inhibit apoptosis
Programmed cell death, i.e., apoptosis, is an energy-dependent series of cellular events that is critical in tissue remodeling during development, e.g., the breast at puberty, during pregnancy/lactation, and involution, and is involved in breast cancer development [Conner, 2007]. Apoptosis can be initiated through either the intrinsic or extrinsic pathway leading to caspase activation [Kaufmann and Hengartner, 2001]. Mitochondria are central to the induction of apoptosis [van Heerde et al., 2000]. Growth factors and trophic hormones reciprocally regulate breast cell apoptosis and proliferation. In the pre-ovulatory phase of the menstrual cycle, when estrogen levels are high, breast cell apoptotic activity is low [Conner, 2007]. The intrinsic, mitochondrial-mediated apoptotic pathway is regulated by Bcl-2 family members that either promote or inhibit apoptosis depending on their interaction with pro-survival family members, e.g., Bcl-2, Bcl-xL and McI-1, or pro-apoptotic , e.g., Bax, Bak, and Bok, Bim, Bid, Puma, and Bad [Labi et al., 2008] E2 protects MCF-7 cells from apoptosis by upregulating transcription of anti-apoptotic/pro-survival Bcl-2 [Perillo et al., 2000]. In neuronal tissues, E2 inhibits apoptosis by increasing mitochondrial sequestration of Ca++ [Nilsen and Brinton, 2003; Nilsen and Brinton, 2004] and by suppressing transcription of pro-apoptotic genes such as nip2 [Belcredito et al., 2000]. In addition, membrane-initiated E2 activation of MAPK and PI3K/pathways is anti-apoptotic [Razandi et al., 2000].
Recent studies have identified microRNAs with anti- or pro- apoptotic functions (reviewed in [Park and Peter, 2008]). Among the miRNA genes identified was let-7c that increased TRAIL-induced caspase 3 activation in MDA-MB-453 breast cancer cells, presumably by targeting CD95L [Ovcharenko et al., 2007]. Of interest in terms of estrogen regulation of apoptosis is the report that let-7c expression was higher in ERα positive than ERα negative human breast tumors [Blenkiron et al., 2007]. On the other hand, transfection of miR-182 inhibited TRAIL-induced caspase 3 activation in MDA-MB-453 breast cancer cells, presumably by targeting caspase 3 and Fas-associated death domain (FADD) protein [Ovcharenko et al., 2007]. Like let-7c, miR-182 was higher in ERα positive than ERα negative human breast tumors [Mattie et al., 2006]. We found that MCF-7 cells treated with 10 nM E2 for 6 hours increased miR-182 (the mature form of this miRNA) by ~ 3-fold as measured by realtime Q-PCR (Klinge, unpublished data). These data are consistent with a potential estrogen-mediated anti-apoptotic function of miR-182. However, remarkably little is known about estrogen regulation of miRNA expression, let alone the targets of miRNA in apoptosis.
Given that E2 increases anti-apoptotic Bcl-2 and mtDNA-encoded MRC expression, and our report that E2 increases NRF-1 [Mattingly et al., 2008], we propose that NRF-1 is also anti-apoptotic. Supporting this idea, knockout of NRF-1 regulated gene Tfam in mouse heart tissue, which results in deficiency in mtDNA transcription, was associated with increased in vivo apoptosis [Wang et al., 2001]. Indeed, one review concluded that the persistent E2/ER-induced ETC protein synthesis and energy metabolism play an important role in apoptotic inhibition [Chen et al., 2005]. Additionally, cellular quiescence has been shown to be maintained by a balance between c-Myc and NRF-1 expression and the overexpression of c-Myc in cancer results in sensitization of cells to apoptosis under serum deprivation by selectively competing with NRF-1 for select NRF-1-target genes triggering apoptosis [Morrish et al., 2003]. Thus, another possible mechanism by which E2 inhibits apoptosis is by increasing NRF-1 expression to achieve a higher ratio of NRF-1/c-Myc.
Conclusions
Estrogens have direct and indirect effects on mitochondrial activity that are mediated by genomic and nongenomic/membrane-initiated activities of ERα and ERβ (reviewed [Chen et al., 2008; Felty and Roy, 2005a; Felty and Roy, 2005b]). E2 increases NRF-1 gene transcription by increasing ERα binding to an ERE in the gene promoter [Mattingly et al., 2008]. An increase in Tfam followed the E2-induced increase in NRF-1 which was followed in time by increased Tfam-regulated mtDNA-encoded COI and NDI genes and increased mitochondrial biogenesis (modeled in Fig. 2). The localization of ERα and ERβ in both nuclear and mitochondrial compartments offers a potential mechanism for regulating coordinate nuclear and mitochondrial gene expression and function that remains to be clarified.
Acknowledgements
Sincere thanks to Dr. Barbara J. Clark for her thoughtful comments and suggestions for the improvement of this review.
Grant sponsor: NIH; Grant numbers: R01 DK53220; R21 CA124811
Literature Cited
- Al-Dhaheri MH, Rowan BG. Protein Kinase A Exhibits Selective Modulation of Estradiol-Dependent Transcription in Breast Cancer Cells that Is Associated with Decreased Ligand Binding, Altered Estrogen Receptor {alpha} Promoter Interaction, and Changes in Receptor Phosphorylation. Mol Endocrinol. 2007;21:439–456. doi: 10.1210/me.2006-0059. [DOI] [PubMed] [Google Scholar]
- Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the Estrogen Receptor and the HER Tyrosine Kinase Receptor Family: Molecular Mechanism and Clinical Implications for Endocrine Therapy Resistance. Endocr Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcredito S, Brusadelli A, Maggi A. Estrogens, apoptosis and cells of neural origin. J Neurocytol. 2000;29:359–365. doi: 10.1023/a:1007161206744. [DOI] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras C, Gambini J, Gomez-Cabrera MC, Sastre J, Pallardo FV, Mann GE, Vina J. Genistein, a soy isoflavone, up-regulates expression of antioxidant genes: involvement of estrogen receptors, ERK1/2, and NF{kappa}B. FASEB J. 2006;20:2136–2138. doi: 10.1096/fj.05-5522fje. [DOI] [PubMed] [Google Scholar]
- Burris TP, Krishnan V. Estrogen: A Mitochrondrial Energizer That Keeps on Going. Mol Pharmacol. 2005;68:956–958. doi: 10.1124/mol.105.017335. [DOI] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A Common Set of Gene Regulatory Networks Links Metabolism and Growth Inhibition. Molecular Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Chen J-Q, Russo PA, Cooke C, Russo IH, Russo J. ER[beta] shifts from mitochondria to nucleus during estrogen-induced neoplastic transformation of human breast epithelial cells and is involved in estrogen-induced synthesis of mitochondrial respiratory chain proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:1732–1746. doi: 10.1016/j.bbamcr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Chen J-Q, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2005;1746:1–17. doi: 10.1016/j.bbamcr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Brown TR, Yager JD. Mechanisms of hormone carcinogenesis: evolution of views, role of mitochondria. Adv Exp Med Biol. 2008;630:1–18. [PubMed] [Google Scholar]
- Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004a;286:E1011–E1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Eshete M, Alworth WL, Yager JD. Binding of MCF-7 cell mitochondrial proteins and recombinant human estrogen receptors alpha and beta to human mitochondrial DNA estrogen response elements. J Cell Biochem. 2004b;93:358. doi: 10.1002/jcb.20178. [DOI] [PubMed] [Google Scholar]
- Cheng X, Hart GW. Alternative O-glycosylation/O-Phosphorylation of serine-16 in murine estrogen receptor beta: Post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O'Brien K, Wang Y, Hilakivi-Clarke LA. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- Coleman KM, Dutertre M, El-Gharbawy A, Rowan BG, Weigel NL, Smith CL. Mechanistic Differences in the Activation of Estrogen Receptor-alpha (ERalpha )- and ERbeta - dependent Gene Expression by cAMP Signaling Pathway(s) J Biol Chem. 2003;278:12834–12845. doi: 10.1074/jbc.M212312200. [DOI] [PubMed] [Google Scholar]
- Conner P. Breast response to menopausal hormone therapy--aspects on proliferation, apoptosis and mammographic density. Ann Med. 2007;39:28–41. doi: 10.1080/07853890601039842. [DOI] [PubMed] [Google Scholar]
- Cuezva JM, Krajewska M, De Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- Denton RR, Notides AC. Estrogen receptor phosphorylation. Hormonal dependence and consequence on specific DNA binding. J. Biol. Chem. 1992;267:7263–7268. [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Nuclear Respiratory Factor 1 Regulates All Ten Nuclear-encoded Subunits of Cytochrome c Oxidase in Neurons. J. Biol. Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng ASL, Li L, Abbosh PH, Huang THM, Nephew KP. Diverse Gene Expression and DNA Methylation Profiles Correlate with Differential Adaptation of Breast Cancer Cells to the Antiestrogens Tamoxifen and Fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- Farhat MY, Lavigne MC, Ramwell PW. The vascular protective effects of estrogen. FASEB J. 1996;10:615–624. [PubMed] [Google Scholar]
- Felty Q, Roy D. Estrogen, mitochondria, and growth of cancer and non-cancer cells. J Carcinog. 2005a;4:1. doi: 10.1186/1477-3163-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felty Q, Roy D. Mitochondrial signals to nucleus regulate estrogen-induced cell growth. Med Hypotheses. 2005b;64:133–141. doi: 10.1016/j.mehy.2003.12.056. [DOI] [PubMed] [Google Scholar]
- Feng W, Webb P, Nguyen P, Liu X, Li J, Karin M, Kushner PJ. Potentiation of estrogen receptor activation function 1 (AF-1) by Src/JNK through a serine 118-Independent pathway. Mol Endocrinol. 2001;15:32–45. doi: 10.1210/mend.15.1.0590. [DOI] [PubMed] [Google Scholar]
- Filardo EJ, Graeber CT, Quinn JA, Resnick MB, Giri D, DeLellis RA, Steinhoff MM, Sabo E. Distribution of GPR30, a Seven Membrane-Spanning Estrogen Receptor, in Primary Breast Cancer and its Association with Clinicopathologic Determinants of Tumor Progression. Clin Cancer Res. 2006;12:6359–6366. doi: 10.1158/1078-0432.CCR-06-0860. [DOI] [PubMed] [Google Scholar]
- Gavrilova-Jordan LP, Price TM. Actions of steroids in mitochondria. Semin Reprod Med. 2007;25:154–164. doi: 10.1055/s-2007-973428. [DOI] [PubMed] [Google Scholar]
- Gigli I, Bussmann LE. Exercise and ovarian steroid hormones: Their effects on mitochondrial respiration. Life Sciences. 2001;68:1505–1514. doi: 10.1016/s0024-3205(01)00954-7. [DOI] [PubMed] [Google Scholar]
- Gottipati S, Cammarata PR. Mitochondrial superoxide dismutase activation with 17 beta-estradiol-treated human lens epithelial cells. Mol Vis. 2008;14:898–905. [PMC free article] [PubMed] [Google Scholar]
- Graber R, Sumida C, Vallette G, Nunez EA. Rapid and long-term effects of 17 beta-estradiol on PIP2-phospholipase C-specific activity of MCF-7 cells. Cell Signal. 1993;5:181–186. doi: 10.1016/0898-6568(93)90069-x. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--A Nexus for Aging, Calorie Restriction, and Sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugneja S, Virbasius CM, Scarpulla RC. Nuclear respiratory factors 1 and 2 utilize similar glutamine- containing clusters of hydrophobic residues to activate transcription. Mol. Cell. Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y-C, Yang S, Choudhry MA, Yu H-P, Rue LW, III, Bland KI, Chaudry IH. PGC-1 upregulation via estrogen receptors: a common mechanism of salutary effects of estrogen and flutamide on heart function after trauma-hemorrhage. Am J Physiol Heart Circ Physiol. 2005;289:H2665–H2672. doi: 10.1152/ajpheart.00682.2005. [DOI] [PubMed] [Google Scholar]
- Hsieh Y-C, Yu H-P, Suzuki T, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. Upregulation of mitochondrial respiratory complex IV by estrogen receptor-[beta] is critical for inhibiting mitochondrial apoptotic signaling and restoring cardiac functions following trauma-hemorrhage. Journal of Molecular and Cellular Cardiology. 2006;41:511–521. doi: 10.1016/j.yjmcc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Mitochondrial DNA Instability and Peri-Implantation Lethality Associated with Targeted Disruption of Nuclear Respiratory Factor 1 in Mice. Mol. Cell. Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Improta-Brears T, Whorton AR, Codazzi F, York JD, Meyer T, McDonnell DP. Estrogen-induced activation of mitogen-activated protein kinase requires mobilization of intracellular calcium. Proc. Natl. Acad. Sci. USA. 1999;96:4686–4691. doi: 10.1073/pnas.96.8.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin RW, Yao J, Hamilton RT, Cadenas E, Brinton RD, Nilsen J. Progesterone and Estrogen Regulate Oxidative Metabolism in Brain Mitochondria. Endocrinology. 2008;149:3167–3175. doi: 10.1210/en.2007-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B-G, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A Topoisomerase II{beta}-Mediated dsDNA Break Required for Regulated Transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D, Gilmore TD. Transcription factor cross-talk: the estrogen receptor and NF-[kappa]B. Trends in Endocrinology and Metabolism. 2005;16:46–52. doi: 10.1016/j.tem.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Hengartner MO. Programmed cell death: alive and well in the new millennium. Trends in Cell Biology. 2001;11:526–534. doi: 10.1016/s0962-8924(01)02173-0. [DOI] [PubMed] [Google Scholar]
- Kauser K, Rubanyi GM. Potential cellular signaling mechanisms mediating upregulation of endothelial nitric oxide production by estrogen. J. Vasc. Res. 1997;34:229–236. doi: 10.1159/000159227. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65:227–251. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen response element sequence as an allosteric regulator of estrogen receptor action. ChemTracts Biochem. Molec. Biol. 2003;16:1–18. [Google Scholar]
- Kuiper GG, Carlsson B, Grandien J, Enmark E, Haggblad J, Nilsson S, Gustafsson J-A. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors a and b. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J-A. Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Shughrue PJ, Merchenthaler I, Gustafsson JA. The estrogen receptor beta subtype: a novel mediator of estrogen action in neuroendocrine systems. Front. Neuroendocrinol. 1998;19:253–286. doi: 10.1006/frne.1998.0170. [DOI] [PubMed] [Google Scholar]
- Labi V, Grespi F, Baumgartner F, Villunger A. Targeting the Bcl-2-regulated apoptosis pathway by BH3 mimetics: a breakthrough in anticancer therapy? Cell Death Differ. 2008;15:977–987. doi: 10.1038/cdd.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of Estrogen Rapid Signaling through Arginine Methylation by PRMT1. Mol Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Leal AM, Ruiz-Larrea MB, Martinez R, Lacort M. Cytoprotective actions of estrogens against tert-butyl hydroperoxide-induced toxicity in hepatocytes. Biochemical Pharmacology. 1998;56:1463–1469. doi: 10.1016/s0006-2952(98)00248-2. [DOI] [PubMed] [Google Scholar]
- Li L, Haynes MP, Bender JR. Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc Natl Acad Sci U S A. 2003;100:4807–4812. doi: 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hisamoto K, Kim KH, Haynes MP, Bauer PM, Sanjay A, Collinge M, Baron R, Sessa WC, Bender JR. Variant estrogen receptor c-Src molecular interdependence and c-Src structural requirements for endothelial NO synthase activation. Proceedings of the National Academy of Sciences. 2007;104:16468–16473. doi: 10.1073/pnas.0704315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. The Expanding Cosmos of Nuclear Receptor Coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of Nuclear Respiratory Factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Monje P, Boland R. Subcellular distribution of native estrogen receptor alpha and beta isoforms in rabbit uterus and ovary. J Cell Biochem. 2001;82:467–479. doi: 10.1002/jcb.1182. [DOI] [PubMed] [Google Scholar]
- Moriarty K, Kim KH, Bender JR. Estrogen Receptor-Mediated Rapid Signaling. Endocrinology. 2006;147:5557–5563. doi: 10.1210/en.2006-0729. [DOI] [PubMed] [Google Scholar]
- Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueck AO, Seeger H, Lippert TH. Calcium antagonistic effect of natural and synthetic estrogens-- investigations on a nongenomic mechanism of direct vascular action. Int. J. Clin. Pharmacol. Ther. 1996;34:424–426. [PubMed] [Google Scholar]
- Nilsen J, Brinton RD. Mechanism of estrogen-mediated neuroprotection: Regulation of mitochondrial calcium and Bcl-2 expression. Proc Natl Acad Sci U S A. 2003;100:2842–2847. doi: 10.1073/pnas.0438041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen J, Brinton RD. Mitochondria as therapeutic targets of estrogen action in the central nervous system. Curr Drug Targets CNS Neurol Disord. 2004;3:297–313. doi: 10.2174/1568007043337193. [DOI] [PubMed] [Google Scholar]
- Nilsen J, Chen S, Irwin RW, Iwamoto S, Brinton RD. Estrogen protects neuronal cells from amyloid beta-induced apoptosis via regulation of mitochondrial proteins and function. BMC Neurosci. 2006;7:74. doi: 10.1186/1471-2202-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen J, Irwin RW, Gallaher TK, Brinton RD. Estradiol In Vivo Regulation of Brain Mitochondrial Proteome. J. Neurosci. 2007;27:14069–14077. doi: 10.1523/JNEUROSCI.4391-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Lone R, Knorr K, Jaffe IZ, Schaffer ME, Martini PGV, Karas RH, Bienkowska J, Mendelsohn ME, Hansen U. Estrogen Receptors {alpha} and {beta} Mediate Distinct Pathways of Vascular Gene Expression, Including Genes Involved in Mitochondrial Electron Transport and Generation of Reactive Oxygen Species. Mol Endocrinol. 2007;21:1281–1296. doi: 10.1210/me.2006-0497. [DOI] [PubMed] [Google Scholar]
- Otto C, Rohde-Schulz B, Schwarz G, Fuchs I, Klewer M, Brittain D, Langer G, Bader B, Prelle K, Nubbemeyer R, Fritzemeier K-H. GPR30 localizes to the endoplasmic reticulum and is not activated by estradiol. Endocrinology. 2008 doi: 10.1210/en.2008-0269. en.2008-0269. [DOI] [PubMed] [Google Scholar]
- Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-Scale MicroRNA and Small Interfering RNA Screens Identify Small RNA Modulators of TRAIL-Induced Apoptosis Pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-M, Peter ME. microRNAs and death receptors. Cytokine & Growth Factor Reviews. 2008;19:303–311. doi: 10.1016/j.cytogfr.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Levin ER. Nature of Functional Estrogen Receptors at the Plasma Membrane. Mol Endocrinol. 2006a;20:1996–2009. doi: 10.1210/me.2005-0525. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Wallace DC, Levin ER. Functional Estrogen Receptors in the Mitochondria of Breast Cancer Cells. Mol. Biol. Cell. 2006b;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Sasso A, Abbondanza C, Palumbo G. 17beta -Estradiol Inhibits Apoptosis in MCF-7 Cells, Inducing bcl-2 Expression via Two Estrogen-Responsive Elements Present in the Coding Sequence. Mol. Cell. Biol. 2000;20:2890–2901. doi: 10.1128/mcb.20.8.2890-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard N, Charbonneau C, Sanchez M, Licznar A, Busson M, Lazennec G, Tremblay A. Phosphorylation of Activation Function-1 Regulates Proteasome-Dependent Nuclear Mobility and E6-Associated Protein Ubiquitin Ligase Recruitment to the Estrogen Receptor {beta} Mol Endocrinol. 2008;22:317–330. doi: 10.1210/me.2007-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras RJ, Marquez-Garban DC. Membrane-Associated Estrogen Receptor Signaling Pathways in Human Cancers. Clin Cancer Res. 2007;13:4672–4676. doi: 10.1158/1078-0432.CCR-07-1373. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk Between Nuclear and Mitochondrial Genomes. Annual Review of Biochemistry. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Prossnitz ER, Oprea TI, Sklar LA, Arterburn JB. The ins and outs of GPR30: A transmembrane estrogen receptor. The Journal of Steroid Biochemistry and Molecular Biology. 2008;109:350–353. doi: 10.1016/j.jsbmb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psarra AM, Sekeris CE. Steroid and thyroid hormone receptors in mitochondria. IUBMB Life. 2008;60:210–223. doi: 10.1002/iub.37. [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG. Isoflavones Promote Mitochondrial Biogenesis. J Pharmacol Exp Ther. 2008;325:536–543. doi: 10.1124/jpet.107.134882. [DOI] [PubMed] [Google Scholar]
- Raz L, Khan MM, Mahesh VB, Vadlamudi RK, Brann DW. Rapid estrogen signaling in the brain. Neurosignals. 2008;16:140–153. doi: 10.1159/000111559. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Levin ER. Plasma membrane estrogen receptors signal to antiapoptosis in breast cancer. Mol Endocrinol. 2000;14:1434–1447. doi: 10.1210/mend.14.9.0526. [DOI] [PubMed] [Google Scholar]
- Razmara A, Sunday L, Stirone C, Wang XB, Krause DN, Duckles SP, Procaccio V. Mitochondrial Effects of Estrogen Are Mediated by Estrogen Receptor {alpha} in Brain Endothelial Cells. J Pharmacol Exp Ther. 2008;325:782–790. doi: 10.1124/jpet.107.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cuenca S, Monjo M, Gianotti M, Proenza AM, Roca P. Expression of mitochondrial biogenesis-signaling factors in brown adipocytes is influenced specifically by 17beta-estradiol, testosterone, and progesterone. Am J Physiol Endocrinol Metab. 2007;292:E340–E346. doi: 10.1152/ajpendo.00175.2006. [DOI] [PubMed] [Google Scholar]
- Rosario GX, D'Souza SJ, Manjramkar DD, Parmar V, Puri CP, Sachdeva G. Endometrial modifications during early pregnancy in bonnet monkeys (Macaca radiata) Reprod Fertil Dev. 2008;20:281–294. doi: 10.1071/rd07152. [DOI] [PubMed] [Google Scholar]
- Sanz A, Hiona A, Kujoth GC, Seo AY, Hofer T, Kouwenhoven E, Kalani R, Prolla TA, Barja G, Leeuwenburgh C. Evaluation of sex differences on mitochondrial bioenergetics and apoptosis in mice. Experimental Gerontology. 2007;42:173–182. doi: 10.1016/j.exger.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. Journal of Cellular Biochemistry. 2005 doi: 10.1002/jcb.20743. n/a. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. Journal of Cellular Biochemistry. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Strub A, Voos W. Protein translocation into mitochondria. Biol Signals Recept. 2001;10:14–25. doi: 10.1159/000046873. [DOI] [PubMed] [Google Scholar]
- Schwend T, Gustafsson J-A. False positives in MALDI-TOF detection of ER[beta] in mitochondria. Biochemical and Biophysical Research Communications. 2006;343:707–711. doi: 10.1016/j.bbrc.2006.02.164. [DOI] [PubMed] [Google Scholar]
- Simpkins JW, Yang S-H, Sarkar SN, Pearce V. Estrogen actions on mitochondria--Physiological and pathological implications. Molecular and Cellular Endocrinology. 2008 doi: 10.1016/j.mce.2008.04.013. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW. Coregulator Function: A Key to Understanding Tissue Specificity of Selective Receptor Modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Song RX, Barnes CJ, Zhang Z, Bao Y, Kumar R, Santen RJ. The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor {alpha} to the plasma membrane. PNAS. 2004;101:2076–2081. doi: 10.1073/pnas.0308334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirone C, Duckles SP, Krause DN, Procaccio V. Estrogen increases mitochondrial efficiency and reduces oxidative stress in cerebral blood vessels. Mol Pharmacol. 2005;7:7. doi: 10.1124/mol.105.014662. [DOI] [PubMed] [Google Scholar]
- Stoica GE, Franke TF, Wellstein A, Czubayko F, List HJ, Reiter R, Morgan E, Martin MB, Stoica A. Estradiol Rapidly Activates Akt via the ErbB2 Signaling Pathway. Mol Endocrinol. 2003;17:818–830. doi: 10.1210/me.2002-0330. [DOI] [PubMed] [Google Scholar]
- Thomas P, Pang Y, Filardo EJ, Dong J. Identity of an Estrogen Membrane Receptor Coupled to a G Protein in Human Breast Cancer Cells. Endocrinology. 2005;146:624–632. doi: 10.1210/en.2004-1064. [DOI] [PubMed] [Google Scholar]
- Tremblay A, Giguere V. Contribution of steroid receptor coactivator-1 and CREB binding protein in ligand-independent activity of estrogen receptor beta. J Steroid Biochem Mol Biol. 2001;77:19–27. doi: 10.1016/s0960-0760(01)00031-0. [DOI] [PubMed] [Google Scholar]
- Tuquet C, Dupont J, Mesneau A, Roussaux J. Effects of tamoxifen on the electron transport chain of isolated rat liver mitochondria. Cell Biol Toxicol. 2000;16:207–219. doi: 10.1023/a:1007695308257. [DOI] [PubMed] [Google Scholar]
- Tzeng DZ, Klinge CM. Phosphorylation of purified estradiol-liganded estrogen receptor by caseine kinase II increases estrogen response element binding but does not alter ligand stability. Biochem. Biophys. Res. Communic. 1996;223:554–560. doi: 10.1006/bbrc.1996.0933. [DOI] [PubMed] [Google Scholar]
- Vallejos RH, Stoppani AO. Site-specific effects of steroids on mitochondrial metabolism. Biochim Biophys Acta. 1967;131:295–309. doi: 10.1016/0005-2728(67)90143-0. [DOI] [PubMed] [Google Scholar]
- van Heerde WL, Robert-Offerman S, Dumont E, Hofstra L, Doevendans PA, Smits JF, Daemen MJ, Reutelingsperger CP. Markers of apoptosis in cardiovascular tissues: focus on Annexin V. Cardiovasc Res. 2000;45:549–559. doi: 10.1016/s0008-6363(99)00396-x. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1{alpha} Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- Vina J, Borras C, Gambini J, Sastre J, Pallardo FV. Why females live longer than males? Importance of the upregulation of longevity-associated genes by oestrogenic compounds. FEBS Letters. 2005;579:2541–2545. doi: 10.1016/j.febslet.2005.03.090. [DOI] [PubMed] [Google Scholar]
- Vivacqua A, Bonofiglio D, Albanito L, Madeo A, Rago V, Carpino A, Musti AM, Picard D, Ando S, Maggiolini M. 17beta-Estradiol, Genistein, and 4-Hydroxytamoxifen Induce the Proliferation of Thyroid Cancer Cells through the G Protein-Coupled Receptor GPR30. Mol Pharmacol. 2006;70:1414–1423. doi: 10.1124/mol.106.026344. [DOI] [PubMed] [Google Scholar]
- Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, Pestell RG. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. PNAS. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson N-G. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. PNAS. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CS, Alyea RA, Jeng YJ, Kochukov MY. Nongenomic actions of low concentration estrogens and xenoestrogens on multiple tissues. Molecular and Cellular Endocrinology. 2007;274:1–7. doi: 10.1016/j.mce.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel JM, Iwen KA, Seitz HJ. Regulation of mitochondrial biogenesis by thyroid hormone. Exp Physiol. 2003;88:121–128. doi: 10.1113/eph8802506. [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Radtke C, Seitz HJ. Two thyroid hormone-mediated gene expression patterns in vivo identified by cDNA expression arrays in rat. Nucl. Acids Res. 2001;29:5148–5155. doi: 10.1093/nar/29.24.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Nuyten DSA, Regev A, Lin M, Adler AS, Segal E, van de Vijver MJ, Chang HY. Revealing Targeted Therapy for Human Cancer by Gene Module Maps. Cancer Res. 2008;68:369–378. doi: 10.1158/0008-5472.CAN-07-0382. [DOI] [PubMed] [Google Scholar]
- Yang S-H, Prokai L, Simpkins JW. Correspondence regarding Schwend and Gustafsson, "False positives in MALDI-TOF detection of ER[beta] in mitochondria". Biochemical and Biophysical Research Communications. 2006;345:917–918. doi: 10.1016/j.bbrc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Zhai P, Eurell TE, Cooke PS, Lubahn DB, Gross DR. Myocardial ischemia-reperfusion injury in estrogen receptor-alpha knockout and wild-type mice. Am J Physiol Heart Circ Physiol. 2000a;278:H1640–H1647. doi: 10.1152/ajpheart.2000.278.5.H1640. [DOI] [PubMed] [Google Scholar]
- Zhai P, Eurell TE, Cotthaus R, Jeffery EH, Bahr JM, Gross DR. Effect of estrogen on global myocardial ischemia-reperfusion injury in female rats. Am J Physiol Heart Circ Physiol. 2000b;279:H2766–H2775. doi: 10.1152/ajpheart.2000.279.6.H2766. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xu J-X. The operation of the alternative electron-leak pathways mediated by cytochrome c in mitochondria. Biochemical and Biophysical Research Communications. 2004;317:980–987. doi: 10.1016/j.bbrc.2004.03.144. [DOI] [PubMed] [Google Scholar]