Abstract
Transcription by the vesicular stomatitis virus (VSV) polymerase has been characterized as obligatorily sequential with transcription of each downstream gene dependant on termination of the gene immediately upstream. In studies described here we investigated the ability of the VSV RNA-dependant RNA polymerase (RdRp) to access mRNA initiation sites located at increasing distances either downstream or upstream of a transcription termination signal. Bicistronic subgenomic replicons were constructed containing progressively extended intergenic regions preceding the initiation site of a downstream gene. The ability of the RdRp to access the downstream sites was progressively reduced as the length of the intergenic region increased. Alternatively, bicistronic replicons were constructed containing a mRNA start signal located at increasing distances upstream of a termination site. Analysis of transcription of these "overlapped" genes showed that for an upstream mRNA start site to be recognized it had to contain not only the canonical 3'-UUGUCnnUAG-5' gene start signal, but that signal needed also to be preceded by a U7 tract. Access of these upstream mRNA initiation sites by the VSV RdRp was proportionately reduced with increasing distance between the termination site and the overlapped initiation signal. Possible mechanisms for how the RdRp accesses these upstream start sites are discussed.
Introduction
Vesicular stomatitis virus (VSV) is the prototype of the non-segmented negative-sense (NNS) RNA viruses, and serves as a model for the many serious pathogens that are classified in this group. The VSV genome is 11,161 nucleotides (nt) in length and contains five genes arranged in the order 3’ N-P-MG-L 5’. Transcription of the 5 VSV genes is best described by the ‘stop-start’ model, which posits that the VSV RNA-dependant RNA polymerase (RdRp) enters the genome only at or near the 3’ end, and then moves towards the 5’ end transcribing a single mRNA from each gene (Whelan, Barr, and Wertz, 2004). UV transcriptional mapping studies have shown that transcription of each successive gene is dependent upon the transcription of all prior genes (Abraham and Banerjee, 1976; Ball and White, 1976), and this has been confirmed by molecular genetic analyses (Barr, Whelan, and Wertz, 1997b; Wertz, Perepelitsa, and Ball, 1998). Thus transcription of the genes of VSV is considered to be obligatorily sequential.
The mechanistic details of RNA synthesis from the VSV nucleocapsid protein-enwrapped RNA genome are largely unknown. However it is known that RdRp activity during transcription is modulated by sequence signals located at the beginning and end of each gene, and also within the intergenic region (IGR) that separates adjoining genes (figure 2A) (Barr, Whelan, and Wertz, 1997a; Barr, Whelan, and Wertz, 1997b; Hwang, Englund, and Pattnaik, 1998; Ogino and Banerjee, 2007; Stillman and Whitt, 1997; Stillman and Whitt, 1998; Wang, McElvain, and Whelan, 2007). A conserved sequence at each gene start 3'-UUGUCnnUAG-5' directs the RdRp to initiate and cap the nascent mRNA, whereas the conserved gene end sequences, 3'-AUACUUUUUUU-5' cause the RdRp to polyadenylate and terminate synthesis of the transcript. However, it has also been shown that the U7 tract at the end of each upstream gene, in addition to being the template for mRNA polyadenylation and critical for termination, is also an important upstream sequence element for synthesis of each downstream gene (Hinzman, Barr, and Wertz, 2002). Following termination of each monocistronic mRNA, the RdRp is thought to migrate downstream across the IGR in a non-transcriptive mode to reach the start of the next gene.
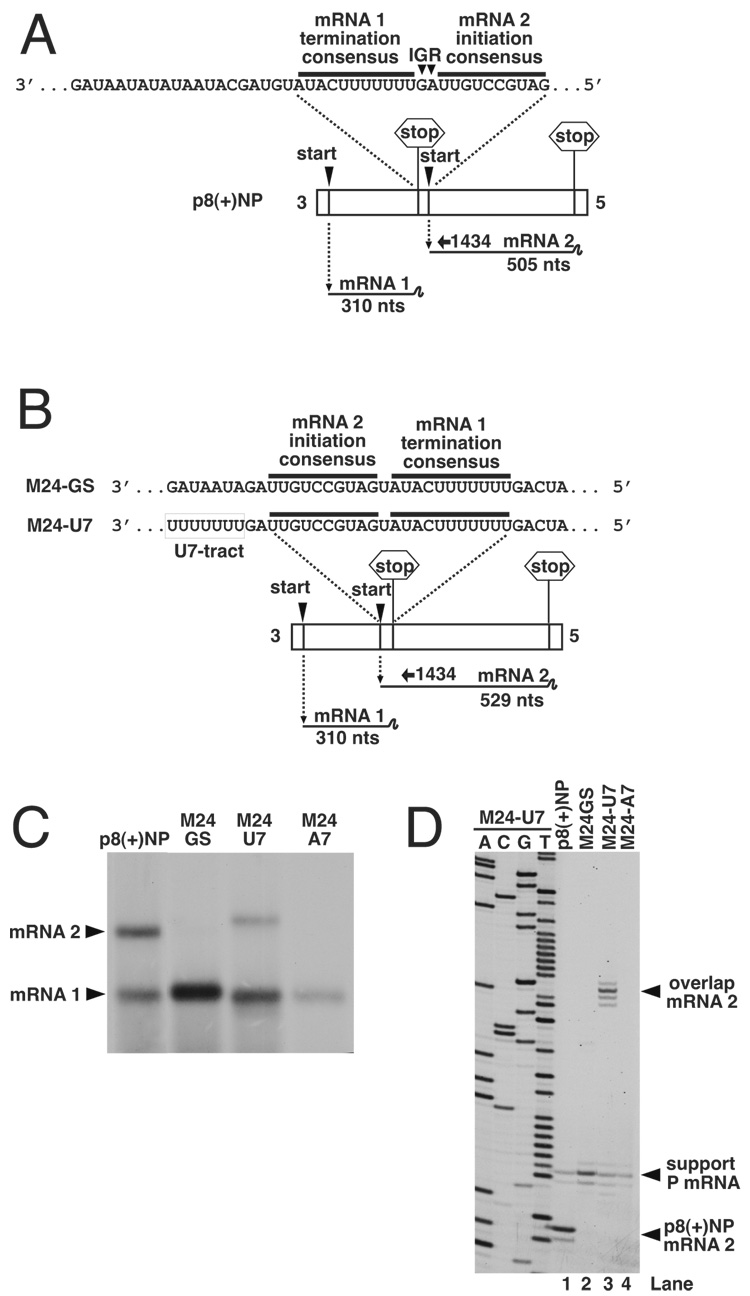
Figure 2. Analysis of the transcription activity of bicistronic genome analogs having genes that overlap by 24 nucleotides.
(A). Schematic representation of genome analog p8(+)NP and the mRNAs that are transcribed from upstream (mRNA1) and downstream (mRNA2) genes separated by the wild-type gene junction. (B). Schematic representation of genome analogs M24-GS and M24-U7, in which the mRNA2 initiation signal is located 24 nts upstream of the mRNA1 termination signal. M24-U7 differs from M24-GS in that it contains an U7 tract upstream of the transcriptional initiation site. Additional template M24-A7 was also generated in which the U7 tract was substituted for A7. (C). These bicistronic genome analogs were expressed in BHK cells, and their RNA synthesis activity compared to that of wild-type template p8(+)NP, which has conventional non-overlapping genes. Actinomyin D-resistant RNAs were metabolically labeled with 3H uridine, harvested, treated with RNase H/oligo d(T) and then electrophoresed on a denaturing 1.75% agarose-urea gel. RNAs were visualized by autoradiography and the positions of mRNAs 1 and 2 are marked with arrowheads. (D). The RNA synthesis activity of bicistronic templates was also analyzed by primer extension analysis using end-labeled oligonucleotide 1434, which anneals within the 5’ end of mRNA 2, shown schematically in panels A and B. Alignment of the 1434 extension product with the adjacent sequence ladder confirms the mRNA2 product of template M24-U7 is initiated at the overlapped initiation site.
At an unknown stage between termination of each upstream gene and initiation of the next gene downstream a process known as polymerase attenuation occurs, which leads to a progressive reduction of transcript initiation at each subsequent downstream gene start by approximately 30% (Iverson and Rose, 1981). It has been postulated that this is due to dissociation of some RdRp molecules from the template. However, little is known about what causes or controls attenuation. Previous studies have shown that changes to the IGR sequence affect attenuation by dramatically reducing transcription initiation (Barr, Whelan, and Wertz, 1997b; Stillman and Whitt, 1997; Stillman and Whitt, 1998), which suggests that the VSV RdRp reacts to sequence signals as it moves across the IGR.
The VSV IGR has also been shown to tolerate modest increases in size. Currently, the longest IGR that has been successfully incorporated into a VSV gene junction is 88 nt in length (Hinzman, Barr, and Wertz, 2002), and this extension from 2 to 88 nts still allowed initiation at the downstream gene start, although at a reduced level. Examination of RNA synthesis as the RdRp crossed this altered gene junction revealed that while the overwhelming majority of initiation events occurred at the consensus gene start signal 88 nts downstream, a low level of initiation events were also detected from within the extended IGR. Primer extension analysis mapped the 5’ ends of these mRNAs to sites that had similarity to, but minor deviations from, the consensus initiation signal and as a consequence exhibited sub-optimal initiation activity.
This finding, and similar findings from a study in which 19 nts were added to a VSV IGR (Stillman and Whitt, 1998) prompted the suggestion that the VSV RdRp migrates across the IGR in a "scanning" mode. In this mode, the polymerase is proposed to non-productively scan the sequence and can initiate transcription at sequences that match or resemble the consensus initiation signal. The extent to which a sub-optimal initiation sequence deviates from the consensus initiation sequence influences not only the likelihood of initiation, but also whether the resulting transcript will possess appropriate 5’ modifications (Stillman and Whitt, 1999).
Interestingly, the RdRps of some NNS RNA viruses are able to access a gene start that lies upstream, rather than downstream, of a termination site. The best-studied example of such RdRp activity is at the junction between the M2 and L genes of human respiratory syncytial virus (HRSV) (Fearns and Collins, 1999). At this gene junction, the initiation signal for the L gene lies 68nts upstream of the M2 gene end. However, transcription from the upstream L gene initiation site was found to be a surprisingly efficient process, such that the level of transcription was comparable to that from an initiation signal located in a conventional downstream position. Another example is the rhabdovirus sigma virus, for which the initiation signal of the glycoprotein gene lies 33 nt upstream of the termination signal for the preceding gene (Teninges, Bras, and Dezelee, 1993).
To better understand how the NNS RNA virus RdRp selects the site of transcription initiation, we examined the ability of the VSV RdRp to transcribe from initiation sequences located at various distances either up-or downstream of a proximal termination signal. Our results show that the VSV RdRp can access initiation signals located over 200 nts up-or downstream from a termination signal, but that it does so with decreasing efficiency as the distance between termination and initiation sites increases.
Materials and Methods
Plasmid constructions
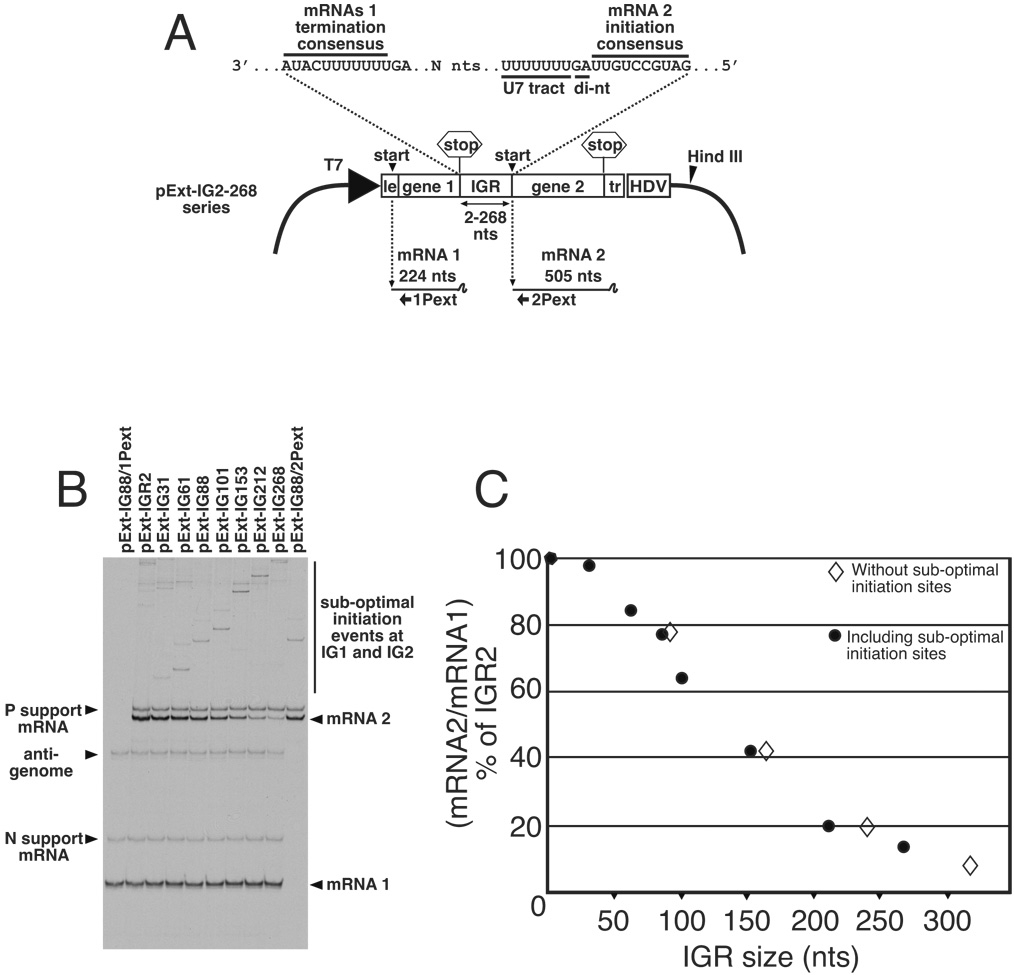
Transcription plasmids expressing VSV genome analog templates having lengthened IGRs were based on previously described plasmids p8(+)NP (Barr, Whelan, and Wertz, 1997b) (figure 2A) and pExt-IG (Hinzman, Barr, and Wertz, 2002) (figure 1A). Plasmid p8(+)NP encodes a VSV anti-genome analog that is subsequently replicated to generate a bicistronic genome analog containing wild-type (WT) leader and trailer regions flanking upstream (gene 1) and downstream (gene 2) transcription units, separated by a wild-type N/P gene junction. This plasmid was modified by the addition of a consensus termination signal 73 nt upstream of the existing gene 1 termination signal, to generate pExt-IG (figure 1A), which possessed an 88 nt-long IGR. PCR-based mutagenesis was then used to cleanly delete the original wild-type N/P gene junction, and then insert Xho I restriction sites at either 15, 45, 85, 137, 196 and 252 nts downstream of the new gene 1 termination signal. The resulting set of 6 plasmids were then each digested with Xho I and Hind III (position shown in figure 1A). The excised fragment was then replaced with an Xho I and Hind III digested fragment amplified by PCR from p8(+)NP using oligonucleotides XhoI-GS (5’-ACGTACTCGAGCAAAAAAACTAACAGATATCATGG-3’) and a generic SP6-primer (figure 1A). This fragment contained a consensus gene start signal including upstream U-tract element followed by the complete sequence of mRNA 2, and all downstream sequences up to the Hind III site, which was located within the multiple cloning site of the original plasmid vector. All 6 resulting plasmids now possessed identical gene 1 (224 nts) and gene 2 (505 nts) sequences separated by IGRs of various lengths, specifically 31, 61, 101, 153, 212 and 268 nts. Due to the strategy of construction, these IGRs comprised VSV N and P gene sequences that are transcribed by the VSV RdRp. To act as a control, we generated template Pext-IG2, which contained the above gene 1 and gene 2 sequences separated by the wild-type N/P gene junction, which has a 2 nt-long IGR. To complete the series of extended IGR templates, we also included previously described template pExt-IG(AUAG), which contains an 88 nt-long IGR (Hinzman, Barr, and Wertz, 2002).
Figure 1. Analysis of the transcription activity of bicistronic genome analogs with lengthened intergenic regions (IGRs).
(A). Schematic representation of plasmid pExt-IG and its derivatives. These plasmids were individually transfected into vTF7-3 infected BHK cells along with support plasmids expressing VSV N, P and L ORFs, where they expressed an RNA representing a VSV anti-genomic analog. Encapsidation and replication of this RNA yielded a genome analog containing upstream (mRNA1) and downstream (mRNA2) transcriptional units separated by IGRs ranging between 2 and 268 nucleotides in length. Locations of U7 tract and intergenic di-nucleotide (di-nt) 3’ of the mRNA 2 initiation signal are shown. (B). Abundance of mRNA1 and mRNA2 transcribed from bicistronic templates was determined by dual extension of end-labeled oligonucleotides 1Pext and 2Pext, respectively. RNAs generated from template pExt-IG88 were also subjected to single primer extension reactions using individually 1Pext and 2Pext, to show position of mRNA 1 and mRNA2 respectively. Extension products were electrophoresed on a denaturing 6% acrylamide sequencing gel, and visualized by autoradiography. (C). The ability of the VSV RdRp to access the downstream gene start was calculated by quantitation of radiolabeled products representing downstream mRNA2 as a fraction of upstream mRNA1. This figure was then expressed as a percentage of that determined for template IGR2, which has the wild-type IGR length of 2 nts, and plotted as closed circles. Templates having the sub-optimal IGR start sites removed by mutagenesis were analyzed in the same way, and the values for mRNA2/mRNA1 % of IGR2 are also plotted as open diamonds.
The panel of transcription plasmids used to generate VSV genome analog templates having overlapped gene junctions were also based on previously described p8(+)NP (Barr, Whelan, and Wertz, 1997b) (figure 2A). Plasmid M24-GS was generated by insertion of a consensus gene start signal preceded by the intergenic di-nucleotide (3’-GAUUGUCCGUAG-5’) 1 nt upstream of the existing termination signal, to create a 24 nt-long overlap (figure 2B). This template was subsequently modified by addition of either A7 or U7 tracts immediately upstream of the newly inserted gene start to give templates M24-A7 and M24-U7, respectively. The overlap of M24-U7 was extended to 35 nts by insertion of the sequence 3’-GGCGAGCTCGC-5’, and all subsequent modifications to increase the length of the gene junction overlap length were engineered at the XhoI site contained within this insert. This strategy was chosen to ensure that the distance between the consensus initiation signal of overlapped gene 2 remained a constant distance from the initiation signal of gene 1. The nucleotide sequence of all overlapped gene junctions are listed in table 1.
Table 1.
Nucleotide sequence of overlapping gene junctions of bi-cistronic genome analogs. All sequences are negative sense and written in 3’ to 5’ orientation. The initiation signal for mRNA2 is shown underlined, and the termination signal for mRNA1 is shown in bold.
| M24 |
| GUUUUUUUGAUUGUCCGUAGUAUACUUUUUUUGA… |
| M35 |
| GUUUUUUUGAUUGUCCGUAGGGCGAGCTCGCUAUACUUUUUUUGA… |
| M46 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAACCGGUAUACUUUUUUUGA… |
| M56 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAAGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M66 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M76 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAAAUAGAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M86 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAAGUAGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M97 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAACCGUAAGUACGACCGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M122 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAACCGUAAGUACGACCGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M150 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAACCUAAGUACGACCGGGGCUCAGAAAUCGUAGGCUUCGCGCGGAGCUCAUUUAACCGUAAGUACGACCGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
| M200 |
| GUUUUUUUGAUUGUCCGUAGUCUUAAGAGCUCAUUUAACCUAAGUACGACUGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAGCUCAUUUAACCUAAGUACGACCGGGGCUCAGAAAGAGCUCAUUUAACCGUAAGUACGACCGGGGCUCAGAAAUCGUAGGCUUCGCGCGCUUUAGCUACCGGUAUACUUUUUUUGA… |
Transfections
Generation of ribonucleocapsid (RNP) complexes competent for RNA synthesis was achieved as described previously (Pattnaik et al., 1992). Briefly, plasmids encoding VSV N, P and L proteins along with plasmids which expressed bicistronic anti-genome analogues were transfected into BHK-21 cells previously infected with vaccinia virus recombinant vTF7-3, which expresses T7 RNA polymerase. Transfections were performed at 37C for a total duration of 22 hrs.
RNA labeling, isolation and detection
VSV RNAs generated from genome analog RNPs were detected by direct metabolic labeling at 16h post-transfection by incubating transfected cells with 3H uridine (33µCi/ml, Perkin Elmer) in the presence of actinomycin D (10µg/ml, Sigma Chemical Co. St. Louis, MI) for 6 hr, as described previously (Pattnaik et al., 1992). After this labeling period, total cellular RNAs were harvested using the RNeasy procedure (Qiagen, Valencia, CA). RNAs to be analyzed by agarose-urea gel electrophoresis were annealed to oligo d(T) and digested by RNase H in order to remove heterogeneous 3’ poly(A) tails. RNAs to be analyzed by primer extension analysis were not digested. RNAs were visualized by subjecting the 3H labeled, actinomycin D-resistant RNAs to electrophoresis in 1.75% agarose -urea gels followed by fluorography and autoradiography, as previously described (Pattnaik et al., 1992).
Primer extension analysis
The 5’ ends of VSV-specific RNAs were identified by primer extension analysis. Briefly, PAGE-purified oligonucleotides (Operon, Valencia, CA) were end-labeled in the presence of γ33P ATP using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA), and purified from unincorporated label using a QIAquick nucleotide removal column. Primers were annealed in excess to VSV-specific RNAs, and extended for 30 minutes at 42°C using superscript III reverse transcriptase (Invitrogen). Sequence ladders to act as size markers were generated using Sequenase modified T7 DNA polymerase (Sequenase 2.0, US. Biochemical Corp., Cleveland, Ohio), according to instructions supplied by the manufacturer. Radiolabeled cDNA fragments were electrophoresed on a standard 6% acrylamide sequencing gel containing 6M urea, and visualized by autoradiography. For detection of downstream initiation events, previously described oligonucleotide 2Pext (Hinzman, Barr, and Wertz, 2002)(5’-ATCTCATCTATCTCTCCTACCGCC-3’) was used, which annealed within the 5’ end of gene 2 mRNAs. To aid in quantitation of 2Pext extension products, previously described oligonucleotide 1Pext (Hinzman, Barr, and Wertz, 2002) (5’-TTTGATTTTCTGAAGTAATCTGCCGGG-3’) was also included in the extension reaction, which annealed within the 5’ end of gene 1 mRNAs. For detection of upstream mRNA2 initiation, either oligonucleotides 1434 (5’-GGACTTGAGATACTCACG-3’), 1365 (5’-AGTTTTTTTCATATGGCCATCGATTT-3’), or T7-14 (5’-TTTTTTTTCATATGG-3’) was used, depending on the template under investigation.
Quantitation
Where appropriate, abundance of radio-labeled cDNA or RNA species was calculated by densitometric analysis of autoradiographs using Quantity One Software and a PDI model 320i densitometer. The abundance of mRNA2 products transcribed from templates with extended IGRs (figure 1B) was expressed as a percentage of mRNA1 levels that were transcribed from the same template. To facilitate comparison of relative mRNA2 transcription levels across the autoradiograph, all mRNA2/mRNA1 levels were shown as a percentage of IGR2. Similarly, abundance of mRNA 2 products initiating from overlapped gene starts tabulated in figure 5B were also expressed as a percentage of mRNA 1 abundance from the same template and shown as a percentage of M97. Abundance of mRNA 2 levels from all other templates with overlapping gene starts (figures 4B and C) were expressed as a percentage of mRNA 2 levels transcribed from the smallest overlap shown on each respective autoradiograph. Quantities shown are an average of at least three independent experiments.
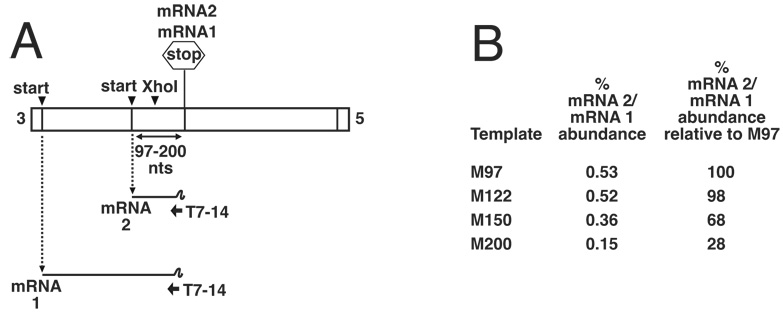
Figure 5. Investigating the upper limit of RdRp access to an overlapped initiation signal using primer extension analysis.
(A). Schematic representation of genome analogs that possess an mRNA 2 initiation signal positioned at increasing distances upstream of the mRNA1 termination signal, up to a distance of 200 nucleotides. The mRNA1 and mRNA2 transcription products are also shown schematically, all of which are predicted to terminate at the mRNA1 termination signal. (B). RNAs generated by templates M97, M122, M150 and M200 were harvested from BHK cells and analyzed by primer extension analysis using end-labeled oligonucleotide T7–14, which anneals within the 3’ end of mRNAs 1 and 2, shown schematically in panel A. Bands representing mRNA2-templated extension products from five separate experiments were quantified and their abundance shown as a percentage of mRNA1 levels from the same template. Percent abundance relative to template M97 was also determined.
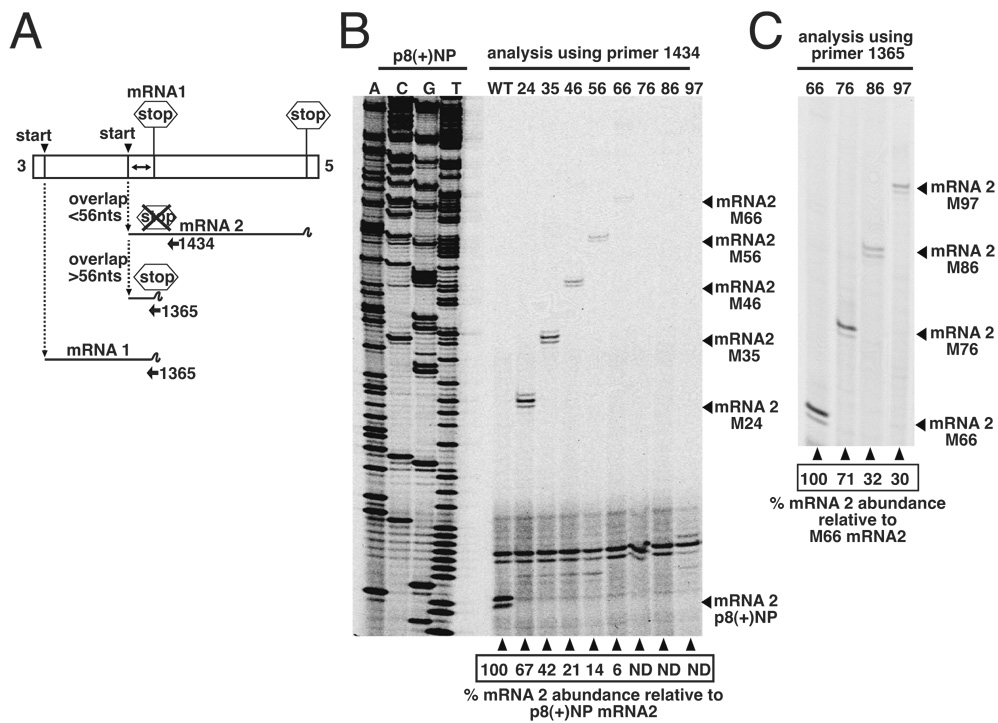
Figure 4. Analysis of RNAs transcribed from bicistronic genome analogs with overlapping genes using primer extension.
(A). Schematic representation of genome analogs in which the mRNA 2 initiation signal was positioned at increasing distances upstream of the mRNA1 termination signal, up to a distance of 97 nucleotides. Transcription products are shown schematically, including the 3’ truncated form of mRNA 2 which is transcribed from templates having overlaps over 56 nts, due to activity of the mRNA1 termination signal. (B). RNAs generated by these bicistronic genome analogs were harvested from BHK cells and analyzed by primer extension analysis using end-labeled oligonucleotide 1434, which anneals within the middle region of mRNA2, shown schematically in panel A. Abundance of each mRNA 2 species is shown as a percentage of mRNA 2 levels generated by template p8+NP and represents the mean of at least 3 separate experiments. The cDNA expressing wild-type anti-genome analog 8(+)NP was sequenced with primer 1434 to act as size marker. (C). RNAs generated from genome analogs having overlaps in excess of 56 nts were also analyzed by primer extension using end-labeled oligonucleotide 1365, which annealed at the 5’ end of mRNA2 and allowed detection of mRNA2 transcripts that terminated at the mRNA1 termination signal, shown schematically in panel A. As above, abundance of each mRNA 2 species is shown as a percentage of mRNA 2 levels generated by template M66, and represents the mean of at least 3 separate experiments.
Results
Access of the RdRp to downstream initiation sites
We first examined the ability of the RdRp to access downstream mRNA initiation sites that were located at progressively greater distances from an upstream termination site. To do this we constructed a panel of 7 plasmids which expressed bicistronic templates in which the up and downstream genes were separated by progressively increased IGR lengths ranging from the wild type 2 nts up to 268 nts. Plasmid construction details are described in materials and methods and shown schematically in figure 1A. Briefly, an additional, functional transcription termination signal (3’-AUACUUUUUUU-5’) was inserted and it was followed by the varied increasing IGRs. A U-tract, shown to be an essential component of the initiation sequence (Hinzman, Barr, and Wertz, 2002) was then positioned to precede the downstream initiation signal (3’-UUUUUUUGAUUGUCnnUAG-5’). VSV-specific RdRp transcription products were analyzed by primer extension using two oligonucleotides: 1Pext, which annealed to mRNA1 (figure 1A) and 2Pext (figure 1A), which annealed to mRNA2, allowing quantitation and comparison of the relative ability of the RdRp to access initiation sites for upstream gene 1 and downstream gene 2 on each of the altered templates.
This analysis showed that genome analogs with wild type or extended IGRs all synthesized mRNA2 in addition to mRNA 1, indicating that in each case, the VSV RdRp was able to access the mRNA 2 initiation signal (figure 1B). However, quantitation of extension products (figure 1C) showed that transcription initiation from the mRNA 2 start-site was progressively diminished with increased IGR length. In comparison to the wild type IGR size of 2 nt, an extended IGR of 268 nts reduced mRNA 2 levels to approximately 14% of that of the wild-type 2 nt IGR. These results show that the ability of the VSV RdRp to access downstream initiation sites is inversely proportional to the distance the RdRp has to travel after terminating transcription of the upstream mRNA.
A low abundance of initiation events from sub-optimal start sequences were generated by templates IGR31, 61, 88, 101, 153, 212 and 268. This was expected, as the IGR within these constructs contains previously identified sub-optimal initiation sequences IG1 (3'-CCGUUCAUAC-5') and IG2 (3'-CUGUUUACUG-5') (Hinzman, Barr, and Wertz, 2002). However, the abundance of these products was both consistent and low for all templates under examination, and so were not responsible for reduced abundance of mRNA 2. To confirm this, these suboptimal starts were eliminated by site-specific mutagenesis and the analyses redone. This new data showed an equivalent reduction in mRNA 2 synthesis as a function of increasing intergenic length (figure 1C).
Access of the RdRp to upstream initiation sites
We next examined the ability of the VSV RdRp to access and transcribe from a gene start positioned at various distances upstream of a termination signal in what has been referred to as an "overlapped" start site. To do this we generated bicistronic genome analog template M24-GS (figure 2B), in which the initiation signal of the second gene (gene 2) was located 24 nt upstream of the first gene (gene 1) termination signal, as described in materials and methods. We have previously shown that access to the second gene start in bicistronic replicon p8(+)NP to generate mRNA2 was strictly-dependant on transcription termination at the upstream gene end (Barr, Whelan, and Wertz, 1997b). Therefore, we reasoned that the VSV RdRp could only access the overlapped gene 2 start signal following termination at the upstream gene end. To analyze the ability of the VSV RdRp to initiate at this upstream site, we examined the VSV-specific RNAs generated by M24-GS by direct metabolic labeling with 3H uridine using agarose-urea gel electrophoresis followed by fluorography. RNAs were also harvested from the parental template p8(+)NP, to provide mRNA1 and mRNA2 size markers (310 and 505 nts, respectively). Although the overlapped mRNA 2 transcriptional unit was 24 nts in length (see Fig 2), we predicted mRNA 2 to be considerably longer, as we have previously shown that the VSV RdRp cannot recognize a termination signal in close proximity (less than approximately 56 nts) to the corresponding upstream initiation site (Whelan, Barr, and Wertz, 2000). RdRp molecules that initiate at the overlapped gene 2 start were therefore expected to by-pass the first (mRNA 1) termination signal and proceed to the termination signal at the end of gene 2, resulting in a 529 nt-long mRNA (505+24 nts) (figure 2B). However, agarose-urea gel analysis showed that template M24-GS did not generate a detectable mRNA 2 of this length (figure 2C).
Role of the U-tract in upstream initiation
We recently showed that in addition to the conserved gene start sequence, initiation of VSV transcription requires an upstream U-tract element that is normally supplied by the termination signal of the gene immediately upstream (3'-AUACUUUUUUU-5’) (Hinzman, Barr, and Wertz, 2002). A consequence of creating the overlap in M24-GS is that the gene start is no-longer immediately downstream of the normal upstream U-tract. To determine whether this element was required for initiation at upstream sites, we inserted a U7-tract in front of the upstream gene start to generate template M24-U7 (figure 2B). As a control, an A7-tract was also positioned in the same location to generate template M24-A7. Agarose-urea gel electrophoresis showed that template M24-U7 signaled abundant synthesis of the downstream mRNA 2 and that the message was of the appropriate size (figure 2C). The template having A7 preceding the overlapped start did not signal initiation. These results show firstly that the RdRp can access upstream initiation sites following termination, and secondly that this initiation is dependant on presence of an upstream U-tract.
The site of upstream initiation
To confirm that the M24-U7 mRNA 2 was initiated at the upstream gene start site we performed primer extension analysis using negative-sense oligonucleotide 1434, which annealed downstream of the mRNA 2 gene start (figure 2A and 2B). Taking into account that we consistently observe reverse transcription of the mRNA 5’ terminal cap structure (as shown by multiple bands as extension product), this analysis showed that the predominant mRNA 2 extension product precisely mapped to the overlapping mRNA 2 gene start (figure 2D, lane 3). In contrast, primer extension products corresponding to mRNA2 were not detected for templates M24-A7 or M24-GS, which corroborated our analysis of these templates using agarose-urea gel analysis (figure 2C), described above.
RdRp access to initiation sites further upstream
We next investigated whether the VSV RdRp was able to recognize initiation sites located further upstream of the gene 1 termination signal. A panel of 8 bicistronic genome analogue templates with increasingly longer gene overlaps was constructed (figure 3A). These templates were generated by sequentially inserting blocks of either 10 or 11 nts within the overlap of template M24-U7, to generate templates M35, M46, M56, M66, M76, M86 and M97, with template numbering representing the total length of the gene overlap. The inserted nucleotides were chosen such that they would not include potential initiation signals, and are shown in table 1. Furthermore, as shown schematically in figure 3A, these nucleotides were inserted into the gene overlap such that the distance between the consensus initiation signal of overlapped gene 2 remained a constant distance from the initiation signal of gene 1.
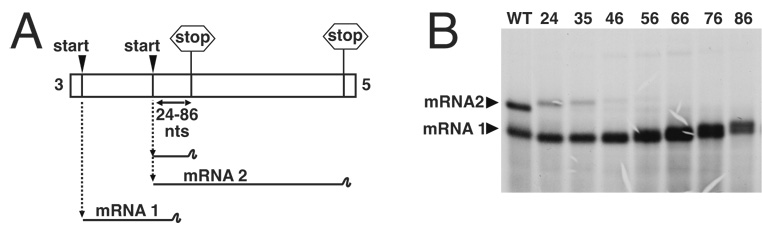
Figure 3. Visualization of RNAs transcribed from bicistronic genome analogs with overlapping genes.
(A). Schematic representation of genome analogs in which the mRNA 2 initiation signal was positioned at increasing distances upstream of the mRNA1 termination signal, up to a distance of 86 nucleotides. Transcription products mRNA1 and mRNA2 are also shown schematically. (B). Bicistronic genome analogs were generated in BHK cells, and their RNA synthesis activity compared to that of wild-type template p8(+)NP, which has conventional nonoverlapping genes. Actinomycin D-resistant RNAs were metabolically labeled with 3H uridine, harvested, treated with RNase H/oligo d(T) and then electrophoresed on a denaturing 1.75% agarose-urea gel. RNAs were visualized by autoradiography and the positions of mRNAs 1 and 2 are marked with arrowheads.
The ability of the RdRp to access the upstream gene start was first analyzed by agarose-urea gel electrophoresis. This analysis showed that as overlap size increased, abundance of mRNA 2 apparently decreased, such that mRNA 2 products were not detected for overlaps greater than 46 nts (figure 3B). However, we postulated that this decline in mRNA 2 transcription could be due to two different mechanisms: Firstly, as described above, the decline could be due to the ability of the VSV RdRp to terminate transcripts over 56 nts in length, leading to termination of small mRNA 2 products at the mRNA 1 termination site (shown schematically in fig 3A). Such small mRNAs would not be resolved in the agarose-urea gel system and would thus be undetectable. Secondly, the decline could represent reduced ability of the VSV RdRp to access the progressively more-distant upstream initiation signals, similar to the results described above wherein polymerase access was proportionally reduced as the distance to a downstream initiation site was increased.
Access to upstream start sites is proportionally reduced with increased size of overlap
To determine the relative contribution of these two mechanisms to the apparent reduction in mRNA 2 initiation with increased overlap size (fig 3B), we performed primer extension analysis using oligonucleotide 1434, which annealed downstream of the mRNA 1 stop site, and also oligonucleotide 1365, which annealed to the mRNA1 gene end (figure 4A). This analysis would allow us to detect all mRNA 2 products, irrespective of whether they terminated at either mRNA 1 or mRNA 2 termination sites (figure 4A). RNAs generated by templates M35, M46, M56, M66, M76, M86 and M97 were first analyzed by primer extension using oligonucleotide 1434. This analysis detected mRNA 2 in quantities that diminished as the overlap size increased, such that no mRNA 2 transcription was detected from templates M76, M86 or M97 (figure 4B). These findings corroborated those of the agarose-urea gel analysis described above (figure 3B).
In contrast, extension of primer 1365 detected transcription of mRNA 2 from all templates tested (figure 4C), including M76, M86 and M97. Taken together, these results show that diminished detection of mRNA 2 by both extension of primer 1434 (figure 4B), and by agarose-urea electrophoresis (figure 3B) was due to the transcribing RdRp now recognizing the proximal termination signal. This confirmed our previous demonstration of a minimal mRNA size requirement for efficient transcription termination.
The detection of extension products corresponding to all upstream start sites using primer 1365 showed the VSV RdRp had the ability to access initiation sites located increasing distances upstream of the termination site. Quantitation of primer 1365 extension products showed that this ability was reduced with increasing overlap size (figure 4C).
To explore an upper limit of upstream RdRp access to transcription start sites, we generated templates M122, M150 and M200, having overlap sizes of 122, 150 and 200 nts respectively. The overlap was increased by inserting sequences derived from annealed oligonucleotides into an unique Xho I site within the overlap region of M97, as described in materials and methods. As previous results have shown that some IGRs can hinder access of the VSV RdRp to initiation signals (Barr, Whelan, and Wertz, 1997b; Hinzman, Barr, and Wertz, 2002; Stillman and Whitt, 1997; Stillman and Whitt, 1998), all inserted cDNA fragments were derived from the overlap region of M97, and are listed in table 1. The ability of the RdRp to initiate at these overlapped gene starts was analyzed by primer extension using oligonucleotide T7–14, which annealed at the 3’ end of both mRNA 1 and 2 (figure 5A). Quantitation of T7–14 extension products that mapped to overlapped mRNA2 gene start sites showed that the VSV RdRp was able to access initiation sequences over 200 nts upstream of the termination signal (figure 5B). However, the abundance of these extension products diminished corresponding to increased overlap size, such that the M200 mRNA 2 extension product was barely detectable. By quantifying the mRNA 2 products generated by each template, we determined that increasing the overlap size from 97 to 200 nts reduced mRNA 2 initiation to approximately 28% of the level of the 97 nt overlap (by approximately 4 -fold), or 15% of corresponding mRNA 1 levels (figure 5B). Therefore, taken together, our results show that the VSV RdRp is able to access upstream initiation sites, but with an ability that decreases in proportion with increased overlap size.
Discussion
In this study, we have shown that the VSV RdRp is able to access initiation sites located both up-and downstream of a proximal termination signal. The efficiency with which these sites are accessed is strongly affected by the distance of the initiation sequence from the gene end termination site, irrespective of whether it is located upstream or downstream. In addition, our data have confirmed two fundamental aspects of VSV transcription: Firstly, these data confirm in a novel way that a U-tract element must be located upstream of an initiation site for it to be efficiently utilized for mRNA transcription. These results show that this requirement applies to initiation sites located either upstream or downstream of the adjacent termination site. Interestingly, we found that non-canonical start sites were recognized, albeit at extremely low levels, (Figure 1B) even when not preceded by a U7 tract during downstream movement of the polymerase. In contrast, a perfect gene start signal was not recognized in the overlapped situation until a U7 tract was positioned in front of it (Figure 2C and D). Therefore, the requirements for efficient recognition of upstream initiation sites may be more stringent than recognition of initiation sites located downstream. Secondly, our results confirm that the ability of the VSV polymerase to recognize and terminate at a gene end termination sequence depends on the mRNA having attained a minimal size (Whelan et al, 2000, and figure 4).
The IGRs of NNS RNA viruses can be classified in two groups: either short and well conserved, or variable in both length and sequence (Whelan, Barr, and Wertz, 2004). The IGRs of VSV fall into the former group, being short and well-conserved as GA or CA, with only the G-L gene junction of the New Jersey strain being an exception at 21 nt-long. Examples of other NNS RNA viruses that exhibit the same IGR format include Hendra virus, Sendai virus, Human parainfluenza virus 3, measles virus and rinderpest virus. Recently, it was reported that extension of the Sendai virus IGR had a similar effect on access of the RdRp to downstream initiation sites as we have described here for VSV, in that RdRp access to downstream start sites was progressively reduced with increasing distance (Plattet et al., 2007). Taking these findings together it is tempting to speculate that the RdRp of viruses with normally short and conserved IGRs may be functionally deficient in scanning long and variable IGRs, and this property has restricted IGR expansion in the corresponding genomes.
Whether the RdRps of NNS RNA viruses that have more variable IGRs, such as HRSV, Ebola virus, rabies virus, simian Virus 5 and mumps virus are more able to tolerate IGR extensions is currently unclear. The only studies performed to date have been for HRSV (subtype-A), which possesses IGRs of between 1–52 nts. The variation in IGR length was suggested not to substantially affect downstream initiation, based on experiments in which the different naturally occurring intergenic regions were compared by placing them between two reporter genes in which the IGRs did not follow their naturally occurring gene end signals, which also vary (Kuo, Fearns, and Collins, 1996). However, these results are difficult to interpret in light of recent functional analyses of the HRSV gene junction, which showed interplay between gene end and IGR sequences (Harmon and Wertz, 2002). More recently, analysis of infectious HRSV recombinants having a single IGR between the M and G genes extended progressively from 65 up to 160 nts in length were shown to display reduced plaque size and replication in single step growth analyses as the length of the IGR increased (Bukreyev, Murphy, and Collins, 2000). Attempts to determine whether these extended IGRs affected virus growth at the level of altered transcription initiation were not conclusive as the Northern blots used to analyze transcription did not measure initiation events. More work is needed to clarify how IGR extension affects gene expression of these viruses.
Our results show the VSV RdRp can traverse substantially extended IGRs to access gene start sites located upstream of a termination site (overlapped) as well as downstream. The ability of the VSV RdRp to access upstream initiation sites is interesting given that VSV does not possess an overlapped gene junction. The related rhabdovirus member sigma virus does possess a transcriptionally active gene overlap (Teninges, Bras, and Dezelee, 1993), as do members of several other NNS RNA virus families such as the pneumoviruses and filoviruses. This raises the possibility that access to upstream start sites may be an ancestral property of NNS RNA virus polymerases that the VSV RdRp has retained. Possibly access to upstream start sites is an activity possessed by all negative strand RdRps, regardless of whether their corresponding RNA templates contain an overlap or not. Alternatively, this feature of polymerase activity may reflect a fundamental mechanism by which the polymerase operates. These possibilities remain to be tested. Surprisingly, the effects of extending the intergenic region on downstream initiations and the effects of extension of an upstream overlap were similiar. Our results suggest that the relative abundance of mRNA 2 in relation to mRNA 1 transcribed from IGR212 is approximately 14% (20% multiplied by 0.7), which compares closely to 15% calculated for overlapped upstream extension construct M200.
Our demonstration that the RdRp can access initiation sites located several hundred nucleotides from the site of termination raises the interesting question of how the RdRp moves to these distant initiation sites. Recent investigation of both bacterial and bacteriophage RNA polymerase (RNAP) activity has shown that the driving force behind downstream RNAP movement during polymerization is the action of Brownian motion coupled with a ‘ratchet’ mechanism, which ensures all movement is uni-directional (Bar-Nahum et al., 2005; Guo and Sousa, 2006). In this model, RNAP movement depends on RNA polymerization, as the ‘ratchet’ component of the machine originates from insertion of NTP molecules into the growing RNA strand.
It is not known whether this model also describes VSV RdRp activity during movement across the IGR. To be compatible, the VSV RdRp would need to transcribe each nucleotide of the IGR into RNA sequences as it moved downstream towards the next consensus gene start. While our data presented here and in previous work suggest that the RdRp responds to the IGR sequence and can transcribe RNAs from suboptimal initiation sequences, we have no evidence that the entire sequence of the IGR is transcribed. One possible reason for failure to detect these sequences could be that these RNAs may be degraded, as they likely will not possess appropriate 5’ end modifications, which are specified by the authentic gene start sequence (3'-UUGUCnnUAG-5') (Stillman and Whitt, 1999).
Alternatively, the movement of the VSV RdRp across an IGR may be independent of RNA polymerization. Both single and multi-subunit RNAPs are thought to locate their respective promoter sequences by diffusion, and it is possible that the VSV RdRp also uses this mechanism to find initiation signals. In contrast, attempts to construct a model to describe how the RdRp accesses upstream initiation sites is more problematical. Any such model must still be consistent with the large body of data that supports the currently favored stop-start model of sequential transcription, and in particular must be consistent with the UV-mapping data from which this model was largely derived (Abraham and Banerjee, 1976; Ball and White, 1976). Within these confines it is difficult to envisage RdRp access to upstream sites by a scanning (polymerizing) RdRp, as upstream movement ratcheted by RNA polymerization would require extension of the RNA strand in the 3’ to 5’ direction, which is unprecedented. In addition, access of upstream sites by diffusion presents another problem in that the RdRp will potentially collide with RdRps that are actively transcribing the same template in the 3' to 5' direction. The likelihood of polymerase collision would presumably depend on the distance between transcribing polymerases on the template, which would pose an upper limit on how far upstream the RdRp could migrate. The distance separating transcribing RdRps is not known, although recent evidence with several negative strand RNA viruses suggests their RdRps comprise oligomeric structures that could conceivably migrate along their corresponding RNA templates in oligomers of adjacent monomers (Sanchez and de la Torre, 2005; Smallwood, Cevik, and Moyer, 2002; Smallwood and Moyer, 2004). If the functional VSV RdRp also formed such oligomers, it is even more difficult to envisage how the RdRp could access upstream initiation sites by diffusion.
However, the concept of the RdRp oligomer allows another possible mechanism for how the RdRp can access upstream start sites. The leading polymerase may terminate transcription at a gene end, allowing a trailing polymerase to initiate at an upstream start site without having to reverse its direction of migration. Whereas this scenario requires the trailing RdRp to violate stop-start transcription, it is nevertheless consistent with the above-mentioned UV mapping data on which the stop-start model is based. The UV-mapping experiments showed that transcription of any of the 5 VSV genes depended on the prior transcription of all preceding genes, however the experiments were unable to address whether the same polymerase that terminated transcription subsequently initiated transcription of the next gene. Thus it is possible that termination of transcription by the leading polymerase allows a trailing polymerase to initiate transcription even though it was not itself responsible for the previous termination event.
The results of our study have highlighted that there are still large gaps in our understanding of NNS RNA virus RdRp function at the most fundamental level.
Acknowledgements
We thank past and present members of the G. W. Wertz laboratory for helpful discussions during the course of this study. In particular we thank Sean P. J. Whelan (Harvard Medical School, Boston, MA) for important contributions to experimental design. This work was supported by NIH grant AI012464 from the NIAID to GWW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham G, Banerjee AK. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A. 1976;73(5):1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LA, White CN. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A. 1976;73(2):442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120(2):183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Barr JN, Whelan SP, Wertz GW. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997a;71(11):8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Whelan SP, Wertz GW. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997b;71(3):1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A, Murphy BR, Collins PL. Respiratory syncytial virus can tolerate an intergenic sequence of at least 160 nucleotides with little effect on transcription or replication in vitro and in vivo. J Virol. 2000;74(23):11017–11026. doi: 10.1128/jvi.74.23.11017-11026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearns R, Collins PL. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73(1):388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358(1):241–254. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Harmon SB, Wertz GW. Transcriptional termination modulated by nucleotides outside the characterized gene end sequence of respiratory syncytial virus. Virology. 2002;300(2):304–315. doi: 10.1006/viro.2002.1541. [DOI] [PubMed] [Google Scholar]
- Hinzman EE, Barr JN, Wertz GW. Identification of an upstream sequence element required for vesicular stomatitis virus mRNA transcription. J Virol. 2002;76(15):7632–7641. doi: 10.1128/JVI.76.15.7632-7641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LN, Englund N, Pattnaik AK. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72(3):1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LE, Rose JK. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23(2):477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- Kuo L, Fearns R, Collins PL. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70(9):6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25(1):85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Pattnaik AK, Ball LA, LeGrone AW, Wertz GW. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69(6):1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- Plattet P, Strahle L, le Mercier P, Hausmann S, Garcin D, Kolakofsky D. Sendai virus RNA polymerase scanning for mRNA start sites at gene junctions. Virology. 2007;362(2):411–420. doi: 10.1016/j.virol.2006.12.033. [DOI] [PubMed] [Google Scholar]
- Sanchez AB, de la Torre JC. Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2005;79(11):7262–7268. doi: 10.1128/JVI.79.11.7262-7268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S, Cevik B, Moyer SA. Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology. 2002;304(2):235–245. doi: 10.1006/viro.2002.1720. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Moyer SA. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318(1):439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71(3):2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72(7):5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. Transcript initiation and 5'-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73(9):7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teninges D, Bras F, Dezelee S. Genome organization of the sigma rhabdovirus: six genes and a gene overlap. Virology. 1993;193(2):1018–1023. doi: 10.1006/viro.1993.1219. [DOI] [PubMed] [Google Scholar]
- Wang JT, McElvain LE, Whelan SP. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J Virol. 2007;81(20):11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz GW, Perepelitsa VP, Ball LA. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci U S A. 1998;95(7):3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J Virol. 2000;74(18):8268–8276. doi: 10.1128/jvi.74.18.8268-8276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]