Abstract
Early B cell factor (EBF) is a critical regulator of B lymphocyte-specific gene transcription. EBF functions, in part, by binding to regulatory sites of genes required for the pre-B- and mature B cell receptors. These DNA targets include the promoters of the mb-1 and Vpreb1 genes that encode Ig-α and one of the components of surrogate light chain, respectively. The biochemical basis of DNA binding and gene activation by EBF is poorly understood. The DNA-binding domain (DBD) of EBF includes a putative zinc-binding motif (HX3CX2CX5C), which we have designated the 'Zn-knuckle'. The Zn-knuckle is required for binding of the mb-1 promoter site in EMSA, but it has not been demonstrated to be important for functional activities of EBF in B cells. Therefore, we expressed EBF with mutations in the Zn-knuckle motif or flanking sequences in plasmacytoma cells in which activation of endogenous mb-1 and Vpreb1 genes is dependent on EBF. EBF with mutations that prevent zinc coordination by the Zn-knuckle did not activate transcription of either target gene. Other mutations affected the sequence preference of DNA binding and differentially inhibited activation of these genes. Our results demonstrate the importance of the Zn-knuckle motif in EBF. These experiments also confirm that EBF can re-activate multiple genes of the early B cell program in plasmacytoma cells, which provide a useful cell-based assay for dissecting mechanisms involving EBF.
Keywords: Early B cell Factor, EBF, mb-1 promoter, DNA-binding specificity, Vpreb1 promoter, zinc-binding motif
1. Introduction
The development of B cells from progenitor cells in the bone marrow is governed by the B cell-specific network of transcriptional regulators (reviewed in Hagman and Lukin, 2006; Medina and Singh, 2005). One of the most important proteins in this network is Early B cell Factor (EBF; also known as EBF1, Olfactory neuronal transcription factor-1 or Olf-1, O/E-1 and Collier-Olf-1-EBF-1 or COE1). EBF is essential for expression of a multitude of genes in the B cell-specific program. Gene targets of EBF include mb-1 (Ig-α; CD79a) and the Vpreb1 surrogate light chain gene (Gisler et al., 1999; Hagman et al., 1993; Hagman et al., 1991). EBF is also required for expression of other genes that are essential for B cell development and function, including B29 (Ig-β), CD19, λ5 and the B lineage commitment factor Pax5 (O'Riordan and Grosschedl, 1999; Åkerblad et al., 1999; Medina et al., 2004; Sigvardsson et al., 2002; Sigvardsson et al., 1997). In the absence of EBF, B cell development is arrested at an early pre-pro-B-like stage of differentiation (Lin and Grosschedl, 1995; Medina et al., 2004). In addition, EBF has been implicated in promoting the accessibility of immunoglobulin (Ig) gene segments for recombination (Goebel et al., 2001; Romanow et al., 2000). The arrest in development is due, in part, to the lack of V(D)J recombination in the absence of Recombination activating gene-1 (Rag1) and -2 (Rag2) expression.
EBF and closely related proteins (e.g. EBF2, EBF3, EBF4, Collier/Knot and Unc-3) constitute a novel transcription factor family (here, termed the EBF family; also referred to as the O/E or COE family). All members of this family possess a highly conserved DNA-binding domain (DBD) that is distinct from that of other known DNA-binding protein families (NCBI protein database). The DBD of murine EBF comprises residues 35–251 (Fig.1) (Hagman et al., 1993). Another conserved feature of EBF family proteins is their homodimerization in the absence of DNA. In addition to protein:protein interactions between EBF DBDs, homodimerization is a function of helix-loop-helix (HLH) domains similar to those of the basic-HLH proteins c-Myc and MyoD1 (Hagman et al., 1993; Wang and Reed, 1993). Domains in EBF that mediate transcriptional activation include a C-terminal serine/threonine/proline-rich domain and additional sequences between residues 18 and 429 (Hagman et al., 1995 and S. F., unpublished data).
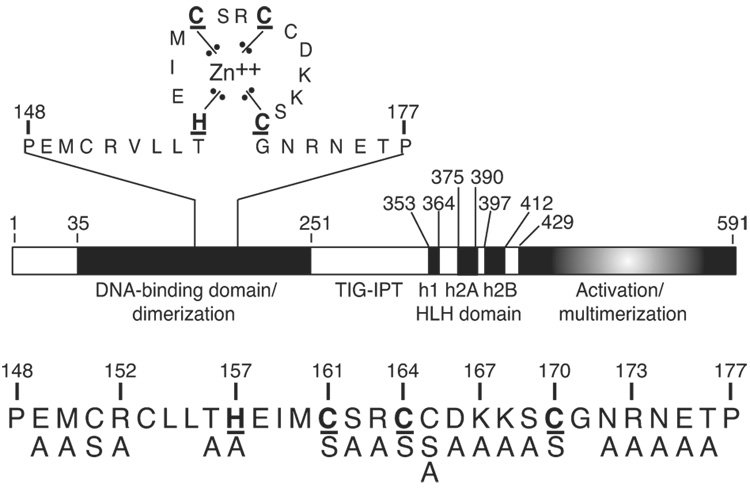
Fig. 1.
The domain structure of EBF. EBF consists of a DNA binding domain that includes the Zn-knuckle (expanded above), the TIG–IPT (Transcription factor Ig-like or Ig-like fold shared by Plexins and Transcription factors) domain, a non-basic HLH dimerization domain and a C-terminal activation domain. The underlined amino acids are proposed to coordinate zinc. The atypical HLH domain of EBF comprises three putative α-helices: h1, h2A and h2B. Mutations tested in these studies are indicated below. Mutations K167A/K168A were tested together.
EBF was first identified as a regulator of the B cell-specific mb-1 promoter ( Feldhaus et al., 1992; Hagman et al., 1991). EBF binds to sequences of the promoter between −180 and −160 that comprise an inverted repeat of 6 bp half-sites separated by 2 bp (5′-AGACTCaaGGGAAT-3′) (Travis et al., 1993). In vivo footprinting (dimethylsulfate protection/ligation-mediated PCR) and EMSA experiments detected the occupancy of EBF binding sites of mb-1 promoters in B lineage cell lines and ex vivo B cells that express mb-1 transcripts (Maier et al., 2004; Sigvardsson et al., 2002). These studies also identified complex interactions between EBF and other factors on the mb-1 promoter, including Runx1/CBFβ, E2A and Pax5. Here, Pax5 recruits Ets family partners to bind composite sites within the mb-1 promoter (Fitzsimmons et al., 1996; Fitzsimmons et al., 2001; Garvie et al., 2001; Wheat et al., 1999). The ability of Pax5 (together with its Ets partners) to activate endogenous mb-1 genes was demonstrated following enforced expression of Pax5 in terminally differentiated µM.2.21 plasmacytoma cells, which stably express the Pax5 paired domain (Maier et al., 2003b). The activation of mb-1 transcription in these cells by Pax5 is dependent on the epigenetic status of the mb-1 promoter (Maier et al., 2003a). However, these epigenetic barriers can be overcome by co-expressing Pax5 and EBF (the Pax5 paired domain is sufficient; Maier et al., 2003b). EBF facilitates mb-1 promoter activation in two ways: 1) EBF initiates demethylation of hypermethylated mb-1 promoters and 2) EBF activates chromatin remodeling, which results in enhanced accessibility of the mb-1 promoter to Pax5 (Maier et al., 2004). Based on these observations, it was proposed that EBF functions as a regulatory 'pioneer' factor by facilitating the binding of other factors necessary for transcriptional activation during early B cell differentiation (Hagman and Lukin, 2005).
Activation of the Vpreb1 gene, which encodes a surrogate light chain component of the pre-BCR, was previously shown to be dependent on EBF. Vpreb1 transcripts are absent in bone marrow B cell progenitors of ebf1−/− mice (Lin and Grosschedl, 1995). Endogenous Vpreb1 gene transcription was activated by the combination of EBF and E47 in human and mouse pro-B and non-lymphoid cell lines (Gisler and Sigvardsson, 2002; Sigvardsson et al., 1997). Activation of the human Vpreb1 promoter was attributed to three functional EBF binding sites (Gisler and Sigvardsson, 2002), which loosely fit a reported consensus site of EBF (5’CCCNNGGG; reviewed in Hagman and Lukin, 2006); however, EBF binding sites were not confirmed in the murine Vpreb1 promoter.
An unusual feature of EBF is a highly conserved sequence in its DNA-binding domain, HEIMCSRCCDKKSC (Hagman et al., 1995). This sequence includes closely spaced histidine and cysteine residues (underlined) similar to those found in known zinc-binding motifs (e.g. the NCp7 nucleocapsid protein of human immunodeficiency virus type I, CX2CX4HX4C; Morellet et al., 1992). The requirement for zinc for EBF DNA binding in biochemical experiments supports the hypothesis that the Zn-kncukle is a zinc-binding motif. DNA binding by purified recombinant EBF 1–429 was abolished following its denaturation in chaotropic agents (Hagman et al., 1995). DNA binding was restored following renaturation of denatured EBF only when renaturation was performed in the presence of zinc ions (or cadmium). To reflect the short length of this motif relative to other types of zinc-binding structures, we have designated this sequence the 'zinc knuckle' (Zn-knuckle) of EBF.
Prior to our current studies, we confirmed that residues involved in zinc coordination within the Zn-knuckle are required for EBF to bind the mb-1 promoter in vitro (Hagman et al., 1995); however, requirements for the Zn-knuckle and flanking residues were not studied in the context of gene transcription. Therefore, we determined the abilities of mutated EBF proteins to activate transcription of the mb-1 and Vpreb1 genes in plasmacytoma cells (558LµM) and measured DNA binding to sites of the mb-1 and Vpreb1 promoters in vitro. Our studies revealed requirements for both the Zn-knuckle motif and a subset of flanking residues for activation of the mb-1 and Vpreb1 promoters in the context of endogenous gene transcription. Interestingly, the abilities of EBF to bind and activate the mb-1 vs. Vpreb1 promoters were dependent on different sets of residues. Our studies presented here extend the understanding of DNA recognition and gene activation by Early B cell Factor.
2. Materials and Methods
2.1. Cell lines and cell culture
The µM.2.21 cell line, which was derived from the terminally differentiated mouse plasmacytoma cell line 558LµM, was described previously (Maier et al., 2003a). The cells were cultured in RPMI medium (GIBCO/Invitrogen) containing 10% FBS (Omega Scientific), 2mM L-glutamine, 50 µg/ml gentamycin, 1X HT media supplement (Hybri-Max; Sigma), 0.3 mg/ml xanthine, 1µg/ml mycophenolic acid and 10 µg/ml puromycin (Sigma). The Phoenix retroviral packaging cell line was cultured in IMDM medium containing 10% FBS, 2 mM L-glutamine and 50 µg/ml gentamycin.
All cell lines were grown at 37°C in 6% CO2.
2.2 Plasmids
All retrovirus constructs were derived using the MSCV2.2-IRES-GFPα vector (Hawley et al., 1994). The plasmid pCL-Eco was described previously (Naviaux et al., 1996). The H157A, C161S, R163A, C164S, K167A/K168A, C170S, N172A, R173A, N174A and E175A mutations were introduced into EBF previously (Hagman et al., 1995). To generate new mutations, the EBF cDNA clone pEBF17 (Hagman et al., 1993) was mutated using the following sense and antisense oligonucleotides (Integrated DNA Technologies): E149A, 5’GCAATGTGCCGAGTATTGCTCACACACGAGATCATG-3’ and 5’-TACTCGGCACATTGCAGGGTTCTTGTCTTGGCCTTC-3’; M150A, 5’-GCGTGCCGAGTATTGCTCACACACGAGATCATGTGC-3’ and 5’-CAATACTCGGCACGCTTCAGGGTTCTTGTCTTGGCC-3’; R152A 5’-GCTGTATTGCTCACACACGAGATCATGTGCAGC-3’ and 5’-TGTGAGCAATACAGCGCACATTTCAGGGTTCTTGTC-3’; T156A 5’-GCACACGAGATCATGTGCAGCCGCTGTTGTGACAAG-3’ and 5’-CATGATCTCGTGTGCGAGCAATACTCGGCACATTTC-3’; S162A, 5′-TTTGTCCACGCTAACTCCAAGCACGG-GCGGA-3′ and 5′-ACAGCGGGCGCACATGATCTCGTGTGTGAGCAA-3′; C165A, 5′-GCCGCTGTGCTGACAAGAAAAGCTGTGGCAACC-3′ and 5′-GCTTTTCTTGTCAGCACAGCGGCTGCACATGATCTCG-3′; D166A, 5′-TGTTGTGCCAAGAAAAGCTGTGGCAACCGA-3′ and 5′-GCTTTTCTTGGCACAACAGCGGCTGCACATGAT-3′; T176A 5’-GCTCCCTCAGATCCAGTGATAATTGACAGGTTCTTC -3’ and 5’-TGGATCTGAGGGAGCCTCATTTCGGTTGCCACAGCT-3’. Each mutated sense oligonucleotide was used in combination with the wild type antisense oligonucleotide 5′-AATCTGCCTGGTGTCCCTTTGC-3’. Each mutated antisense oligonucleotide was used in combination with the wild type sense oligonucleotide 5′-GTGGCCGGATCCACTACCGGCTCCAG-3′. All reactions employed Pfu DNA polymerase (Stratagene, La Jolla, CA). For each mutation, the two overlapping fragments were gel purified, combined and PCR amplified using the wild type sense and antisense oligonucleotides. Resulting fragments were digested with BamHI and XhoI, gel purified and ligated into the plasmid MSCV-FLAG-EBF(18–429)-GFP cut with BamHI and SalI. Previously generated mutations were similarly shuttled into this vector as BamHI-XhoI fragments. All mutated EBF constructs were sequenced.
For expression of recombinant murine EBF (18–429), we introduced the EBF sequence into pDW464, which adds a biotinylated N-terminal peptide to enable purification following over-expression in insect cells (Duffy et al., 1998). We first introduced a flexible linker sequence encoded by oligonucleotides 5’-AATTGTGGGTGCACTTACAGGAGCTGTCGACGAG-3’ and 5’-AATTCTCGTCGACAGCTCCTGTAAGTGCACCCAC-3’ into the EcoRI site of pDW464. The EBF(18–429) fragment was prepared by PCR amplification of pEBF17 using primers 5’-GATGTGCTCGAGCCGCTGGGCAGCGGCATGAA-3’ and 5’-AACCTAGAGCTCAGACCGAAGTGTTAGCAAGGGC-3’ and cut with XhoI and Ecl136II for insertion into modified pDW464, which was cut with SalI and Ecl136II for ligation with the EBF fragment. The resulting plasmid, pDW464/EBF(18–429) was sequenced.
2.3 Production of retroviruses, infection and flow cytometry
Production of ecotropic retroviruses and infection of cells were performed as described previously (Maier et al., 2003a; Maier et al., 2004). Four days after infection, µM.2.21 cells were stained with goat anti-IgM antibodies conjugated with phycoerythrin (anti-IgM:PE; Caltag Laboratories) for detection of membrane IgM (mIgM). Labeled cells and GFP were detected using a FACScalibur™ flow cytometer (BD Biosciences) and data was analyzed using FloJo™ software (Tree Star, Inc.). Virally infected cells were sorted using a MoFlo™ cell sorter (Cytomation, Inc.). Statistical significance was assessed using Student’s t test.
2.5 Quantitative PCR
Total cellular RNA was isolated from sorted GFP+ cells (>90% purity) using the PicoPure™ RNA Isolation Kit (Arcturus). Preparation of cDNA and quantitative real-time PCR (qPCR) for detection of mb-1 and control β-actin transcripts using SYBR Green (Applied Biosystems) was performed as described previously (Maier et al., 2004). Primers for qPCR detection of Vpreb1 transcripts were 5’-GCTCATGCTGCTGGCCTATC-3’ and 5’-TCCAAGGGAAGAAGATGCTAATG-3’. Statistical significance was assessed using Student’s t test.
2.6 Over-expression and purification of recombinant EBF(18–429)
Recombinant murine EBF(18–429) was overexpressed in Spodoptera frugiperda cells and purified similar to the previously reported production of androgen receptor (Juzumiene et al., 2005). Plasmid pDW464/EBF(18–429) was transformed into DH10Bac E. coli for transposition into bacmids using the Bac-to-Bac® system (Invitrogen, Madison, WI). Sf9 cells were transfected with bacmids as described to make recombinant baculovirus. Hi5 cells were infected with amplified baculovirus at M.O.I.=2 in spinner flasks. At two days post-infection, cells were washed with ice cold PBS and whole cell lysates were prepared by lysing cells in 50mM Tris HCl, pH 8.0/10mM 2-mercaptoethanol/100mM KCl/1% Nonidet P-40/0.5% protease inhibitor cocktail set III (Calbiochem, La Jolla, CA)/1µM ZnOAc/50mM NaF/50mM β-glycerophosphate. Lysates were clarified by centrifugation at 20,000 rpm in a Beckman J2-21M centrifuge for 30 min at 4°C. For purification, a BioRad (Hercules, CA) poly-prep column was packed with 900 µl of 50% monomeric Softlink Avidin beads (Promega, Madison, WI), pre-equilibrated with 0.1M NaPO4, pH 7.0, and pre-absorbed with 5mM biotin in the same buffer. To denature and renature avidin, the column was sequentially washed with eight volumes 10% HOAc, eight volumes of 0.1M NaPO4 (pH 7.0) and two volumes 50mM Tris-HCl (pH 8.0). Lysate was absorbed onto the column and washed with 20 volumes buffer A (20mM Tris HCl, pH 8.0/10mM 2-mercaptoethanol/100mM KCl/10% glycerol/1µM ZnOAc) and 20 volumes buffer B (20mM Tris HCl, pH 8.0/10mM 2-mercaptoethanol/1M KCl/10% glycerol/1µM ZnOAc). The column was washed with five volumes of elution buffer without biotin (50mM Tris HCl, pH 8.0/200mM KCl/10% glycerol/1µM ZnOAc/4mM DTT) and eluted in 500µl fractions of elution buffer + 5mM biotin. Protein concentrations were determined using the BioRad Protein assay and purity was established (>95%) using SDS-PAGE. Aliquots were stored at −80°C.
2.7 Cell extracts, immunoblotting and EMSA
To generate whole cell extracts (WCE), retrovirally infected GFP+ cells were harvested and washed twice with cold 1X PBS. The cells were lysed in buffer C (20mM HEPES, pH 7.9/0.4M NaCl/1.5mM MgCl2/0.5% Nonidet-P40/1mM dithiothreitol/10µg/ml of leupeptin/10µg/ml aprotinin/1mM phenylmethlysulfonyl fluoride). Twenty µg of extracts were resolved on a 10% Tris-HCl SDS-polyacrylamide gel. Retrovirally expressed, FLAG-tagged proteins were transferred to membranes and probed with rabbit polyclonal anti-FLAG peptide antibody (affinity purified rabbit anti-FLAG; Rockland Immunochemicals, Inc.). As a loading control, blots were also probed with a rabbit polyclonal anti-Sp3 antibody (Santa Cruz Biotechnology). Binding of primary antibodies was detected using IRDye™-700DX–conjugated donkey-anti-rabbit IgG [H and L] secondary antibody (Rockland Immunochemicals, Inc.) and membranes were analyzed using an Odyssey™ infrared imaging system (LI-COR). Oligonucleotide probe sequences, annealing and labeling of DNA probes and EMSA using the mb-1 promoter EBF site probe were performed as described previously (Maier et al., 2004). The sequences of the oligonucleotides annealed to make the murine Vpreb1 site probes were: site 2, 5’-AGTTGGCTCAGCCTCTCAAGGGGAGTTGAGGTCAC-3’ and 5’-AGTTGTGACCTCAACTCCCCTTGAGAGGCTGAGCC-3’ and site 3, 5’-AGTTGTCACTCTCCCTCAGGGAGACAAGCCCGCAG-3’ and 5’-AGTTCTGCGGGCTTGTCTCCCTGAGGGAGAGTGAC. To estimate apparent KD’s of mutant EBF proteins, EMSA was performed using constant amounts of WCE’s as described above and excess unlabeled double-stranded oligonucleotides as indicated. Data were quantitated using a Typhoon 9200 PhosphorImager system (Molecular Dynamics). Plots and estimates of apparent KD’s were produced using GraphPad Prism 4.0b (GraphPad Software, Inc., San Diego, CA).
3. Results
3.1. The Zinc knuckle motif is required for activation of endogenous mb-1 genes in µM.2.21 cells
To determine effects of alterations in EBF on its functions in intact cells, we used retroviruses to express EBF containing mutations in our plasmacytoma cell system and evaluated the abilities of the mutated proteins to activate the transcription of endogenous gene targets. EBF(18–429) was used in these experiments because it reproduces essential functions of full length EBF(1–591), but is expressed more stably over the course of time. The first seventeen amino acids of EBF, which were shown previously to be dispensable for its function (Hagman et al., 1995), were replaced with the twelve–residue FLAG epitope tag to enable detection of virally expressed EBF in cell extracts by Western blotting. The retroviruses also expressed enhanced GFP to estimate the frequency of infection.
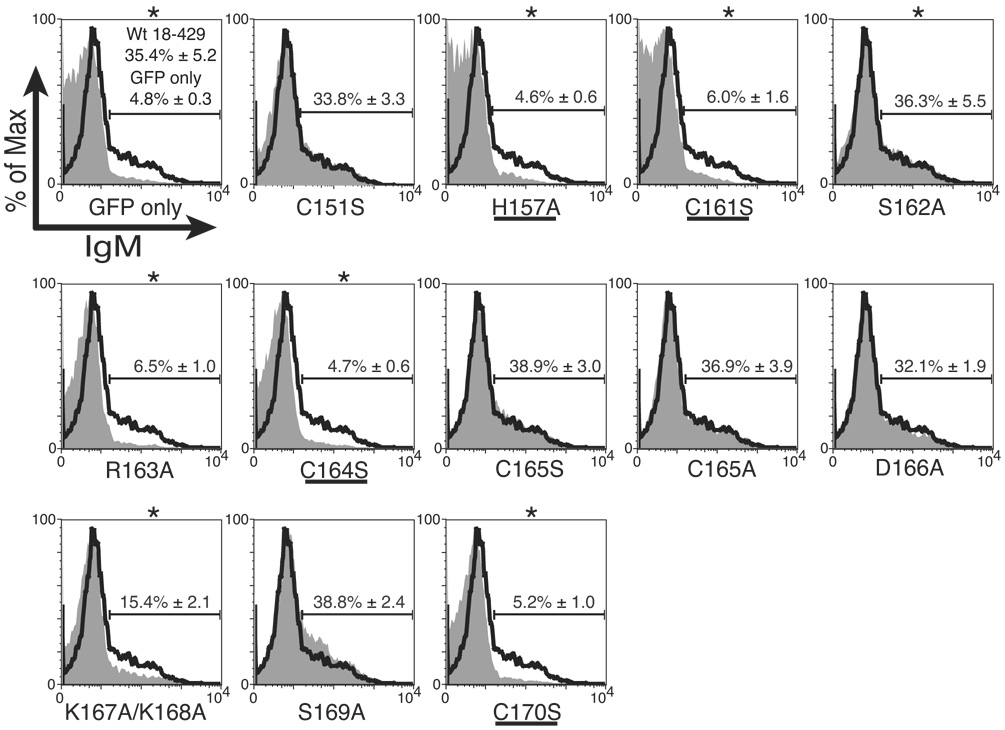
µM.2.21 cells, a subclone of 558LµM cells (Hombach et al., 1988), stably express the Pax5 DBD (1–149) fused with the nuclear localization signal sequence of SV40 large T antigen, but do not express endogenous EBF (Maier et al., 2004). Retrovirally expressed EBF activates mb-1 transcription in µM.2.21 cells, which results in the display of mIgM on the cell surface. To determine requirements for mb-1 promoter activation by EBF, µM.2.21 cells were infected with retroviruses expressing control GFP alone or GFP together with wild type or mutant (Fig.1) EBF(18–429). Four days after infection, the cells were stained with anti-IgM antibodies and analyzed by flow cytometry. Cell surface IgM (mIgM) expression was determined for GFP+ populations only (generally 40–50% of total cells). Fig.2A shows histograms depicting representative patterns of mIgM expression (mIgM+ cells were gated as shown) in GFP+ cells. In each panel, mIgM levels on cells expressing mutated EBF or control GFP alone (grey shaded areas) were compared with wild type EBF (solid black line). Approximately 30–40% of the cells consistently expressed mIgM on the cell surface in response to enforced wild type EBF(18–429) expression. In contrast, the reduced level of mIgM on cells expressing mutated EBF demonstrated the importance of select residues. To examine requirements for putative metal coordinating residues within the Zn-knuckle motif, we replaced the putative zinc-coordinating histidine (H157) with the neutral amino acid alanine. We also replaced each of the three putative coordinating cysteine residues with serine, which maintains biochemical properties of cysteine, but cannot coordinate metal ions. Mutation of each of these Zn coordination residues (H157A, C161S, C164S and C170S) resulted in nearly complete absence of mIgM relative to wild type EBF. Each of these observations was highly significant (p ≤ 0.008). In contrast, mutations of other nearby cysteines that are not suspected to coordinate zinc (C151S, C165S or C165A) stimulated mIgM expression similarly to that observed with wild type EBF. The nearly complete loss of function following replacements of putative coordinating or ligand residues is similar to the loss of function observed following similar replacements in conventional zinc finger proteins (reviewed in Wolfe et al., 2000).
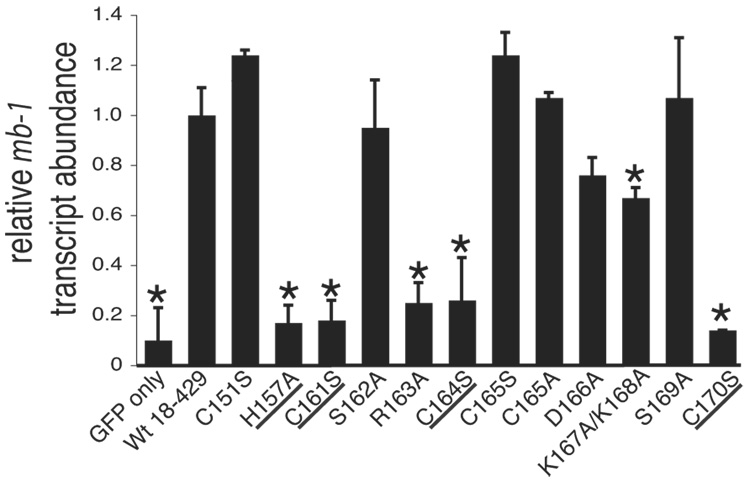
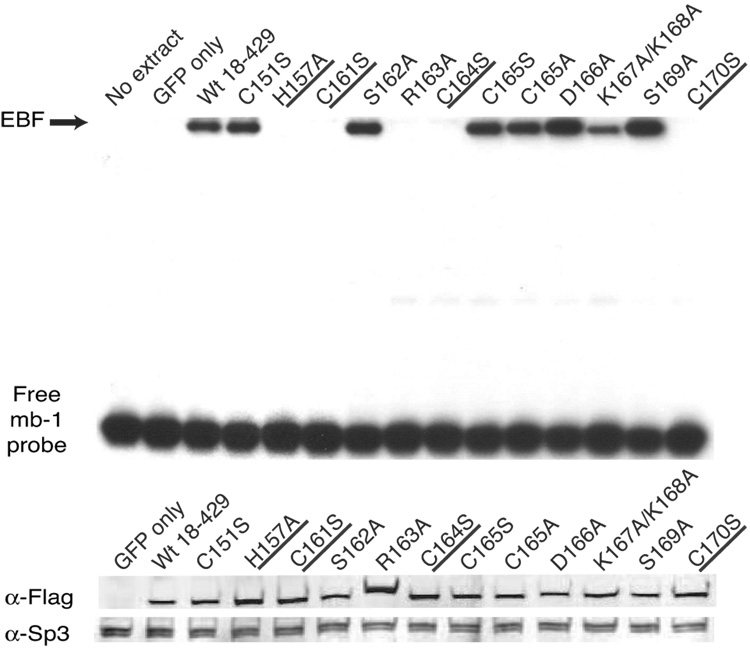
Fig. 2.
Analysis of mb-1 gene expression and EBF DNA binding: Zn knuckle mutations. (A) Representative mIgM expression on µM.2.21 cells retrovirally infected to express empty vector/mutant EBF. Histograms indicate the percentage of GFP+ cells that display mIgM (% of max versus mIgM expression). Mean detection of mIgM+ cells (gated relative to GFP only controls) of three independent experiments is indicated in each box. Mutations that disrupt zinc binding are underlined. In this and subsequent figures, flow data marked with an * indicate a Student’s t test of p ≤ 0.05. (B) Quantitative real-time PCR (qPCR) of mb-1 mRNA isolated from infected GFP+ µM.2.21 cells infected to express wild type or mutated EBF proteins. Data include mean +/− s.d. of three independent experiments. All values were normalized to endogenous β-actin mRNA expression. Amounts of mb-1 transcripts from cells expressing empty vector were set to one for these experiments. (C) Binding of the mb-1 probe by wild type and mutated EBF proteins in nuclear extracts of infected cells. Top: EMSA was performed using the previously reported murine mb-1 promoter EBF site (mb-1) probe (Maier et al., 2004). Bottom: Western blotting of FLAG-tagged EBF and endogenous Sp3 (loading control) expressed in µM.2.21 cells.
Additional mutations revealed the importance of other residues within the Zn knuckle. Expression of EBF with the R163A substitution resulted in only background numbers of mIgM+ cells. These data demonstrate that R163 is essential for activating mb-1 gene transcription, although it is not predicted to participate in zinc coordination. In contrast, activity of EBF with mutation of the adjacent residue, S162A, was similar to that of wild type EBF. Introduction of the D166A mutation did not significantly reduce EBF function. Mutation of the adjacent lysines K167A and K168A resulted in less than half the activity of wild type EBF (p ≤ 0.02). Mutation of the adjacent residue (S169A) resulted in activity that was elevated slightly when compared to wild type EBF.
To further quantitate effects of these mutations, we measured the relative abundance of mb-1 transcripts. GFP+ cell populations were sorted and mb-1 mRNA transcripts were detected using quantitative real-time PCR (qPCR). For comparisons between samples, we normalized amounts of mb-1 transcripts to β-actin transcripts. In general, amounts of mb-1 transcripts (Fig.2B) correlated well with the detection of mIgM by flow cytometry (Table I). As expected, EBF with mutations in the putative zinc coordinating residues of the Zn-knuckle motif did not induce mb-1 transcript levels significantly above that induced by empty vector. The R163A EBF mutant also activated mb-1 transcription only weakly. Reduced activity was obtained with EBF containing the D166A or K167A/K168A mutations. Other mutations (C151S, S162A, C165S, C165A and S169A) had only minor effects on mb-1 transcription. These results demonstrate the importance of residues within the Zn-knuckle motif for activating mb-1 gene transcription in B cells.
Table 1.
Summary of mIgM expression and qPCR results for mb-1 and Vpreb1 expression.
| mb-1 |
Vpreb1 |
||
|---|---|---|---|
| mIgM | qPCR | qPC | |
| Wild type | +++ | +++ | +++ |
| E149A | +++ | ++ | ++++ |
| M150A | ++ | ++ | +++ |
| C151S | +++ | +++ | +++ |
| R152A | +/− | + | + |
| T1566A | ++ | ++ | + |
| H157A | − | − | − |
| C161S | − | − | − |
| S162A | +++ | +++ | +++ |
| R163A | − | − | + |
| C164A | − | − | − |
| C165S | +++ | +++ | +++ |
| C165A | +++ | +++ | +++ |
| D166A | +++ | +++ | +++ |
| K167A/K168A | +/− | ++ | − |
| S169A | +++ | +++ | ++++ |
| C170S | − | − | − |
| N172A | +/− | + | + |
| R173A | +++ | +++ | ++ |
| N174A | ++ | ++ | + |
| E175A | ++ | +++ | ++++ |
| T176A | +++ | +++ | ++ |
| GFP only | − | − | − |
Scores for mIgM expression are as follows: (+++)>30%, (++) 29% to 25%, (+) 24% to 20%, (+/−) 19% to 15%, and (−) <10%. Wild type is set to +++ (mIgM expression of > 30%). Scores for mb-1 and Vpreb1 qPCR expression are as follows: (++++) > 1, (+++) 1 to 0.75, (++) 0.74 to 0.50, (+) 0.49 to 0.25, (−) <0.24. Wild type is set to 1 (+++).
To determine whether mutated EBF proteins expressed in µM.2.21 cells can bind to the mb-1 promoter, we performed an electrophoretic mobility shift assay (EMSA) using whole cell extract (WCE) proteins isolated from purified GFP+ cells (Fig.2C). For each EBF test protein, EMSA results are shown from one representative WCE (one of the three cell populations tested in Fig.2B). Equal amounts of extract were incubated with a 32P-labeled double-stranded mb-1 promoter probe (mb-1 probe) prior to electrophoresis on a non-denaturing polyacrylamide gel. Immunoblotting of WCEs demonstrated the presence of similar amounts of EBF and of the nuclear protein Sp3, indicating the loading of equivalent total protein (Fig.2C).
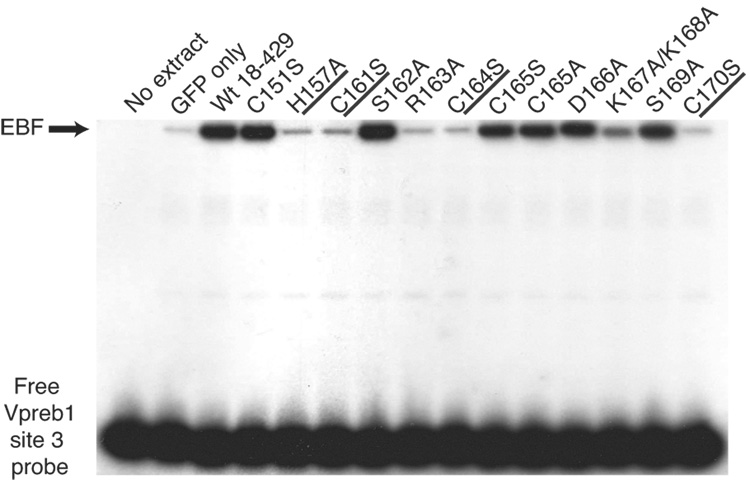
As expected from the lack of mb-1 gene activation in µM.2.21 cells containing EBF H157A, C161S, R163A, C164S or C170S, these EBF proteins failed to bind the mb-1 probe significantly in vitro. In parallel with the activation of mb-1 genes in the cells, EBF with the K167A/K168A mutations bound DNA approximately 60% as well as wild type EBF. These results confirm that DNA binding by EBF with mutations in the putative zinc coordination motif and the R163A mutation is impaired, resulting in the failure of these proteins to activate the mb-1 promoter in plasmacytoma cells.
3.2. Amino acid requirements for activation of the Vpreb1 promoter by EBF in plasmacytoma cells
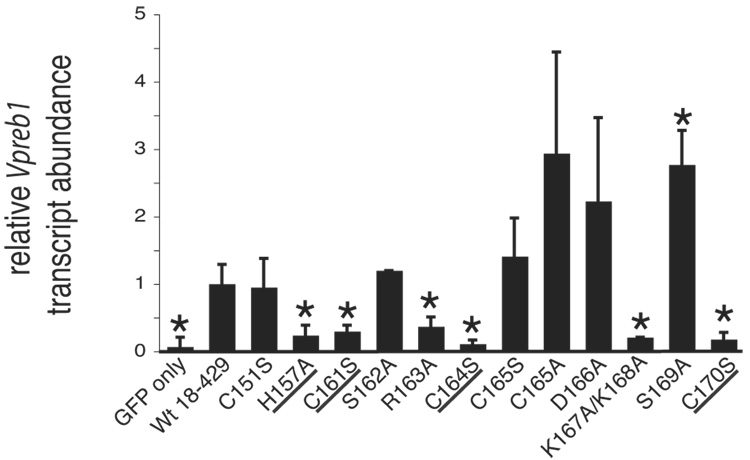
The question arose as to whether EBF activates B cell-specific genes in addition to mb-1 in plasmacytoma cells. Previous studies demonstrated that the surrogate light chain genes λ5 and Vpreb1 are activated by EBF when it is ectopically expressed (together with endogenous E47) in non-lymphoid cells (Sigvardsson et al., 1997). Therefore, we examined whether enforced expression of EBF activates surrogate light chain gene transcription in µM.2.21 cells, which express E47. λ5 transcription was not activated significantly by EBF(18–429) in µM.2.21 cells (data not shown); however, activation of the Vpreb1 gene by wild type EBF (Fig.3A) was robust (>13-fold relative to ‘GFP only’ control). Therefore, to compare relative requirements for the activation of Vpreb1 and mb-1 transcription by EBF, we analyzed the activation of Vpreb1 transcription by wild type and mutated EBF.
Fig. 3.
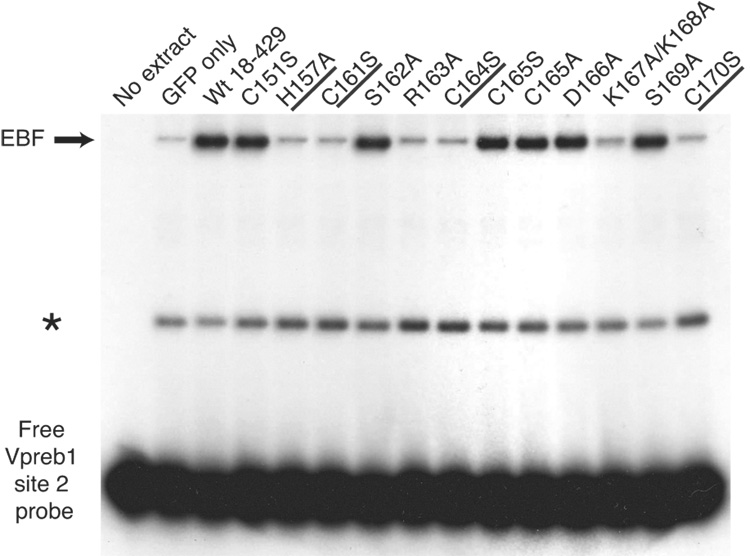
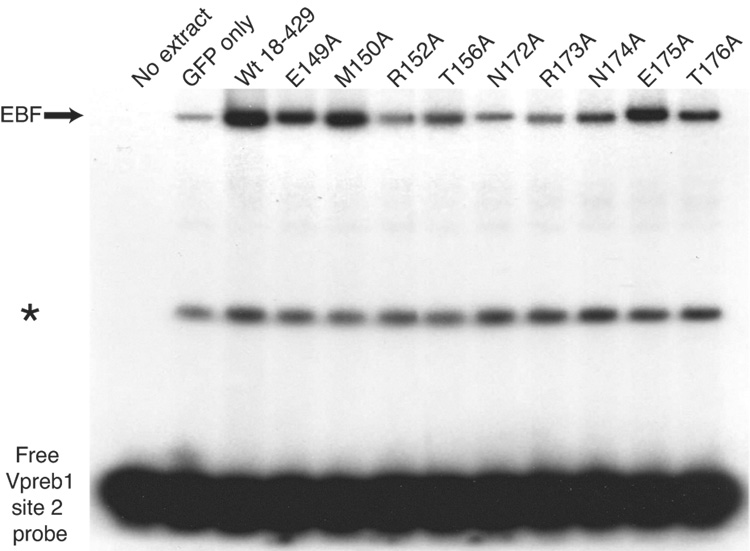
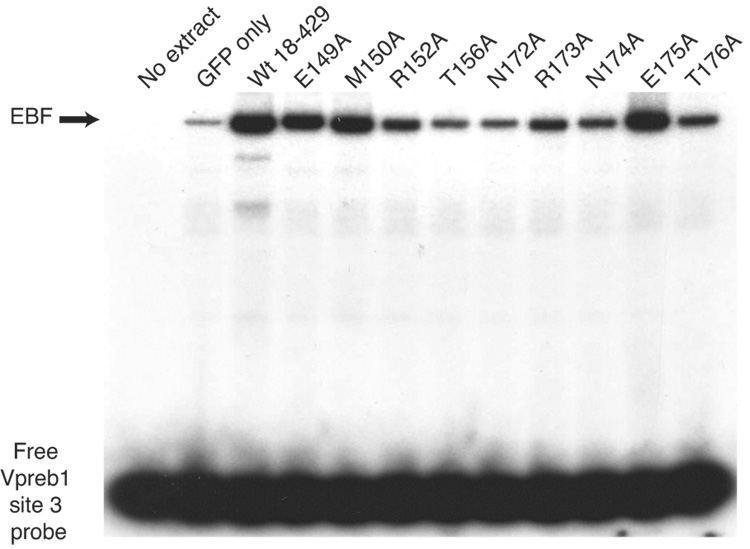
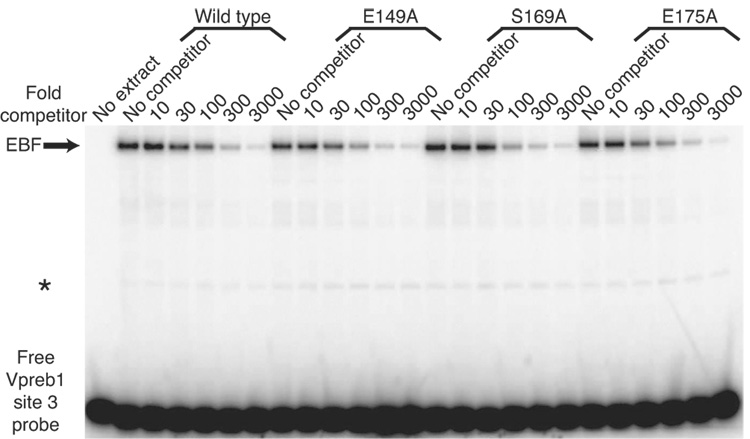
Quantitation of Vpreb1 transcripts and DNA binding of the Vpreb1 promoter by wild type and mutated EBF: Zn knuckle mutations. (A) qPCR of Vpreb1 mRNA isolated from µM.2.21 cells infected to express wild type and mutated EBF proteins. Data include mean +/− s.d of three independent experiments. Amounts of Vpreb1 expression from cells expressing wild type EBF were set to one for this experiment (n=3). (B–C) Binding of Vpreb1 site 2 (B) or site 3 (C) by recombinant EBF(18–429). Recombinant EBF (18–429) was incubated with the 32P-labeled Vpreb1 site 2 probe and site 3 (at 3nM or 1.2nM, respectively) in the presence or absence of increasing amounts of Vpreb1 or control mb-1 competitors as shown. (D–E) EMSA of binding of the murine Vpreb1 site 2 (D) or site 3 (E) probe by whole cell extracts of cells infected to express wild type or mutated EBF proteins. *Endogenous complex unrelated to EBF. In (D) and (E), DNA binding and EMSA were performed exactly as in Fig.2C.
Similar to activation of the mb-1 gene by EBF, mutations of the putative zinc coordinating residues greatly reduced the activation of the Vpreb1 gene. Thus, the zinc knuckle motif is required for binding of EBF to different promoter sites. EBF with the R163A mutation also failed to activate Vpreb1 gene expression. In contrast, EBF mutants with the C151S, S162A or C165S mutations were able to activate Vpreb1 transcription in a similar manner to activation with wild type EBF. Interestingly, activation of Vpreb1 transcription by EBF with the S169A mutations was enhanced (2.8-fold) relative to wild type EBF. Thus, the S169A mutation significantly (p ≤ 0.03) increases the ability of EBF to activate Vpreb1, but not mb-1 transcription. Another notable difference between the mb-1 and Vpreb1 genes was illustrated by the inability of EBF with the K167A/K168A mutations to activate Vpreb1 transcription. These residues are more critical for activating Vpreb1 (79% decrease relative to wild type EBF) than mb-1 transcription (33% decrease).
3.3 EBF binding to the murine Vpreb1 promoter
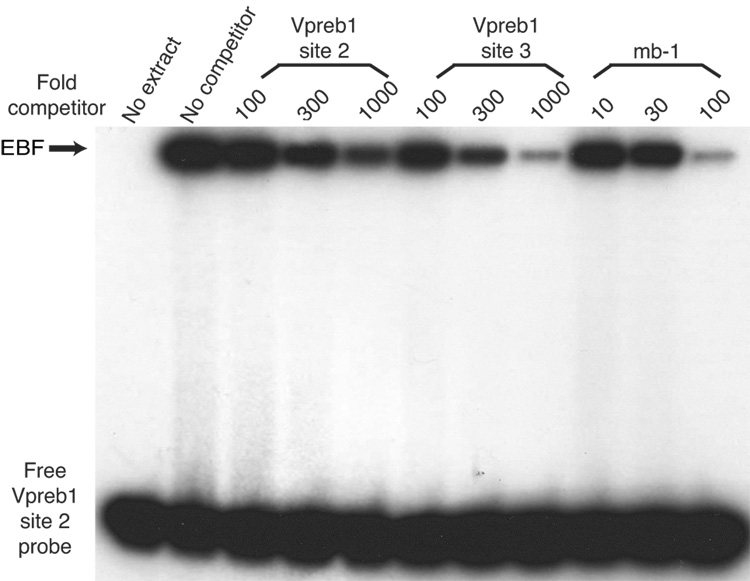
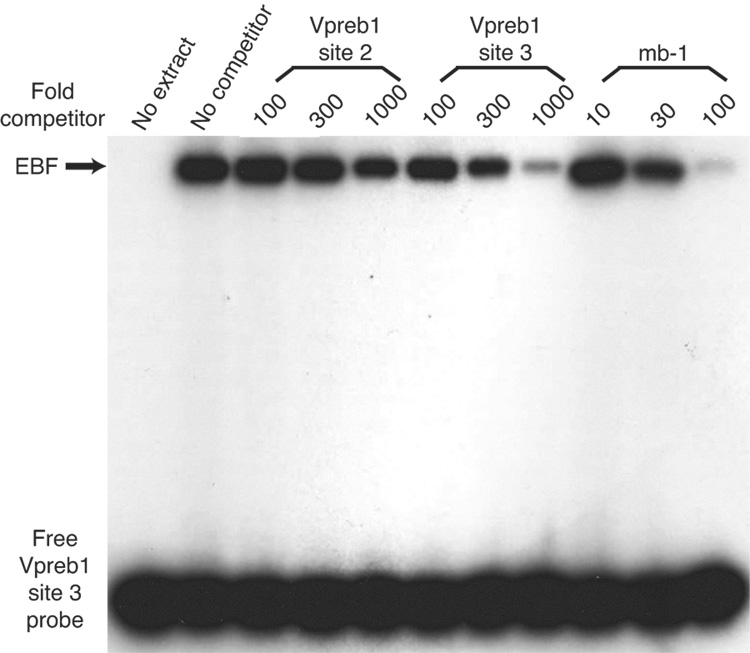
Although the human Vpreb1 promoter includes three independent EBF binding sites, only two of these sites are conserved with similar spacing between the human and mouse Vpreb1 promoters (Gisler and Sigvardsson, 2002). A sequence (5’-CCTCTCaaGGGGAG-3’) within the mouse promoter possesses significant identity (underlined) with human promoter site 2 (5’-TTCCTCagGGGGAA) and to experimentally optimized EBF recognition sites (5’-ATTCCCnnGGGAAT-3’; Travis et al., 1993). The murine promoter also includes a sequence (5’-CTCTCCctGAGGGA-3’) that is conserved relative to the sequence of human Vpreb1 site 3 (5’-CGACCCctGAGGTA-3’). To confirm that these sequences are bona fide EBF binding sites, we measured EBF binding to these sites or to the high affinity EBF site of the mb-1 promoter in a competitive DNA binding assay. The binding of highly purified recombinant EBF(18–429) was measured quantitatively (using a Phosphorimager) to probes comprising site 2 or site 3 of the murine Vpreb1 promoter in the absence or presence of increasing amounts of unlabeled competitors (Figs.3B–C). Each of the sites bound EBF specifically, but the site 2 and site 3 sequences competed approximately one tenth and one third as well as the mb-1 probe sequence, respectively. Interestingly, the individual Vpreb1 binding sites have different affinities for EBF because the site 3 competitor nearly abolished binding to either probe while the box 2 competitor competed only modestly for EBF binding. We conclude that EBF binds the three sites with different relative affinities (mb-1>site3>site2).
To determine whether differences in abilities to bind the two Vpreb1 sites account for the relative activation of transcription by the mutant proteins, we measured DNA binding of mutated EBF proteins to labeled probes comprising the Vpreb1 promoter site 2 (Fig.3D) or site 3 (Fig.3E). Excellent concordance was noted between the binding of EBF proteins to the probes and functional activities in µM.2.21 cells (Table I). EBF with the H157A, C161S, R163A, C164S or C170S mutations did not bind either probe appreciably. In explanation of the lack of activation by EBF K167A/K168A, binding of this mutant protein to either probe was greatly diminished.
3.4. Effects of mutations in Zn-knuckle-proximal residues on mb-1 promoter activation by EBF
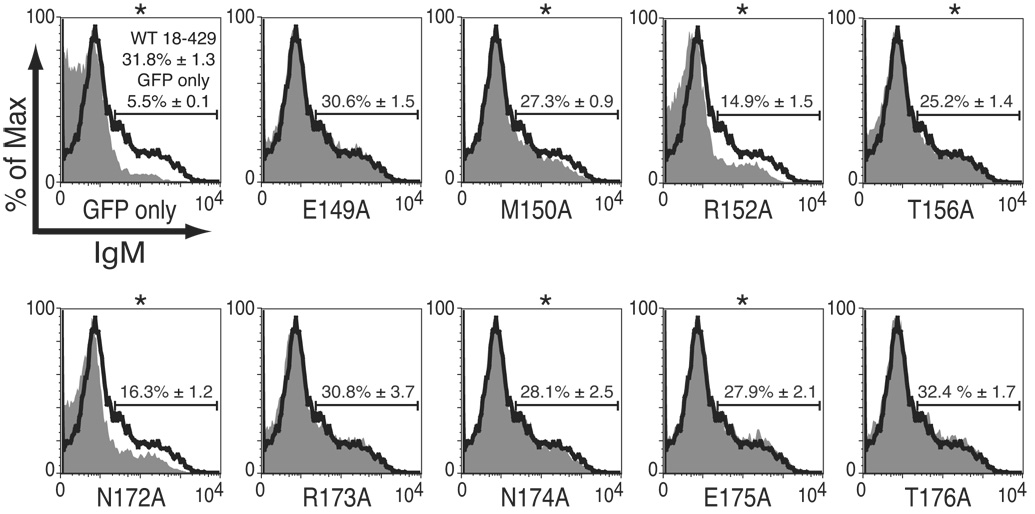
DNA recognition by zinc finger motifs involves contacts with DNA by residues between and adjacent to the zinc coordinating residues (reviewed in Wolfe et al., 2000). To determine whether residues flanking the Zn-knuckle of EBF are functionally important, we introduced point mutations into the sequences immediately N-terminal to H157 (residues 149 to 156) and C-terminal to C171 (amino acids 172–176) of EBF and tested their properties in µM.2.21 cells. Mutations inserted N-terminal to the Zn-knuckle had variable effects on the cell surface expression of mIgM (Fig.4A). The M150A, N174A, and E175A reduced the frequency of mIgM+ cells significantly relative to wild type EBF (by as much as 20%). The R152A and N172A mutations each reduced mIgM+ cells by more than half, suggesting the importance of these residues for EBF function.
Fig. 4.
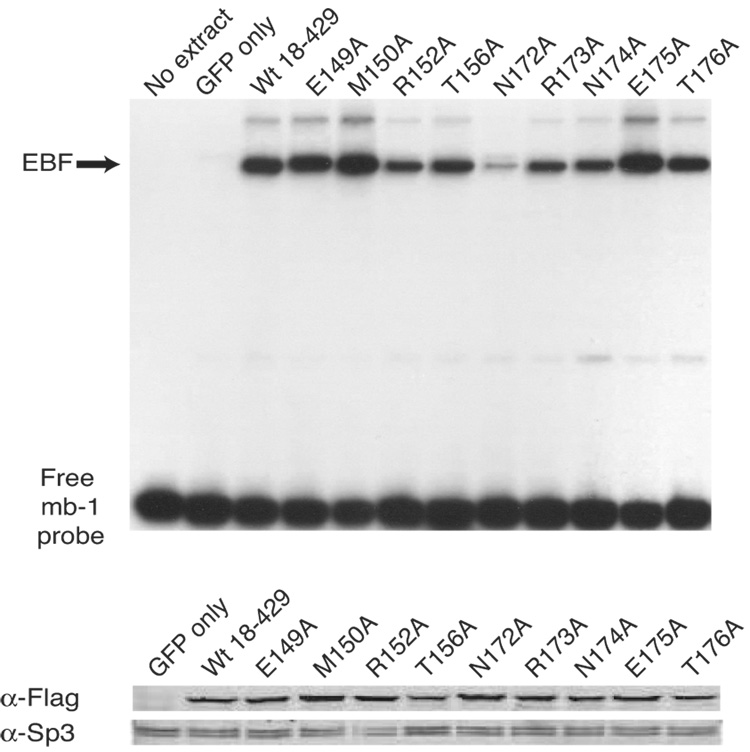
Analysis of mb-1 gene expression and EBF DNA binding: mutations of flanking residues. (A) mIgM expression on retrovirally infected βM.2.21 cells. Presentation is identical with Fig.2A. Mean activation of mIgM expression (relative to GFP only control) from three independent experiments is indicated in each box. (B) qPCR of mb-1 mRNA isolated from µM.2.21 cells infected to express wild type or mutated EBF in (A). Other aspects of these experiments (n = 3) are similar to Fig.2B. (C) EBF DNA binding. Top: Binding of the mb-1 probe by wild type and mutated EBF. Bottom: Western blotting of FLAG-tagged EBF or endogenous Sp3 (loading control) expressed in µM.2.21 cells. Other aspects of these experiments are similar to Fig.2C.
The data suggest that residues flanking the Zn-knuckle are important for mb-1 promoter activation by EBF. Therefore, similar to our analysis of other residues in EBF we measured the effects of mutations on mb-1 transcripts (Fig.4B) and binding of EBF to the mb-1 promoter probe (Fig.4C). Notably, effects of the R152A and N172A mutations mirrored their reduced abilities to generate mIgM+ cells (Fig.4A). These mutations also greatly reduced binding of the mb-1 probe by EBF.
3.5. Effects of mutations of Zn-knuckle-proximal residues on Vpreb1 activation by EBF
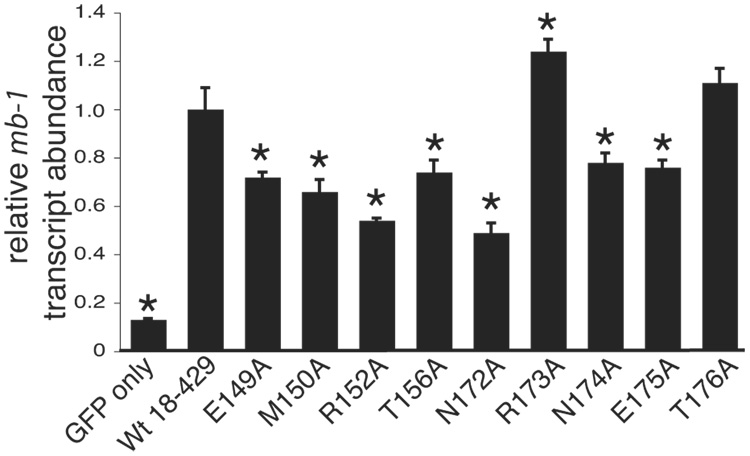
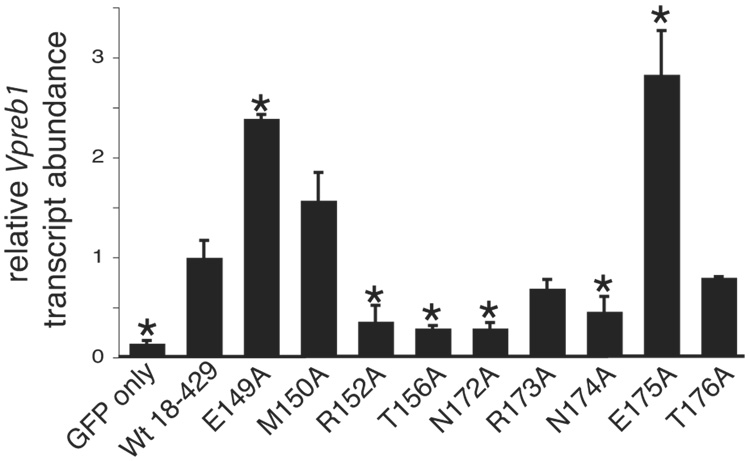
The activation of Vpreb1 genes was also selectively reduced by mutations in the regions flanking the Zn knuckle (Fig.5A). The R152A, T156A, N172A and N174A mutations each reduced Vpreb1 transcripts to less than half the level of transcripts produced in response to wild type EBF (all with significance p ≤ 0.04). The E149A and E175A mutations increased Vpreb1 transcription by 2.4-fold (p ≤ 0.004) or 2.8-fold (p ≤ 0.05), respectively, relative to wild type EBF. These results were reflected by the strength of DNA binding to sites 2 and site 3 by the mutated EBF proteins (Figs.5B–C). Interestingly, mutations including R152A, T156A, N172A and N174A had significantly greater effects on the activation of Vpreb1 transcription relative to mb-1 transcription. E149A and E175A enhanced activation of Vpreb1 transcription by more than 2-fold, but they had little effect if any on the expression of mb-1 transcripts (compare with Fig.4B).
Fig. 5.
Quantitation of Vpreb1 transcripts and DNA binding of the Vpreb1 promoter by wild type and mutated EBF: mutations of flanking residues. (A) qPCR of Vpreb1 mRNA isolated from µM.2.21 cells infected to express wild type or mutated EBF. Other aspects of these experiments (n = 3) are similar to Fig.3A. (B–C) EMSA of binding of the murine Vpreb1 site 2 (B) or site 3 (C) probe by wild type and mutated EBF proteins. *Endogenous complex unrelated to EBF. All other aspects of these experiments were performed as in Fig.3D–E.
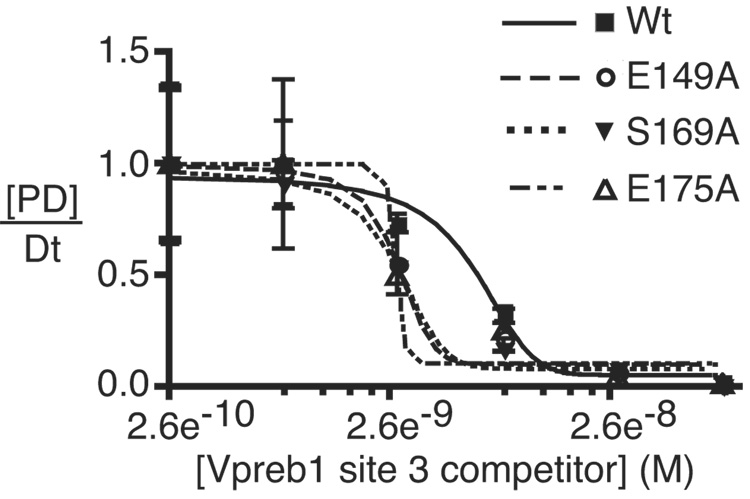
3.6 Mutations selectively enhance binding of EBF to Vpreb1 site 3
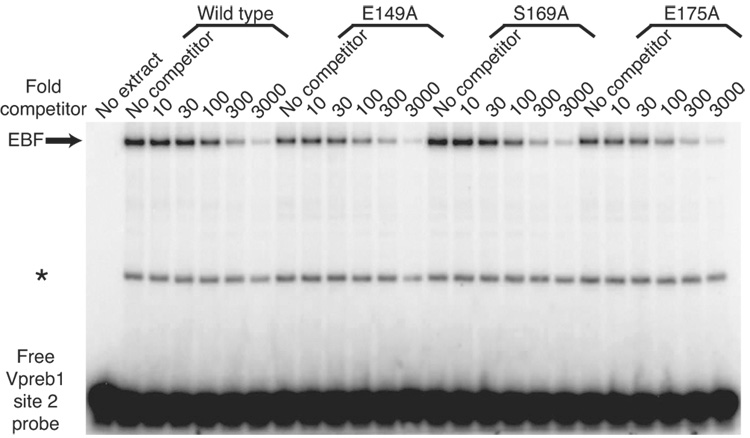
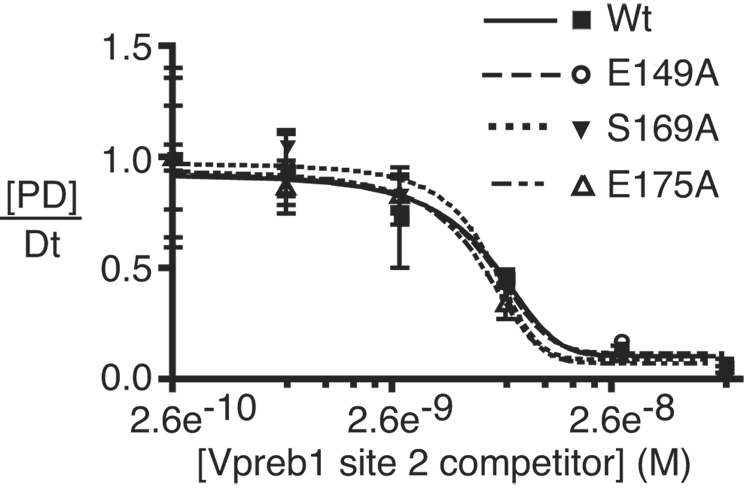
To confirm that the increased activation of Vpreb1 genes by EBF with the E149A, S169A and E175A mutations reflects their increased affinities for Vpreb1 promoter sites, we measured their binding to Vpreb1 box 2 or 3 probes in the presence of excess unlabeled competitor DNAs (Fig.6A–B). No significant differences were observed between the binding of EBF wild type, E149A, S169A and E175A to the Vpreb1 box 2 probe (Fig.6C; apparent KD’s all ≅ 7 nM). In contrast, enhanced binding of the three mutant EBF proteins to Vpreb1 box 3 was observed (Fig.6D; wild type, KD = 6.3 nM; E149A, S169A, E175A, KD‘s all ≅ 2.8 nM). These data support the hypothesis that EBF utilizes different sets of residues, or adopts different conformations, to bind different nucleotide sequences.
Fig. 6.
Enhanced DNA binding by mutant EBF proteins. (A–B) EMSA of EBF binding in WCE of infected cells. A representative EMSA is shown using Vpreb1 site 2 (A) or site 3 (B) probes, WCE of wild type or mutant EBF and unlabeled competitors as shown. Other aspects of these experiments are identical with Figs.3B–C. (C–D) Apparent KD’s of EBF wild type, E149A, S169A or E175A binding to Vpreb1 promoter sites 2 (C) or 3 (D) were calculated using EMSA data from A and B. Bound and free DNA were quantitated using a Typhoon 9200 PhosphorImager system (Molecular Dynamics) and plotted as the concentration of competitor DNA versus the fraction of DNA in complexes with protein [PD]/Dt. For each curve, KD‘s are estimated (from three independent experiments) as the concentration of competitor resulting in 50% inhibition of DNA binding.
4. Discussion
EBF is essential for B cell development. It stimulates the transcription of multiple genes of the B lineage-specific program and is essential for the expression of proteins that assemble the pre-B- and mature B cell-receptors (Hagman and Lukin, 2005). In addition to its functional significance, EBF is a novel type of DNA binding protein. Due to the lack of structural information, very little is understood concerning how EBF binds DNA and activates transcription. Chromatin remodeling activities of EBF were described only recently (Maier et al., 2004). A better understanding of these mechanisms is essential for determining how EBF functions in the B cell-specific regulatory network (Medina et al., 2004; Medina and Singh, 2005).
To better understand how transcription factors activate gene transcription in B cells, we developed an assay that measures the activation of endogenous mb-1 genes in plasmacytoma cells. In previous studies we used this assay to identify functionally important residues in Pax5, which activated mb-1 genes in 558LµM plasmacytoma cells (Maier et al., 2003b). However, in a subset of these cells, Pax5 was unable to activate mb-1 transcription by itself due to the epigenetic state of mb-1 promoter chromatin (Maier et al., 2003a). We concluded that the transcription of mb-1 genes in this context requires additional proteins. Further studies suggested that one of these proteins is EBF (Maier et al., 2004). The assignment of this function to EBF is logical. The onset of EBF expression, which precedes that of Pax5 during early B lymphopoiesis, parallels the progressive demethylation of mb-1 promoters. Moreover, the mb-1 gene fails to become hypomethylated in B cell progenitors from EBF-deficient mice. EBF can initiate CpG demethylation of mb-1 genes in plasmacytoma cells. Furthermore, EBF enhances chromatin accessibility of mb-1 promoters, which, in turn, facilitates synergy between EBF and Pax5. Thus, mb-1 gene activation is dependent on a hierarchy of transcription factors in which EBF acts upstream of Pax5.
In our current studies we introduced mutations into EBF and measured mb-1 and Vpreb1 gene activation. Transactivation of these genes in plasmacytoma cells expressing the Pax5 DBD was robust in response to wild type EBF. Notably, endogenous Ebf1 and Pax5 genes in µM.2.21 cells were not stimulated by enforced expression of EBF and the Pax5 DBD (S.F., data not shown); therefore, the observed gene activation was due to the ectopically expressed factors. Mutations predicted to reduce zinc binding by EBF (H157A, C161S, C164S and C170S) each reduced mb-1 and Vpreb1 transcripts to nearly background levels. These mutations revealed the importance of the Zn-knuckle in functional DNA binding by EBF. R163 is also essential for DNA binding. Other residues, including R152, T156, K167/K168 and N172 are important for binding and activating the mb-1 promoter, but mutations of these residues had stronger effects on binding and activation of the Vpreb1 promoter. R173A significantly reduced activation of Vpreb1 transcription, but it had only minor effects on mb-1 transcription. Although residues including lysines 167 and 168 are not predicted to coordinate zinc, they may be essential for maintaining the Zn-knuckle in the proper orientation for DNA binding or for contacting DNA directly. Interestingly, E149A and E175A had opposing effects on mb-1 (decreased) vs. Vpreb1 (increased) transcription. We conclude that EBF utilizes different sets of amino acid side chains for binding different DNA sequences. It is likely that this flexibility of DNA recognition contributes to the degenerate DNA-binding specificity of EBF.
The Zn-knuckle and surrounding residues between prolines 148 and 177 of EBF are conserved between EBF family proteins of diverse species (Altschul et al., 1997; Schäffer et al., 2001). The four zinc coordination residues and flanking residues are perfectly conserved between EBF (EBF1), EBF3 and other EBF orthologs and paralogs of many vertebrates including mice and humans. Identical sequences are present in EBF proteins of more distant species including sea urchins (S. purpuratus) and flour beetles (T. castaneum). Nearly perfect matches are also notable between murine EBF and the ortholog Collier/Knot proteins of insects. Collier/Knot regulates cell fates during development of head structures of D. melanogaster (Crozatier et al., 1996). Intriguingly, Collier/Knot also regulates the innate immune system in flies (Crozatier et al., 2004; Crozatier et al., 1996). Vertebrate EBF2 and the Unc-3 protein of C. elegans feature only conservative changes, including variation of an aspartic acid (D166 of EBF) to glutamic acid. Zn-knuckles within vertebrate EBF4 proteins exhibit conservative changes of a single lysine (K168 of EBF) to arginine. Interestingly, one cysteine (similar to C151 of EBF) in the Zn-knuckle region varies extensively in EBF-like proteins. Our studies demonstrated that C151 is not required for functions of murine EBF. Overall, the very high degree of conservation of the Zn-knuckle suggests its importance for functional activities of EBF family proteins in organisms as diverse as humans, fish and insects.
The configuration of metal coordinating residues in the EBF Zn-knuckle, HX3CX2CX5C, is highly unusual. The most common zinc fingers are similar to repeated elements in the TFIIIA transcription factor and conform to the conserved sequence Y/FXCX2–5CX3Y/FX5LX2HX3–5H. Zinc finger motifs in nuclear hormone receptors have a different configuration of cysteines, CX2CX13CX2C (C2C2 zinc fingers). Other zinc binding motifs include the C6-zinc clusters (as in Gal4 in S. cerevisiae) and the CHCC motifs of retroviral gag proteins. Only two other groups of proteins possess a configuration of zinc coordinating residues similar to the HCCC motif of EBF. Highly conserved proteins that regulate sex determination in metazoans possess a cyteine-rich DNA-binding domain, termed the Doublesex–MAB-3 (D–M) domain (Lints and Emmons, 2002; Volff et al., 2003). The D–M motif comprises intertwined zinc-binding structures featuring CCHC and HCCC configurations of metal coordinating residues (Zhu et al., 2000). Similar to EBF, D–M proteins exhibit zinc-dependent DNA binding. HCCC motifs have also been reported in the CREB Binding Protein (CBP)/p300 co-activators (De Guzman et al., 2000; De Guzman et al., 2005). CBP and p300 possess three tandem HCCC motifs (TAZ1–3) that are required for interactions with other proteins (e.g. Hypoxia-Inducible transcription Factor-1α). The three-dimensional structures of these domains (TAZ1 and/or TAZ2) have been determined to include zinc ions. It is not known whether the Zn-knuckle of EBF adopts a fold similar to that of TAZ1 and TAZ2. However, it is notable that the Zn-knuckle of EBF is similar in overall length (fourteen residues) to TAZ2 (sixteen residues). It is not known whether, similar to the TAZ motifs, the Zn-knuckle of EBF mediates protein:protein interactions in addition to its DNA binding functions.
In conclusion, our data demonstrate that the Zn-knuckle of EBF is important for DNA recognition and transcriptional regulation of early B cell-specific genes. However, it is currently unknown whether Zn-knuckles strictly function as: 1) DNA recognition motifs, 2) scaffolding of the DNA-binding domain and/or 3) for recruitment of accessory proteins, such as chromatin remodeling complexes necessary for the ‘pioneer’ activity of EBF. Further studies, including determination of the three-dimensional structure of EBF, are important for resolving these questions.
Acknowledgements
The authors would like to thank Kara Lukin and Yosef Refaeli for their helpful comments. The authors wish to thank members of the Flow Cytometry Laboratory at National Jewish for their excellent technical assistance and Philippa Marrack and Inder Verma for providing plasmids. We also thank Donald P. McDonnell, John Kappler and Francis Crawford for assistance with generating recombinant EBF. This work was supported by National Health Service Grants R01 AI54661, R01 AI56322 and P01 AI22295 and by the Rocky Mountain Chapter of the Arthritis Foundation. S. F. was generously supported by post-doctoral training grant 5 T32 AI007405.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Åkerblad P, Rosberg M, Leanderson T, Sigvardsson M. The B29 (immunoglobulin β-chain) gene is a genetic target for early B cell factor. Mol. Cell. Biol. 1999;19:392–401. doi: 10.1128/mcb.19.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:2289–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M, Ubeda J-M, Vincent A, Meister M. Cellular immune response to parasitization in Drosophila requires the EBF orthologue Collier. PLOS Biology. 2004;2:1107–1113. doi: 10.1371/journal.pbio.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Liu HY, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP. J. Mol. Biol. 2000;303:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry. 2005;44:490–497. doi: 10.1021/bi048161t. [DOI] [PubMed] [Google Scholar]
- Duffy S, Tsao K-L, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. anal. Biochem. 1998;262:122–131. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- Feldhaus A, Mbangkollo D, Arvin K, Klug C, Singh H. BlyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol. Cell. Biol. 1992;12:1126–1133. doi: 10.1128/mcb.12.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons D, Lutz R, Wheat W, Chamberlin HM, Hagman J. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucl. Acids Res. 2001;29:4154–4165. doi: 10.1093/nar/29.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax-5 complex formation on DNA. Molec. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- Gisler R, Åkerblad P, Sigvardsson M. A human early B-cell factor-like protein participates in the regulation of the human CD19 promoter. Mol. Immunol. 1999;36:1067–1077. doi: 10.1016/s0161-5890(99)00092-9. [DOI] [PubMed] [Google Scholar]
- Gisler R, Sigvardsson M. The human V-preB promoter is a target for coordinated activation by early B cell factor and E47. J. Immunol. 2002;168:5130–5138. doi: 10.4049/jimmunol.168.10.5130. [DOI] [PubMed] [Google Scholar]
- Goebel P, Janney N, Valenzuela JR, Romanow WJ, Murre C, Feeney AJ. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 2001;194:645–656. doi: 10.1084/jem.194.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of Early B-cell Factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Early B cell Factor ‘pioneers’ the way to B cell development. Trends Immunol. 2005;26:455–461. doi: 10.1016/j.it.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Hagman J, Travis A, Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 1991;10:3409–3417. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- Hombach J, Sablitzky F, Rajewsky K, Reth M. Transfected plasmacytoma cells do not transport the membrane form of IgM to the cell surface. J. Exp. Med. 1988;167:652–657. doi: 10.1084/jem.167.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juzumiene D, Chang C-y, Fan D, Hartney T, Norris JD, McDonnell DP. Single-step purification of full-length human andogen receptor. Nuclear Receptor Signaling. 2005;3:1–5. doi: 10.1621/nrs.03001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lints R, Emmons SW. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 2002;16:2390–2402. doi: 10.1101/gad.1012602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B cell-specific mb-1 (Ig-α) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol. Cell. Biol. 2003a;23:1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immun. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Parenti S, Fitzsimmons D, Abraham LJ, Garvie CW, Hagman J. Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-α gene transcription by Pax-5 (BSAP) Nucl. Acids Res. 2003b;31:5483–5489. doi: 10.1093/nar/gkg785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Pongubala JMR, Reddy KL, Lancki DW, DeKoter RP, Kieslinger M, Grosschedl R, Singh H. Defining a regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Medina KL, Singh H. Genetic networks that regulate B lymphopoiesis. Curr. Opin. Hemat. 2005;12:203–209. doi: 10.1097/01.moh.0000160735.67596.a0. [DOI] [PubMed] [Google Scholar]
- Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high titer recombinant retroviruses. J. Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero ILM, van Dongen JJM, Feeney AJ, Murre C. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucl. Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Åkerblad P, Breslin T, Bilke S, Liu Y-F, Yeamans C, Zhang G, Hagman J. Early B cell Factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol. Cell. Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- Travis A, Hagman J, Hwang L, Grosschedl R. Purification of Early-B-cell Factor and characterization of its DNA-binding specificity. Mol. Cell. Biol. 1993;13:3392–3400. doi: 10.1128/mcb.13.6.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Zarkower D, Bardwell VJ, Schartl M. Evolutionary dynamics of the DM domain gene family in metazoans. J. Mol. Evol. 2003;57 Suppl 1:S241–S249. doi: 10.1007/s00239-003-0033-0. [DOI] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor OLF-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Wheat W, Fitzsimmons D, Lennox H, Krautkramer SR, Gentile LN, McIntosh LP, Hagman J. The highly conserved β-hairpin of the paired DNA-binding domain is required for the assembly of Pax:Ets ternary complexes. Mol. Cell. Biol. 1999;19:2231–2241. doi: 10.1128/mcb.19.3.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;3:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]