Abstract
Mycobacterium avium subsp. paratuberculosis causes paratuberculosis, a chronic granulomatous enteritis. Detecting animals with paratuberculosis infections is difficult because the currently available tools have low sensitivity and lack specificity; these tools are prone to generating spurious positive test results caused by exposure to environmental M. avium complex organisms. To generate candidate antigens for incorporation into a specific test for paratuberculosis, subspecies-specific proteins were determined by proteomic comparison of M. avium subsp. paratuberculosis and M. avium subsp. avium. Analysis was aimed at revealing proteins only expressed (or predominant) in the protein profile of M. avium subspecies paratuberculosis. Two-dimensional gel electrophoresis resolved approximately 1,000 protein spots from each subspecies. Proteome analysis identified protein spots whose expression profile appeared markedly increased in M. avium subsp. paratuberculosis, and 32 were identified by analysis of their tryptic peptide profile by matrix-assisted laser desorption ionization-time of flight analysis. Thirty of these proteins were cloned, and their recombinant proteins were expressed. Ovine paratuberculosis sera were used to assess their immunoreactivity by enzyme-linked immunosorbent assay (ELISA), Western blotting, and dot blot analysis. Seventeen proteins were detected in at least one of the immunoassays, and eleven proteins were detected by ELISA with an optical density in excess of the cutoff of 0.1 in four of six sera tested. The immunoreactivity of these proteins indicates their potential as unique diagnostic antigens for the development of a specific serological detection of paratuberculosis.
Mycobacterium avium subspecies paratuberculosis is a pathogen of domestic and wild ruminants, causing paratuberculosis or Johne's disease. Paratuberculosis, a chronic granulomatous enteritis, is characterized by severe emaciation and ultimately death. It is believed that animals are infected at an early stage in their life through the fecal-oral route, but clinical symptoms only become apparent much later. In the intervening period, these animals that appear healthy can be shedding vast quantities of M. avium subsp. paratuberculosis in their feces, allowing subliminal intraherd transmission.
Paratuberculosis places a severe economic burden on the global livestock industry and impacts herd productivity heavily. The disease is endemic within the United States and the United Kingdom. A study carried out by U.S. Department of Agriculture, “The Dairy '96 Study” (18), estimated that at least 22% of U.S. dairies were infected with paratuberculosis. In the United Kingdom the prevalence of this disease is unknown in sheep and goat flocks, but it has been estimated that 17% of dairy herds are infected in England (9), although this value may not reflect the current status.
Control initiatives advocate the use of management techniques to avoid exposure of vulnerable animals to paratuberculosis. More aggressive control measures, such as the removal of high-shedding animals, the major source of infective material, is desirable and may be necessary to break the cycle of infection. Once achieved, protecting disease-free status by monitoring purchased animals to detect early-stage paratuberculosis infections before introduction into the herd is as yet only aspirational, because available diagnostic tools have limitations.
Serological tests, or those that rely on fecal culture can only confirm clinical paratuberculosis and may detect less than one-third of all infected cattle (12, 25); hence, these tests fail to agree in the diagnosis of an individual animal (1). It has been suggested that serological tests fail to detect preclinical animals because the humoral response does not develop until the onset of fecal shedding (16); this may be true, but it is also a problem of sensitivity (12). Sensitive and specific tests are required for the detection of early infections to eliminate animals that have the potential to shed and transmit disease to uninfected livestock. Recently, the EVELISA (13) has been developed; this test is able to reliably detect antibodies in the sera of low-shedding animals and uses antigens extracted from the surface of M. avium subsp. paratuberculosis. Classical serological tests and the EVELISA rely on mixtures of undefined M. avium subsp. paratuberculosis antigens, and thus their specificity is in question. There is a high degree of genetic similarity between members of the M. avium complex (MAC), which makes it difficult to differentiate between bona fide paratuberculosis cases and spurious positive test results caused by exposure to shared antigens of environmental MAC organisms. The improved specificity of the enzyme-linked immunosorbent assay (ELISA) by preabsorption of test sera with M. phlei antigens has been achieved (7, 11), but this has a negative impact on the ELISA response of the test (7), which inevitably affects sensitivity.
Construction of an ELISA using antigens from MAC subspecies-specific proteins would be a clear improvement. The drive to produce specific diagnostic reagents has focused work to identify polymorphic genomic regions unique to M. avium subsp. paratuberculosis K10 strain (3, 4, 21). However, the ability of proteins encoded in these regions to act as diagnostic reagents has been brought into question. Many of the long sequence polymorphisms unique to paratuberculosis described by Semret et al. (23) have characteristics of mobile genetic elements, and their presence has been found to be variable within the circulating population of strains, leading these researchers to conclude that “… in silico identifications of a species-specific sequence in a prototype genome does not guarantee utility as a diagnostic marker.”
Here we report the detection of M. avium subsp. paratuberculosis-specific proteins obtained by comparison of its proteome to that of M. avium subsp. avium. Although these organisms share more than 95% identity at the sequence level (5), the protein profile they express has marked differences. We have identified 32 proteins whose expression profile is markedly increased in M. avium subsp. paratuberculosis compared to M. avium subsp. avium when they are grown in Middlebrook 7H9 broth culture to either exponential or stationary phase. Where possible, these proteins were cloned and expressed in Escherichia coli and purified by affinity chromatography. Their ability to be recognized by the sera of sheep naturally infected with M. avium subsp. paratuberculosis is assessed here as an indication of their potential as unique diagnostic antigens in the development of a specific serological method for detecting paratuberculosis.
MATERIALS AND METHODS
Diagnosis of paratuberculosis.
Animals with suspected clinical paratuberculosis were euthanized with an intravenous injection of 100 mg of pentobarbital/kg. Approximately 30 cm of terminal ileum was removed postmortem and flushed with water to remove contaminating debris. Small sections of the tissue were removed for bacteriological and histopathological examination to confirm the diagnosis and determine whether the multibacillary or paucibacillary form was present. For histopathology, tissues were fixed in 10% (vol/vol) Formol-saline for a minimum of 24 h, trimmed, dehydrated through graded alcohols, and embedded in paraffin wax. Sections 5 μm thick were cut and stained with hematoxylin and eosin. Serial sections were stained by the Ziehl-Neelsen method.
Preparation of sera from sheep.
Blood samples from sheep with multibacillary paratuberculosis (n = 6) or healthy controls (n = 3) were obtained by venipuncture into a BD Vacutainer serum tube and allowed to clot at 4°C overnight. The blood was then centrifuged 2,000 × g for 15 min at 4°C. The serum was removed by aspiration and again centrifuged at 13,000 × g for 5 min at room temperature. The serum was stored at −80°C until required.
Source of microbial strains and in vitro growth of mycobacteria.
M. avium subsp. avium strain (JD88/118) was originally isolated from a deer and is well characterized with regard to its IS901-positive and mycobactin-independent status. Since deer and sheep share pasture in Scotland, this strain is likely to be representative of the environmental organisms encountered by sheep (although they rarely show clinical symptoms, making it difficult to find ovine M. avium subsp. avium isolates). M. avium subsp. paratuberculosis strain (JD88/107) was originally isolated from a deer with clinical paratuberculosis. It has a type II pulsed-field profile, which is commonly observed in strains isolated from clinical cases of ovine paratuberculosis in Scotland (our unpublished data) and is mycobactin dependent and IS900 positive.
Routine propagation and maintenance of mycobacteria were carried out by using modified Middlebrook 7H11 medium composed of 2.1% (wt/vol) 7H11 agar (Becton Dickinson, Oxford, United Kingdom), 2.5% (vol/vol) glycerol, 2.3 mM l-asparagine supplemented with 2 μg of mycobactin J (Allied Monitor, Fayette, MO) ml−1, 10% (vol/vol) Middlebrook oleic acid-albumin-dextrose-catalase enrichment supplement (OADC; Becton Dickinson), Selectatabs (two per liter; Mast Laboratories, Merseyside, United Kingdom), and 20% (vol/vol) heat-inactivated newborn calf serum (Invitrogen, Ltd., Paisley, United Kingdom). For propagating mycobacteria for proteome analysis, starter cultures were initiated by inoculating a loopful of mycobacteria (from a modified 7H11 slope with confluent growth) into a 10-ml volume of Middlebrook 7H9 medium supplemented with 10% (vol/vol) Middlebrook OADC, 0.1% (wt/vol) Tween 80, 2 μg of mycobactin J ml−1, and 2.5% (vol/vol) glycerol. The cultures, contained in 50-ml flasks, were stirred continuously using magnetic stirrer bars on a multiple-position low-heat transmission magnetic stirrer. After 48 h of incubation, 3 ml of the starter culture was transferred to 300 ml of prewarmed medium in liter flasks. The cultures were incubated at 37°C with continuous stirring.
The optical density at 600 nm (OD600) was measured at appropriate time points throughout the growth of the 300-ml liquid cultures. Small aliquots of the culture (0.2 ml) were removed aseptically, and the ODs were ascertained. The ODs of the liquid cultures were plotted against time. It was possible to observe three phases of growth: (i) the initial lag phase with little change in OD with respect to time; (ii) the exponential phase, in which the OD increased linearly with respect to time, generally peaking at an OD of 2.5 to 3; and (iii) the stationary phase, in which the growth rate slowed and the OD of the culture declined.
When cultures were deemed to have reached either exponential or stationary phase and achieved an OD of at least 1, they were assessed for contamination by Ziehl-Neelsen staining and light microscopy. Aliquots (50 ml) were removed aseptically, cooled on ice, and then centrifuged (4,000 × g, 30 min, 4°C). The supernatant was decanted, and the cell pellet was carefully resuspended in 25 ml of ice-cold phosphate-buffered saline (PBS). The cell suspension was then centrifuged again (4,000 × g, 30 min, 4°C), the supernatant was removed, and the pellet was stored at −20°C. Cell pellets were stored for no longer than 1 week before further processing for proteomic analyses.
Protein preparation for two-dimensional (2-D) electrophoresis.
Protein extraction was performed essentially as described previously (14). Briefly, cells (0.2 to 0.3 g [wet weight]) were suspended in 2% (wt/vol) sodium dodecyl sulfate (SDS), 0.04 M Tris, and 0.06 M dithiothreitol. The suspension was layered onto washed 0.1 mm zirconium-silica beads (Biospec Products, Bartlesville, OK). The mixture was lysed six times by using a Fastprep 120 (Qbiogene, Cambridge, United Kingdom) at 6.5 beats/s with 1 min of cooling on ice between each round. The resulting suspension was centrifuged (500 × g, 30 s, room temperature) to reduce frothing. The supernatant was pipetted into microcentrifuge tubes and heated to 100°C for 5 min. It was then rapidly cooled and centrifuged (21,000 × g, 20 min, at room temperature). The protein extract was stored at −70°C until required.
Protein clean-up was performed as described in the protocol accompanying a PlusOne 2-D Clean-Up kit (GE Healthcare, Bucks, United Kingdom). The pellet obtained was prepared for isoelectric focusing (IEF) as described previously (14). Soluble protein extracts were used immediately or stored at −70°C.
The concentration of protein extracts was determined by using a PlusOne Quant kit (GE Healthcare) and was performed essentially as described in the protocol accompanying the product.
Resolution of proteins, visualization, and proteomic analysis.
IEF of proteins (150 to 200 μg for silver-stained gels or 300 to 500 μg for colloidal Coomassie brilliant blue [CBB]-stained gels) was performed as described previously (14). Strips were either stored at −70°C until required or equilibrated prior to electrophoresis in the second dimension.
IEF strips were incubated in SDS equilibration buffer (0.05 M Tris, 6 M urea, 0.065 M dithiothreitol, 30% [vol/vol] glycerol, 2% [wt/vol] SDS, and 0.002% [wt/vol] bromophenol blue) for 15 min with gentle shaking, followed by a similar incubation in a fresh aliquot of equilibrium buffer containing iodoacetamide (0.135 M). SDS-polyacrylamide gel electrophoresis (PAGE) gels (26 by 20 cm, 12.5% [wt/vol]; GE Healthcare) were electrophoresed by using an Ettan Dalt system (GE Healthcare) at 1 W per gel overnight at 15°C. The power was then increased to 2 to 4 W per gel until the dye front was within 1 cm of the bottom of the gel.
Gels were stained with either silver using the method described by Morrissey (17), except that the glutaraldehyde fix (step 2) was omitted, or with colloidal CBB using the method described by Neuhoff et al. (20).
Scanned gel images were obtained by using a Umax PowerLook III Imagescanner coupled with Imagemaster Labscan v3.01 software (Amersham Bioscience, Bucks, United Kingdom). Image analyses and gel comparisons were subsequently performed using Imagemaster Progenesis software (Nonlinear Dynamics, Newcastle-on-Tyne, United Kingdom).
For each individual spot, the differences in median spot volume between M. avium subsp. paratuberculosis and M. avium subsp. avium were assessed by using a Mann-Whitney test. Because a large number of tests were carried out, it was necessary to adjust the P value from each test to allow for this multiple testing. The false discovery rate (FDR) approach of Benjamini and Hochberg (8) was used; with an FDR of 5% it would be expected that ca. 5% of the spots identified as differentially expressed would be false positives.
Identification of proteins and mass spectrometry.
Proteins of interest were excised from gels (preferentially CBB-stained gels) with either a 15- or 30-mm “spot picker” (The Gel Company, San Francisco, CA) and cut into small pieces approximately 1 to 2 mm in diameter; no excess gel from around the spot was taken. Gel pieces were prepared for analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) as described previously (14) using a Voyager-DE Pro mass spectrometer (PerSeptive Biosystems, Inc., Framingham, MA) selecting for a mass range of 600 to 5,000 Da.
Silver-stained spots were first destained by using a commercial kit (SilverQuest; Invitrogen) prior to the first dehydration step. All subsequent steps were identical to those used for CBB-stained spots as described above.
Data Explorer was used to create the peak list from the raw data with the smoothing function applied; the signal-to-noise correlation factor was set at 0.7, and the data were baseline corrected with the following parameters: peak width = 32, flexibility = 0.5, and degree = 0.1. The peak height at which centroids were calculated was 50%, and peaks were deisotoped. The resolution for the mass spectrometry was greater than 10,000, with a mass accuracy of ±0.01%. A close-external means of calibrating each spectrum and no means of exclusion of known contaminant ions (such as keratin) were used.
Bioinformatics.
The proteome of M. avium subsp. paratuberculosis strain K10 (accession numbers NC_002944 and AE016958) contains 4,350 predicted open reading frames (15), which were used to compile a protein database, and this was queried by using Mascot 2.0 (Matrix Science, Ltd., London, United Kingdom) (22). Searches for trypsin cleavage patterns used a fragment ion mass accuracy of 100 ppm, carbamidomethyl modification was selected, and up to one missed cleavage site was permitted.
Proteins identified using this procedure were characterized by using the Entrez nr Peptide Sequence database (National Center For Biotechnology Information) using the protein-protein BLAST program. The NCBI Conserved Domain Search service was used to identify domains present in protein query sequences and the Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.com) was used to identify relevant metabolic pathways.
Cloning and expression of M. avium subsp. paratuberculosis-specific sequences.
Maltose-binding protein (MBP) fusions of M. avium subsp. paratuberculosis predicted coding sequences were produced in E. coli using the pMAL-c2 vector (New England Biolabs). Primers were designed from the reading frame of each coding sequence and contained an XbaI site in the 5′ primer and a HindIII site in the 3′ primer for cloning purposes. Amplifications were performed by using AmpliTaq Gold polymerase (Applied Biosystems) and M. avium subsp. paratuberculosis genomic DNA as the template under conditions described previously (3). The vector and amplification product were digested with XbaI and HindIII. Ligation of these restricted DNA fragments resulted in an in-frame fusion between the malE gene in the vector and the reading frame of interest. After ligation, the products were transformed into E. coli DH5α and selected on LB agar plates containing 100 μg of ampicillin/ml (4).
Each MBP fusion protein was overexpressed in E. coli by induction with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma, St. Louis, MO) and purified by affinity chromatography using amylose resin supplied by New England Biolabs. A detailed protocol has been published previously (2). Expression of mycobacterial fusion proteins was monitored by using GelCode Blue (Pierce Biotechnology, Inc., Rockford, IL)-stained SDS-PAGE gels. The same approach was used for the production and purification of all MBP fusion proteins. E. coli DH5α harboring the parental plasmid pMAL-c2 was expressed, purified, and used as a control in all experiments. Purified protein from this control strain consists of an MBP fusion of the LacZ alpha peptide. (4).
M. avium subsp. paratuberculosis-specific protein ELISA.
Portions (0.5 μg) of M. avium subsp. paratuberculosis-specific protein in coating buffer (0.2 M carbonate-bicarbonate buffer [pH 9.4]; Pierce) were added to wells in an ELISA plate, sealed, and incubated overnight at 4°C. Seals and residual coating solution were removed, and the bound antigen was washed three times with PBS buffer containing 0.05% (vol/vol) Tween 80 (PBST). Plates were then incubated with PBST containing 10% (wt/vol) Infasoy (Cow & Gate, United Kingdom) at 37°C for 1 h and then washed three times with PBST. Ovine serum (diluted 1,000-fold in PBST containing 5% [wt/vol] Infasoy) was added to the well, followed by incubation at 37°C for 1 h. Plates were washed three times in PBST. Donkey anti-sheep immunoglobulin G (IgG)-horseradish peroxidase (HRP) conjugate (Sigma-Aldrich, Dorset, United Kingdom) diluted 1,000-fold in 5% (wt/vol) Infasoy-PBST was added to each well, and then the plates were resealed and incubated at 37°C for 1 h. The plates were washed three times with PBST. Next, 40 μg of o-phenylenediamine dihydrochloride peroxidase (OPD) substrate, 40 μg urea-hydrogen peroxide in 0.05 M urea hydrogen phosphate-citrate buffer obtained from Sigma FAST OPD tablets (Sigma-Aldrich) was added to each well, followed by incubation for 10 min at room temperature (avoiding sunlight). The reactions were stopped after 10 min by the addition of sulfuric acid (1 M final concentration), and the OD492 values were read within 5 min by using a Labsystems iEMS plate reader (MTX Labsystems).
Dot blots.
A total of 500 ng of purified recombinant protein in PBS containing 0.5% (vol/vol) Tween 80 and 0.5 M NaCl (PBST2) were spotted onto Hybond ECL membrane (GE Healthcare). The membrane was allowed to dry and then incubated for 1 h with shaking in PBST2 containing 10% (wt/vol) Infasoy at room temperature. The membrane strips were incubated in sera obtained from the six clinical cases of ovine paratuberculosis (diluted 1:1,000 in PBST2 containing 5% [wt/vol] Infasoy) for 1 h at room temperature with shaking before being washed five times in PBST2. Strips were incubated in secondary antibody (donkey anti-sheep IgG-HRP conjugate (Sigma-Aldrich) diluted 1:1,000 in PBST2 containing 5% (wt/vol) Infasoy for 1 h at room temperature with shaking before being washed in PBST2 as before. Strips were incubated for 1 min in ECL detection reagents (GE Healthcare) and then visualized by using Fluorchem SP Alpha imager (Alpha Innotech).
Western blots.
Proteins were separated by SDS-PAGE and then transferred electrophoretically to nitrocellulose membranes essentially using the methods described in the Mini-Protean II electrophoresis cell instruction manual and the Mini-Trans-Blot electrophoretic transfer cell instruction manual (Bio-Rad Laboratories, Herts, United Kingdom). Membranes were incubated in PBST2 containing 10% (wt/vol) Infasoy overnight at 4°C without shaking. Sheep serum diluted 1:1,000 or monoclonal anti-MBP (M6295; Sigma-Aldrich) diluted 1:4,000 in PBST2 containing 5% (wt/vol) Infasoy was added as the primary antibody. Membranes were incubated 1 h at room temperature with shaking and washed extensively in PBST2. HRP-conjugated donkey anti-sheep IgG or goat anti-mouse IgG peroxidase conjugate (both from Sigma-Aldrich) diluted (1 in 1,000 and 1 in 2,000, respectively) in PBST2 containing 5% (wt/vol) Infasoy was added, followed by incubation for 1 h at room temperature with shaking, followed in turn by another washing step as described previously. Reactivity was detected by enhanced chemiluminescence (GE Healthcare) imaged on a Fluorchem SP Alpha imager.
Ethical considerations.
All experimental procedures and management protocols were examined and approved by the Moredun Research Institute Experiments and Ethics Committee and conducted within the framework of the United Kingdom Animals (Scientific Procedures) Act 1986 administered by the United Kingdom government Home Office.
RESULTS
Proteomes of M. avium subsp. paratuberculosis and M. avium subsp. avium.
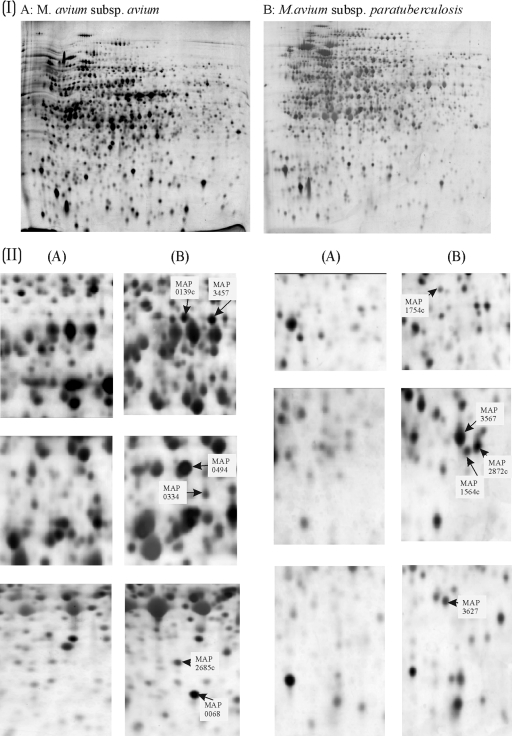
The proteomes were generated by extraction and separation of organisms grown under optimal unrestrictive conditions in the laboratory. The proteomes generated from M. avium subsp. paratuberculosis and M. avium subsp. avium grown in liquid culture are presented Fig. 1IA and IB. The 2-D electrophoretograms were stained by using the modified silver-staining protocol (17). The proteomes obtained have approximately 1,000 well-resolved protein spots each. The parameters of separation gave an even spread of protein across the electrophoretogram, enabling the picking of individual spots. The location of the spots was sufficiently consistent between gels of M. avium subsp. paratuberculosis and M. avium subsp. avium to allow recognition of patterns and matching of spots between gels. Variation of spot intensities between analytic replicates is inherent in the technical procedure, and therefore analysis of several biological and analytical replicates was required.
FIG. 1.
(I) Proteomic comparison of M. avium subsp. paratuberculosis and M. avium subsp. avium cultured in Middlebrook 7H9 broth. Silver-stained 2-D electrophoretograms of M. avium subsp. avium (A) and M. avium subsp. paratuberculosis (B) are shown. The organisms used to generate proteomes were grown in Middlebrook 7H9 broth and harvested during exponential phase. (II) Magnification of areas of the proteomic profiles showing proteins that are specific or predominant in M. avium subsp. paratuberculosis. Composites of analogous regions of 2-D electrophoretograms of either M. avium subsp. avium (A) or M. avium subsp. paratuberculosis (B) are presented. Regions of the gels shown in part I are magnified to demonstrate the differences observed. This figure was compiled by using Phoretix 2D Elite (part I) and CorelDraw 9 software (parts I and II).
Analysis of proteomes and identification of proteins upregulated and/or only present in M. avium subsp. paratuberculosis.
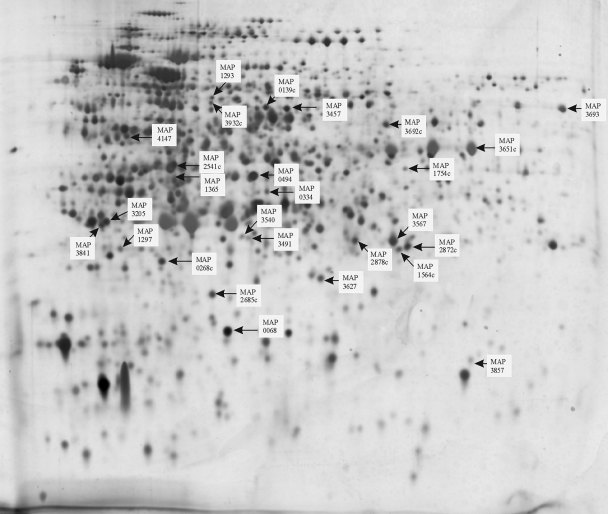
Eight samples of M. avium subsp. paratuberculosis were analyzed by 2-D electrophoresis (six samples were harvested during the exponential phase, and two were harvested during the stationary phase). Five samples of M. avium subsp. avium were also analyzed (three samples were harvested during the exponential phase, and two were harvested during the stationary phase). Proteomes of each organism harvested in both exponential and stationary phases were analyzed to preclude growth-phase-specific differences. Computer-assisted analysis using Progenesis PG200 software was carried out that allowed analogous small sections of the proteomes to be viewed simultaneously. Breaking down the complex proteome into smaller regions facilitated the manual detection of protein spots, whose expression appeared to be consistently increased in the proteomes of M. avium subsp. paratuberculosis. Protein spots that aligned on the proteomes of both M. avium subsp. paratuberculosis and M. avium subsp. avium were deemed to be present in both subspecies and therefore not marked for further analysis. However, proteins on the M. avium subsp. paratuberculosis proteome that appeared considerably more abundant or that had no obvious counterpart on the M. avium subsp. avium proteome were picked. Spot volumes of these proteins did not appear to be affected by the growth phase of the cultures from which they were harvested. Normalized spot volumes of all gels analyzed can be accessed at http://www.moredun.ac.uk/supplements, and the median spot volumes of the selected M. avium subsp. paratuberculosis proteins are recorded in Table 1, along with the relative percentage differences between the spot volume medians of the M. avium subsp. paratuberculosis and M. avium subsp. avium groups and their corresponding statistical significance. Masses obtained by MALDI-TOF analysis of tryptic digests were used to query the M. avium subsp. paratuberculosis and M. avium 104 in silico tryptic-digest peptide databases using Mascot (22). Thirty-two proteins were identified with a significant hit to the K10 M. avium subsp. paratuberculosis database and are numbered and indicated by black arrows in Fig. 2. Expanded views of areas containing the selected spots in the M. avium subsp. paratuberculosis proteome alongside the analogous area in the M. avium subsp. avium proteome are shown in Fig. 1II. Mascot search data for these proteins are presented in Table 1. Some spots that were analyzed by tryptic peptide profiling gave multiple significant hits, suggesting that the spots were a mixture of a number of proteins. For example, MAP4147, MAP1012c, MAP3175c, and MAP4233 were identified in a single spot. An additional two spots generated double identifications: (i) MAP3385 and MAP3205 and (ii) MAP1160c and MAP2878c.
TABLE 1.
2-D and MALDI-TOF analysis of 32 spots selected from the proteome of M. avium subsp. paratuberculosisa
| Annotationb | Protein | Nominal mass | 2-D expression analysis
|
Mass spectrometry identification
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median spot vol | Relative difference in medians (%) | P (Mann-Whitney) | Mascot score | PI | Peptide count | No. of masses searched | Sequence coverage (%) | |||
| MAP0068 | SsB | 17.5 | 0.51 | 99 | 0.002 | 149 | 5.12 | 11 | 28 | 66 |
| MAP0139c | 23.6 | 0.05 | 100 | 0.002 | 56 | 9.95 | 10 | 107 | 54 | |
| MAP0268c | 23.8 | 0.26 | 100 | 0.002 | 106 | 4.96 | 7 | 16 | 39 | |
| MAP0334 | 34.5 | 0.09 | 100 | 0.002 | 147 | 5.34 | 10 | 17 | 33 | |
| MAP0494* | 38.5 | 0.36 | 89 | 0.002 | 180 | 5.85 | 11 | 16 | 47 | |
| MAP1012c | 37.5 | 0.07 | 100 | 0.003 | 118 | 4.6 | 13 | 57 | 48 | |
| MAP1160c | 29.4 | 0.38 | 56 | NS | 84 | 5.44 | 7 | 27 | 33 | |
| MAP1293 | HisD | 49.4 | 0.04 | 89 | 0.002 | 94 | 4.92 | 12 | 33 | 27 |
| MAP1297 | HisA | 25.4 | 0.09 | 100 | 0.002 | 106 | 4.73 | 8 | 29 | 58 |
| MAP1365 | ArgF | 33.6 | 0.04 | 100 | 0.002 | 71 | 4.9 | 6 | 26 | 30 |
| MAP1564c | 23.1 | 0.12 | 100 | NS | 110 | 5.66 | 8 | 23 | 43 | |
| MAP1754c | Usp | 30.8 | 0.03 | 100 | 0.002 | 131 | 5.72 | 10 | 24 | 38 |
| MAP2541c | MDH | 34.5 | 0.09 | 100 | 0.002 | 208 | 4.87 | 13 | 18 | 44 |
| MAP2685 | 21.3 | 0.20 | 39 | NS | 130 | 4.84 | 7 | 11 | 57 | |
| MAP2872c | FabG5_2 | 26.7 | 0.37 | 75 | NS | 86 | 5.65 | 10 | 71 | 40 |
| MAP2878c | DapB | 25.6 | 0.38 | 58 | NS | 89 | 5.52 | 7 | 27 | 39 |
| MAP3175c | PrfB | 41.5 | 0.07 | 100 | 0.003 | 68 | 4.73 | 8 | 57 | 28 |
| MAP3205 | NuOE | 26.9 | 0.11 | 39 | NS | 66 | 4.66 | 6 | 31 | 31 |
| MAP3385 | 32.3 | 0.11 | 39 | NS | 57 | 4.62 | 5 | 18 | 24 | |
| MAP3457 | MetC | 47.6 | 0.07 | 68 | NS | 153 | 5.25 | 12 | 28 | 46 |
| MAP3491 | 28.2 | 0.05 | 100 | NS | 124 | 5.45 | 8 | 17 | 49 | |
| MAP3540c | 25.1 | 0.03 | 100 | 0.002 | 124 | 5.18 | 9 | 23 | 38 | |
| MAP3567 | 30.1 | 0.40 | 88 | 0.002 | 217 | 5.7 | 14 | 33 | 66 | |
| MAP3627 | 23.1 | 0.18 | 100 | 0.002 | 92 | 5.4 | 7 | 22 | 34 | |
| MAP3651c | FadE3_2 | 43.9 | 0.47 | 93 | 0.002 | 273 | 6.15 | 19 | 29 | 49 |
| MAP3692c | FabG4 | 47.1 | 0.14 | 23 | NS | 102 | 5.79 | 8 | 19 | 30 |
| MAP3693* | FadA2 | 46.7 | 0.11 | 73 | NS | 165 | 6.21 | 14 | 31 | 42 |
| MAP3841 | GrpE | 23.7 | 0.23 | 71 | NS | 151 | 4.58 | 11 | 24 | 42 |
| MAP3857 | UmpA | 18.7 | 0.06 | 100 | 0.002 | 103 | 6.06 | 7 | 19 | 49 |
| MAP3932c | MoA3 | 41.4 | 0.04 | 89 | 0.002 | 56 | 5.01 | 6 | 28 | 24 |
| MAP4147 | 42.2 | 0.07 | 100 | 0.003 | 125 | 4.73 | 15 | 57 | 49 | |
| MAP 4233 | Rpo | 0.07 | 100 | 0.003 | 125 | 4.73 | 15 | 57 | 49 | |
The median spot volume is that of the M. avium subsp. paratuberculosis population. The relative difference in medians is the difference between the median spot volume of the M. avium subsp. paratuberculosis population and that of the M. avium subsp. avium population as a percentage of the median spot volume recorded for M. avium subsp. paratuberculosis. NS, the difference is not significant using an FDR of 10%. PI, isoelectric point.
Clones containing the sequence of proteins indicated by an asterisk were unobtainable after three attempts to amplify or clone the sequence.
FIG. 2.
Location of identified M. avium subsp. paratuberculosis proteins excised from 2-D protein gels. A silver-stained 2-D electrophoretogram of M. avium subsp. paratuberculosis grown to exponential phase in modified Middlebrook 7H9 broth is presented. Protein spots that were selected as being in greater abundance or uniquely present by comparison to the electrophoretogram of M. avium subsp. avium are highlighted, and their identifications as determined by MALDI-TOF are indicated. Some spots were apparently mixtures of proteins. The spot indicated as MAP4147 also had significant hits to MAP1012c, MAP3175c, and MAP4233. Also, the spot indicated as MAP3205 had a significant hit to MAP3385, as did MAP2878c to MAP1160c. This figure was compiled by using CorelDraw 9 software.
For each of the 1,146 spots that could be distinguished on the reference gel, a Mann-Whitney test was carried out to determine whether there was evidence that the median volumes were different in the two strains, and the corresponding P value was noted. Because of the large number of tests carried out, it was necessary to allow for multiple testing. The FDR approach of Benjamini and Hochberg (8) was applied. An FDR of 5% applied to spots that had a P value of ≤0.002 (an FDR of 10% applied to spots with P values of 0.006), which could be regarded as being differentially expressed in the two strains. Table 1 shows the M. avium subsp. paratuberculosis spot volume medians, and their corresponding percentage increase in the expression of proteins that appeared to be less abundant or were not present in the M. avium subsp. avium proteome.
Although a few of the proteins listed appeared to be considerably less abundant on average on the M. avium subsp. avium proteome, the differences between the subtypes were not statistically significant. This was due to the variability of the data recorded and may reflect the errors in spot matching and spot identification which are inherent in the analysis. However, it is possible that these proteins are worthy of further investigation and may be useful in the generation of M. avium subsp. paratuberculosis-specific antigens.
Investigation of the immunogenicity of M. avium subsp. paratuberculosis-specific recombinant proteins.
Sequences of all but two proteins (MAP0494 and MAP3693) were obtained by amplification and then cloned into the vector pMAL-c2x as described in Materials and Methods. MBP fusions of the remaining 30 M. avium subsp. paratuberculosis-specific proteins were expressed and purified by affinity chromatography and were used in three immunological assays. ELISAs and dot blots were both performed to determine whether native recombinant proteins could be detected with sera from clinical ovine paratuberculosis cases. Dot blots, although yielding poorer quantitative data, may augment the number of immunoreactive proteins detected if ELISAs of such proteins are negative due to their failure to adhere to the ELISA plates. Western blots were also performed to determine whether denatured proteins could be detected with sera from clinical ovine paratuberculosis cases.
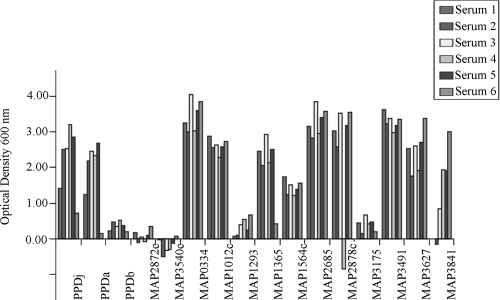
ELISAs were performed for all 30 recombinantly expressed, affinity-purified M. avium subsp. paratuberculosis-specific fusion proteins and also for the purified protein derivatives (PPDs) from M. avium subsp. paratuberculosis (PPDj), M. avium subsp. avium (PPDa), and M. bovis (PPDb). Averaged ODs of the ELISAs carried out in triplicate, corrected for background and MBP, were recorded for the six sera from infected sheep. M. avium subsp. paratuberculosis proteins that showed ODs in excess of the cutoff of 0.1 OD units in at least four of six sera tested are shown in Fig. 3. Eleven M. avium subsp. paratuberculosis proteins fit these criteria (MAP0334, MAP1012c, MAP1293, MAP1365, MAP1564c, MAP2685, MAP2878c, MAP3175c, MAP3491, MAP3627, and MAP3841). The other 19 M. avium subsp. paratuberculosis-specific proteins did not meet the criteria, and 2 are shown as examples (MAP2872c and MAP3540c).
FIG. 3.
ELISA tests with recombinant proteins. The immunoreactivity of ovine sera to recombinant proteins in six sheep with clinical paratuberculosis, as determined by ELISA, was charted. The results shown are averaged from ELISAs carried out in triplicate (corrected for background and MBP). The results for all 11 recombinant proteins that met the criteria of having ODs in excess of the cutoff of 0.1 OD units in at least four of six sera are shown, along with the results for 2 proteins (MAP2872c and MAP3540c) that did not. PPDs from M. bovis source (PPDb), M. avium subsp. avium (PPDa), and M. avium subsp. paratuberculosis (PPDj) are also included for comparison.
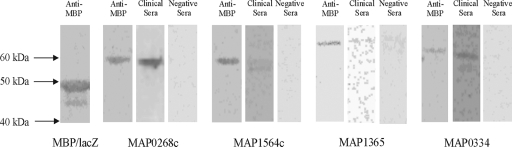
The immunoreactivities of the recombinant proteins as assessed by ELISA, Western blots, and dot blots are recorded in Table 2. In addition to the 11 proteins with a positive ELISA response, a number of other proteins were positive by dot blot analysis (these were MAP0268c, MAP1297, MAP3857, and MAP4147); presumably, these antigens did not adhere to the ELISA plate using the standard conditions and would be worthy of further investigation in order to optimize the binding of the protein to the ELISA plate. Western blotting was carried out with both monoclonal anti-MBP antibody and one serum sample selected either from the serum panel of the clinical cases of paratuberculosis or the healthy individuals as the primary antibody. All fusion proteins gave a positive signal with anti-MBP with a retarded running position with respect to the MBP-LacZ fusion product, as expected, and no signal with control sera from a healthy sheep. Of the 11 recombinant, ELISA-positive proteins, two (MAP2878c and MAP3491) did not react by Western blotting with antisera from a clinical paratuberculosis case, presumably because the antigenic epitope was disrupted by denaturation with SDS in the Western blotting procedure. The positive result of the Western blotting of MAP0268c, MAP1297, MAP3932c, and MAP4147 agreed with the dot blot result, strengthening the idea that ELISA with this protein had probably failed only for technical reasons. MAP3457 showed immunogenicity on Western blots even though both the ELISA and the dot blot results had proved negative. This indicates that the antigen of this protein is only revealed upon denaturation. Figure 4 shows three examples of ELISA-positive proteins that were also positive by immunoblotting with paratuberculosis sera from a case of ovine paratuberculosis (MAP1564c, MAP1365, and MAP0334). MAP0268c, which was determined to be negative by ELISA, is also included. There is some evidence of proteolysis of the recombinant proteins in the figure shown.
TABLE 2.
M. avium subsp. paratuberculosis-specific proteins responding to clinical sera from cases of ovine paratuberculosisa
| Protein | Positive (+) or negative (−) immunoassay result
|
||
|---|---|---|---|
| ELISA | Dot blot | Western blot | |
| MAP0268c | - | + | + |
| MAP0334 | + | + | + |
| MAP1012c | + | + | + |
| MAP1293 | + | + | + |
| MAP1297 | - | + | + |
| MAP1365 | + | + | + |
| MAP1564c | + | + | + |
| MAP2685 | + | + | + |
| MAP2878c | + | + | - |
| MAP3175c | + | + | + |
| MAP3457 | - | - | + |
| MAP3491 | + | + | - |
| MAP3627 | + | - | + |
| MAP3841 | + | - | + |
| MAP3857 | - | + | - |
| MAP3932c | - | + | + |
| MAP4147 | - | + | + |
Only data from proteins with immunoreactivity as detected by ELISA, Western blot, or dot blot analyses are recorded.
FIG. 4.
Immunoblots of recombinant proteins. Immunoblots of recombinant proteins detected with anti-MBP monoclonal antibody, with clinical sera from cases of ovine paratuberculosis, or with negative sera from a healthy control sheep are shown. The MBP-LacZ fusion protein detected with anti-MBP monoclonal antibody has been included for comparison.
Sequence comparison of proteins in the two subspecies.
Blastp analysis showed that MAP0268c, MAP1160c, MAP1365, MAP2541c, MAP2685, MAP2872c, MAP3205, MAP3385, MAP3457, MAP3491 MAP3567, MAP3627, MAP3651c, MAP3692c, MAP3693, MAP3841, MAP3857, MAP3932c, and MAP4233 had no or limited amino acid differences in both subspecies. Therefore, the absence of these proteins on the proteome of M. avium subsp. avium cannot be explained by sequence changes causing a variation in position on the 2-D electrophoretogram. Genes encoding some of the proteins (i.e., MAP0334, MAP1297, MAP1754c, and MAP3540c) had become altered by either point mutation, insertion, or deletion in their nucleotide sequence. These changes may have resulted in altered migration positions on the 2-D electrophoretogram (due to an altered molecular-weight isoelectric point). Other proteins were annotated with extended N termini in M. avium subsp. paratuberculosis compared to M. avium 104 (MAP0139c, MAP0494, MAP1012c, MAP1293, MAP1564c, MAP2878c, and MAP3175c). However, examining the gene sequence of these proteins indicated that there were no obvious variations between the two subtypes, and therefore these apparent differences in the translational starts of these proteins were due to the way they had been annotated.
DISCUSSION
Using a comparative proteomic approach, we have identified 32 M. avium subsp. paratuberculosis proteins that are absent (or very much diminished) in the proteome of M. avium subsp. avium. These proteins are not derived from sequences specific to M. avium subsp. paratuberculosis (none of the proteins reported lie in the LSP regions (23) but are either differentially expressed or accumulated to a greater extent by comparison to M. avium subsp. avium.
Comparison of proteomes of M. avium subsp. paratuberculosis and M. avium subsp. avium revealed a number of proteins that appeared to be absent or present to a much lesser degree in M. avium subsp. avium. Of these proteins, 32 were identified by MALDI-TOF peptide profiling with a significant hit to the K10 M. avium subsp. paratuberculosis protein in the in silico tryptic-digest database. Proteomic analysis confirmed that all proteins had higher median values for these identified spots in the M. avium subsp. paratuberculosis group, and most were significant (P < 0.003).
Proteome-determined M. avium subsp. paratuberculosis-specific proteins described in the present study have been defined by using organisms grown in vitro in Middlebrook 7H9 broth. Whether these proteins are expressed by M. avium subsp. avium under different conditions (for example, during colonization of its host) remains to be determined. However, it is unlikely that the specificity of these proteins for the paratuberculosis subtype will be altered by the growth of M. avium subsp. avium within the host, since most proteins identified in the present study do not appear to be upregulated in M. avium subsp. paratuberculosis isolated from pathological lesions (14). Only one such protein, MAP1754c, a universal stress protein, was upregulated in vivo; however, its specificity is not compromised by growth conditions since its absence on the M. avium subsp. avium proteome is due to an interruption of the open reading frame.
The sequences of all but two proteins (MAP0494 and MAP3693) were obtained by amplification and then cloned into an E. coli expression vector. In order to assess whether these recombinant proteins had any potential as diagnostic reagents, they were subjected to ELISA tests using the sera from six cases of ovine paratuberculosis. Eleven of the proteins were classified as having positive ELISA results, as defined by an OD of >0.1 in at least four of six sheep. Some of these proteins (MAP0334, MAP1012c, MAP1365, MAP1564c, MAP2685, MAP3491, and MAP3627) were recognized by all six clinical sera. MAP1293, MAP2878c, MAP3175c, and MAP3841 were recognized by four or five of the clinical sera. Interestingly, one individual sheep failed to respond to three of our antigens MAP1293, MAP3175c, and MAP3841. It is possible that this individual was at a different clinical stage than the others and that these antigens were not recognized as efficiently at this phase of the disease. Monitoring the performance of these ELISAs over the course of an experimental infection would determine whether the proteins are antigenic at all stages of infection or only in sheep at terminal stages of Johne's disease. Of the 11 recombinant, ELISA-positive proteins, 2 did not produce a signal on Western blots with antisera from a clinical paratuberculosis case (MAP2878c and MAP3491). The immunoreactivity of MAP1297 observed in the Western blot agreed with a dot blot result (data not shown), strengthening the idea that ELISA with this protein had only failed for technical reasons. Two additional proteins, MAP3457 and MAP3932c, showed immunoreactivity on Western blots even though both the ELISA and the dot blot results had proved negative. This indicates that antigens of these proteins are only revealed upon denaturation.
M. avium subsp. paratuberculosis and M. avium subsp. avium have a high degree of similarity with >95% sequence identity. Single nucleotide polymorphisms, although rare, may be responsible for some of the changes observed by altering elements that control the levels of expression; alternately, single nucleotide polymorphisms occurring within open reading frames may result in stability changes, particularly if the protein is truncated or extended as a result. This may impact the steady-state level of the protein within the cell.
The high level of sequence identity between the two organisms belies the major genomic rearrangements that have occurred in the evolution of the subspecies. Comparison of the genome organization of the organisms reveals two major inversions that span almost 2 Mb, plus numerous minor inversions. The context of a gene may be very important in defining its level of expression, and even basic physical properties such as the strand of DNA from which they are expressed may have an impact. Fortuitous positioning by random events such as inversions may cause genes to enter “neighborhoods” that give them an expression advantage. In addition, genes may become juxtaposed to elements that may influence their expression. It is not possible to accurately predict the steady-state concentrations of proteins by analyzing sequence data, but the proteomics approach we have used is an empirical way to find proteins whose level of expression is altered and thus define a set of M. avium subsp. paratuberculosis-specific proteins broader than that defined by a sequence-specific approach alone. Some of the proteins identified here are key to metabolic processes and are therefore likely to be expressed by all strains of the subtype. This is important for a diagnostic reagent that should detect all strains.
In the scope of the present study the recombinant antigens have been only tested with six antisera, too few to make any comment on the sensitivity of the assay. To address this, the recombinant antigens are currently being tested against a panel of sera from sheep and cattle in the framework of a European Union funded consortium to further validate their use as diagnostic reagents. The paratuberculosis strain used here is type II as described by Stevenson et al. (24), the same category as the strains categorized as C type by Collins et al. (10), and sera were obtained from animals with clinical paratuberculosis caused by type II strains. Type I organisms (which may correspond to the S type as defined by Collins et al. [10]) have differences in their proteome (unpublished observations) and therefore sera from animals infected by type I organisms may be different serologically and not respond as well to the antigens thus described.
If recombinant antigenic proteins were used in an ELISA, their quality and quantity could be easily controlled, and this would give an advantage over ELISAs that rely on undefined cell extracts. Two proteins whose sequence is unique to M. avium subsp. paratuberculosis have shown potential as antigens for use in an ELISA test for ovine paratuberculosis but had a temporally variable response (6). It is becoming clear that a cocktail of specific antigens will be required in an ELISA, and those reported here will increase the available specific antigens from which a final selection may be chosen. Such an ELISA built on M. avium subsp. paratuberculosis-specific antibodies should fulfill the wishes of the Committee on Diagnosis and Control of Johne's Disease: “exposure to other mycobacteria such as M. avium subsp. avium is likely to be common in cattle, so it is essential that any test to identify animals in the early stages of infection be highly specific for M. avium subsp. paratuberculosis” (19).
Acknowledgments
This study was funded by the Rural and Environmental Research and Analysis Directorate, the European Union (contract number QLK2-CT-2001-01420), Genecom (Concept Definition Fund), the USDA-CSREES-CAP grant (JDIP), and the USDA-Agricultural Research Service.
We thank the staff at the Moredun Research Institute, Linda May for expertise in bacteriological culture, the staff of the Moredun Proteomics Facility for MALDI-TOF analysis, Alex Lainson and Raja Yaga for preparing the Mascot database, and the staff of the Clinical Division for animal husbandry. We also thank the local farmers for providing the clinical cases of ovine paratuberculosis used in this study.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Antognoli, M. C., H. L. Hirst, F. B. Garry, and M. D. Salman. 2007. Immune response to and faecal shedding of Mycobacterium avium ssp. paratuberculosis in young dairy calves, and the association between test results in the calves and the infection status of their dams. Zoonoses Public Health 54:152-159. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York, NY.
- 3.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome-scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannantine, J. P., J. K. Hansen, M. L. Paustian, A. Amonsin, L.-L. Li, J. R. Stabel, and V. Kapur. 2004. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., Q. Zhang, L. L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannantine, J. P., V. Rosu, S. Zanetti, S. Rocca, N. Ahmed, and L. A. Sechi. 2007. Antigenic profiles of recombinant proteins from Mycobacterium avium subsp paratuberculosis in sheep with Johne's disease. Vet. Immunol. Immunopathol. 122:116-125. [DOI] [PubMed] [Google Scholar]
- 7.Bech-Neilson, S., J. B. Jorgensen, P. Ahrens, and N. C. Feld. 1992. Diagnostic accuracy of a Mycobacterium phlei-absorbed serum enzyme linked immunosorbent assay for diagnosis of bovine paratuberculosis in dairy cows. J. Clin. Microbiol. 30:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 9.Cetinkaya, B., H. M. Erdogan, and K. L. Morgan. 1998. Prevalence, incidence and geographical distribution of Johne's disease in cattle in England and the Welsh borders. Vet. Rec. 143:265-269. [DOI] [PubMed] [Google Scholar]
- 10.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, M. T., and D. C. Sockett. 1993. Accuracy and economics of the USDA licensed enzyme linked immunosorbent assay for bovine paratuberculosis. J. Am. Vet. Med. Assoc. 203:1456-1463. [PubMed] [Google Scholar]
- 12.Collins, M. T., S. J. Wells, K. R. Petrini, J. E. Collins, R. D. Schultz, and R. H. Whitlock. 2005. Evaluation of five antibody detection tests for diagnosis of bovine paratuberculosis. Clin. Diagn. Lab. Immunol. 12:685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eda, S., J. P. Bannantine, W. R. Waters, Y. Mori, R. H. Whitlock, M. C. Scott, and C. A. Speer. 2006. A highly sensitive and subspecies-specific surface antigen enzyme-linked Immunosorbent assay for diagnosis of Johne's disease. Clin. Vaccine Immunol. 13:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, V., S. Smith, A. Garcia-Sanchez, J. Sales, and K. Stevenson. 2007. Proteomic comparison of Mycobacterium avium subspecies paratuberculosis grown in vitro and isolated from clinical cases of ovine paratuberculosis. Microbiology 153:196-205. [DOI] [PubMed] [Google Scholar]
- 15.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subsp. paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milner, A. R., A. W. D. Lepper, W. N. Symonds, and E. Gruner. 1987. Analysis by ELISA and Western blotting of antibody reactivities in cattle with Mycobacterium paratuberculosis after absorption of serum with M. phlei. Res. Vet. Sci. 42:140-144. [PubMed] [Google Scholar]
- 17.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 18.NAHMS. 1997. Johne's disease on U.S. dairy operations. Document N245.1097. USDA/APHIS/VS, CEAH, National Animal Health Monitoring System, Fort Collins, CO.
- 19.National Research Council. 2003. Diagnosis and control of Johne's disease, p. 129. National Academies Press, Washington, DC. [PubMed]
- 20.Neuhoff, V., R. Stamm, and H. Eibl. 1985. Clear background and highly sensitive protein staining with Coomassie blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6:427-448. [Google Scholar]
- 21.Paustian, M. L., A. Amonsin, V. Kapur, and J. P. Bannantine. 2004. Characterisation of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis. Implications for diagnosis of Johne's disease. J. Clin. Microbiol. 42:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence data bases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 23.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 40:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitlock, R. H., S. J. Wells, R. W. Sweeney, and J. Van Tiem. 2000. ELISA and faecal culture for paratuberculosis (Johne's disease) sensitivity and specificity of each method. Vet. Microbiol. 77:387-398. [DOI] [PubMed] [Google Scholar]