Abstract
Q fever is a widespread zoonosis caused by Coxiella burnetii. Diagnosis of Q fever is usually based on serological testing of patient serum. The diagnostic antigen of test kits is formalin-fixed phase I and phase II organisms of the Nine Mile reference strain. Deficiencies of this antigen include (i) potential for cross-reactivity with other pathogens; (ii) an inability to distinguish between C. burnetii strains; and (iii) a need to propagate and purify C. burnetii, a difficult and potentially hazardous process. Consequently, there is a need for sensitive and specific serodiagnostic tests utilizing defined antigens, such as recombinant C. burnetii protein(s). Here we describe the use of a C. burnetii protein microarray to comprehensively identify immunodominant antigens recognized by antibody in the context of human C. burnetii infection or vaccination. Transcriptionally active PCR products corresponding to 1,988 C. burnetii open reading frames (ORFs) were generated. Full-length proteins were successfully synthesized from 75% of the ORFs by using an Escherichia coli-based in vitro transcription and translation system (IVTT). Nitrocellulose microarrays were spotted with crude IVTT lysates and probed with sera from acute Q fever patients and individuals vaccinated with Q-Vax. Immune sera strongly reacted with approximately 50 C. burnetii proteins, including previously identified immunogens, an ankyrin repeat-domain containing protein, and multiple hypothetical proteins. Recombinant protein corresponding to selected array-reactive antigens was generated, and the immunoreactivity was confirmed by enzyme-linked immunosorbent assay. This sensitive and high-throughput method for identifying immunoreactive C. burnetii proteins will aid in the development of Q fever serodiagnostic tests based on recombinant antigen.
Coxiella burnetii is a gram-negative obligate intracellular bacterium and the etiological agent of the zoonosis Q (“query”) fever. Human populations most at risk for infection are those routinely exposed to infected animals and their products. The organism has a diverse animal reservoir that includes domestic livestock such as dairy cattle, goats, and sheep. Chronically infected dairy cattle shed C. burnetii in milk and other secretions, and the products of livestock parturition can deposit tremendous numbers of the organisms into the environment. The insidious nature of C. burnetii is further exacerbated by the organism's aerosol route of infection, low infectious dose, and pronounced extracellular stability. Q fever most commonly manifests as a self-limiting but debilitating influenza-like illness that includes signs and/or symptoms of prolonged high fever, headache, and malaise. Chronic infection can occur, normally in predisposed individuals, that typically presents as a life-threatening endocarditis (reviewed in reference 17).
Two advancements that would aid in control of Q fever are (i) a specific and sensitive serodiagnostic test based on recombinant antigen and (ii) an efficacious and safe vaccine that does not require prevaccination skin testing. Human Q fever is currently diagnosed by clinical presentation and supporting serological responses against fixed, whole-cell phase I and phase II forms of the C. burnetii Nine Mile reference strain. Platforms for serological testing include immunofluorescence, complement fixation, enzyme-linked immunosorbent assay (ELISA) and microagglutination (21). Unfortunately, the complex nature of the whole-cell antigen results in lack of uniformity and specificity in test results. There is currently not a Food and Drug Administration-approved Q fever vaccine for use in the United States, although a killed-cellular vaccine (Q-Vax) is licensed in Australia (15). Q-Vax, along with other investigational vaccines based on formalin-inactivated organisms, are highly efficacious in prevention of Q fever by inducing both robust humoral and cell-mediated immune responses to C. burnetii antigens (33). However, these vaccines can cause severe local and occasionally systemic reactions in individuals previously sensitized to C. burnetii. Thus, skin testing for preexisting immunity against C. burnetii is required prior to vaccination (16). Since both current commercial serological tests and vaccines rely on intact organisms, a biosafety level 3 laboratory is required for antigen production.
With the ready availability of pathogen genome sequences, reverse approaches to vaccine and diagnostic antigen discovery are feasible (18). Pathogen antigen screens using E. coli-expressed recombinant protein have used bioinformatic tools to prioritize proteins for screening. Reductive strategies are used to remove housekeeping proteins and other proteins present in nonpathogens, while retaining proteins predicted to interact with the host immune system, such as known or suspected virulence-associated proteins that are surface exposed or secreted (11). For example, by screening 156 bioinformatically selected Chlamydia trachomatis recombinant proteins with sera from urogenitally infected patients, novel antigens were revealed, some of which are secreted during the course of infection (24). Unbiased genome-scale antigen discovery using E. coli-expressed antigen has also been performed. A near-total proteome screen using purified recombinant protein was conducted with Treponema pallidum, wherein ca. 85% of the organism's predicted proteins were tested for serological reactivity in a 96-well plate platform (4). That study identified novel antigens that are differentially recognized by sera from patients in primary, secondary, and latent disease stages of syphilis.
Although informative, genome-scale antigen discovery using purified recombinant protein is labor intensive and E. coli expression of heterologous proteins can be problematic due to toxicity and insolubility. To circumvent these problems and to provide a more high-throughput platform, protein microarrays utilizing recombinant protein expressed by in vitro transcription and translation (IVTT) have been developed (2, 7, 8). In this procedure, whole-genome DNA libraries consisting of either individual plasmid-cloned genes or gene-specific transcriptionally active PCR (TAP) products are used as templates in IVTT reactions (7, 20). Nanoliter amounts of crude IVTT lysates containing synthesized protein are then spotted onto nitrocellulose-coated glass slides in a microarray format and screened for antibody reactivity using a fluorescence scanner (7). Davies et al. (7, 8) were first to describe an unbiased “immunoproteome” serological screen using IVTT synthesized protein. Using high-throughput methods of PCR amplification and in vivo recombinational cloning into a T7 promoter expression plasmid, these researchers identified 14 proteins within the vaccinia proteome that strongly react with vaccinia virus immun.globulin. This technology was later used to identify 48 and 103 immunodominant antigens of Francisella tularensis and Borrelia burgdorferi, respectively, from >80% of these pathogen's predicted proteomes (2, 10).
A more thorough understanding of the humoral response to C. burnetii infection is necessary for development of a new generation of Q fever diagnostics and vaccines based on recombinant antigen. To this end, we developed a C. burnetii protein microarray to comprehensively identify immunodominant antigens recognized in the context of C. burnetii infection or vaccination. A subset of these immunodominant antigens was expressed as recombinant proteins, purified, and examined for reactivity in a standardized ELISA format to correlate the predictive ability of the array-identified proteins to serve as defined antigen diagnostic reagents.
MATERIALS AND METHODS
Organism cultivation and chromosomal DNA isolation.
The C. burnetii Nine Mile isolate (RSA493) in phase I was used in the present study. Organisms were propagated in African green monkey kidney (Vero) fibroblasts (CCL-81; American Type Culture Collection) grown in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 2% fetal bovine serum. Organisms were purified by Renografin density gradient centrifugation as previously described (5, 23). Total genomic DNA was isolated directly from purified C. burnetii using an UltraClean microbial DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA). An additional heating step (85°C for 30 min) was added before physical disruption of the bacterial cells. All DNA was resuspended in distilled H2O and frozen at −20°C.
Generation of TAP products.
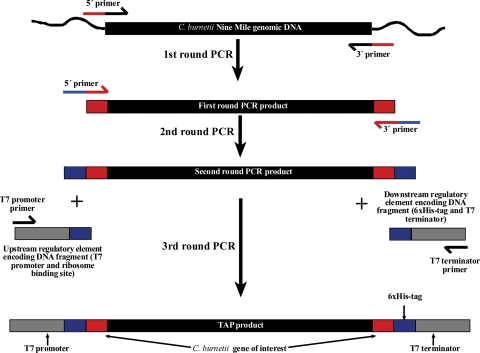
TAP products were generated by using a modification of the Roche rapid translation system (RTS) E. coli linear template generation set, the His6-Tag system (Roche Applied Science, Indianapolis, IN). This system generates TAP products compatible with Roche's RTS. The 5′ and 3′ gene-specific primers for first-round PCR were optimally designed by Sigma-Genosys (St. Louis, MO) to specifically amplify each C. burnetii ORF. Up to five codons were removed from the 5′ and/or the 3′ end of some ORF sequences to allow specific amplification of similar genes (e.g., paralog gene families). The 5′ end of each 5′ primer also contains the sequence CACCATGGGCGGC, which encodes tandem glycine codons (GGC), an ATG start codon, and the sequence CACC to allow potential directional TOPO cloning (Invitrogen) for downstream applications. The GGC codons were added to extend the length of common sequence on 5′ primers to allow annealing of primers for second-round PCR. The 5′ end of each 3′ primer contains the sequence TCCAGCAATAGTTGGGTTAAG, which encodes the epitope tag LNPTIAG. This peptide sequence constitutes an immunodominant epitope of the C. trachomatis major outer membrane protein and reacts with the monoclonal antibody L2I-10 (1). PCR products of C. burnetii ORFs were generated by using 12 pmol of ORF-specific primers, 20 ng of C. burnetii genomic DNA, and Pfu DNA polymerase (Stratagene, La Jolla, CA). Reactions were conducted in 96-well plates, and the resulting PCR products were analyzed on a 0.8% agarose gel for the correct size. Aliquots of both positive and negative PCRs were transferred by using a multichannel pipetter to new 96-well plates for second-round PCR. Complementary second-round primers were used that incorporate unique overlap regions onto amplicon ends to allow subsequent annealing of DNA fragments encoding T7 regulatory elements and a C-terminal His6 tag. Aliquots of second PCRs were transferred by using a multichannel pipetter to new 96-well plates for third-round PCR. Regulatory element and His6 tag-encoding DNA fragments were added to wells and incorporated in the final TAP product by a third round of PCR using complementary primers. Second- and third-round PCR were conducted using Accuprime Pfx DNA polymerase (Invitrogen) and 2 μl of the previous round PCR as a template. Final PCR products were purified by using a Roche HighPure 96 UF cleanup kit to remove nucleotides and primer dimers. TAP products were analyzed on a 0.8% agarose gel for the correct size, and aliquots of all final PCR products (both positive and negative for TAP product) were used in subsequent IVTT reactions. A schematic of the TAP product synthesis is shown in Fig. 1.
FIG. 1.
Schematic of TAP product synthesis. C. burnetii Nine Mile ORFs were amplified in a first round of PCR using genomic DNA as a template and PCR primers consisting of gene-specific sequence and common 5′ overlap regions. TAP products were generated by an two additional rounds of PCR. Second-round primers were used that incorporate unique overlap regions onto amplicon ends to allow subsequent annealing of DNA fragments encoding T7 regulatory elements and a C-terminal His6 tag that were incorporated into the final TAP by a third round of PCR using complementary primers.
Generation of plasmid expression library.
A revised version of a previously published high-throughput recombination cloning method (7) was used to generate plasmids expressing a subset of C. burnetii antigens. In this revised method, recombination was accomplished in vitro using the In-Fusion recombinase enzyme from Clontech. Custom PCR primers comprising 20 bp of gene-specific sequence with 33 bp of “adapter” sequences were used in PCRs with C. burnetii genomic DNA as a template. The adapter sequences, which become incorporated into the termini flanking the amplified gene, were homologous to the cloning site of the linearized T7 expression vector pXT7 (7). A total of 40 ng of PCR-generated linear vector was mixed with 10 to 50 ng of PCR-generated ORF fragment (molar ratio, 1:1; vector, 1-kb ORF fragment) and 0.1 U of In-Fusion enzyme (In-Fusion CF liquid PCR cloning kit; Clontech, Mountain View, CA) in a total volume of 10 μl. The mixture was incubated at 25°C for 30 min. For transformation, 2 μl of this mixture was added to 10 μl of competent E. coli DH5α cells. The mixture was incubated on ice for 45 min, heat shocked at 42°C for 1 min, and then chilled on ice for 1 min. Transformed cells were mixed with 250 μl of super optimal catabolizer medium (2% tryptone, 0.55% yeast extract, 10 mM NaCl, 10 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose), incubated at 37°C for 1 h, and then diluted into 3 ml of LB medium supplemented with 50 μg of kanamycin per ml. Cultures were incubated overnight with shaking. Plasmids were isolated and purified from cultures without colony selection. We have found that this method using In-Fusion can reduce the amount of linear vector and the amount of ORF PCR product needed for transformation and greatly reduce the background level of empty vector in the plasmid preparation. It also enables shorter “adapter” sequences to be used, which reduces the cost of the primers.
IVTT expression of C. burnetii proteins.
To produce IVTT protein from expression plasmids or TAP fragments, 10 μl of DNA template was added to individual wells of a 96-well Roche RTS 100 E. coli HY kit. IVTT reactions were incubated for 8 h at 30°C with shaking (600 rpm) in a Roche RTS Proteomaster. Samples were analyzed for the presence of C-terminal His6-tagged protein by dot immunoblotting. Briefly, 3 μl of each IVTT reaction was spotted onto a nitrocellulose membrane (Bio-Rad, Hercules, CA), which was blocked for 1 h in PBST (10 mM sodium phosphate, 150 mM sodium chloride [pH 7.4], and 0.1% Tween 20) containing 5% nonfat dry milk. One microgram of Penta-His mouse monoclonal antibody (Qiagen, Valencia, CA) was then added, and the membrane was incubated for 1 h with rocking. The membrane was washed three times in PBST and then probed for 1 h with a peroxidase-conjugated anti-mouse immunoglobulin G (IgG) secondary antibody diluted 1:5,000 in PBST. Bound antibodies were detected by chemiluminescence using Supersignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) and Hyperfilm ECL (Amersham, Piscataway, NJ).
Human sera.
Serum samples from acute and chronic Q fever patients, Q-Vax vaccinees, and naive individuals are part of an archived Australian serum bank and were obtained from B. Marmion. Additional naive serum samples were obtained from donors at the University of California-Irvine. Seroreactivity (IgG) of serum samples against C. burnetii was determined by using an indirect fluorescent antibody (IFA) kit (Focus Diagnostic, Cypress, CA) according to the manufacturer's instructions. Fifty-five acute Q fever patient sera collected between 38 and 172 days after the onset of clinical symptom were confirmed as having phase II IgG IFA titers ranging from 1:160 to 1:5,120. Five chronic Q fever endocarditis patient sera had both phase I and phase II IFA IgG titers ranging from 160 to 1,280 and from 640 to 5,120, respectively. Thirty-two serum samples from naive individuals were confirmed as IFA negative. Human serum samples were used according to Institutional Review Board protocol 2003-0419 (J. E. Samuel).
Microarray detection of immunoreactive C. burnetii antigens.
Portions (10 μl) of 0.125% Tween 20 were mixed with 15 μl of each IVTT reaction (both His tag-positive and -negative reactions) to give a final concentration of 0.05% Tween 20. Then, 15-μl portions of IVTT reaction-Tween mixtures were then transferred to 384-well plates that were centrifuged at 1,600 × g to pellet any precipitated material. The supernatant was printed without further purification onto nitrocellulose-coated FAST glass slides (Schleicher & Schuell, Keene, NH) by using an OmniGrid 100 microarray printer (Genomic Solutions, Ann Arbor, MI). Protein microarray chip printing was conducted by Douglas Molina at Antigen Discovery, Inc., Irvine, CA. Prior to use with arrays, human sera (1:200 dilution) were incubated for 30 min with constant mixing in protein array blocking buffer (Whatman, Florham Park, NJ) that was supplemented with a lysate of E. coli at a final protein concentration 5 mg/ml (7). The arrays were rehydrated in blocking buffer for 30 min, incubated with the pretreated sera for 12 h at 4°C with constant agitation, washed in 10 mM Tris (pH 8.0)-150 mM NaCl containing 0.05% Tween 20 buffer, and then incubated with biotin-conjugated goat anti-human IgG (Fc-γ fragment-specific) serum (Jackson Immunoresearch, West Grove, PA) that was diluted 1:200 in blocking buffer. After the array slides were washed in 10 mM Tris (pH 8.0)-150 mM NaCl, bound antibodies were detected with streptavidin conjugated with the dye PBXL-3 (Martek, Columbia, MD). The washed and air-dried slides were scanned with a Perkin-Elmer ScanArray Express HT apparatus at a wavelength of 670 nm and with an output of RGB format TIFF files that were quantitated by using ProScanArray Express software (Perkin-Elmer, Waltham, MA) with correction for spot-specific background.
Cloning and expression of recombinant proteins.
ORFs corresponding to C. burnetii immunoreactive proteins were amplified by PCR and cloned into the pBAD/TOPO ThioFusion expression vector (Invitrogen). Recombinant proteins were expressed as His6-tagged fusion proteins in E. coli Top10 and purified by nickel affinity chromatography (Invitrogen).
Production of C. burnetii soluble antigen.
A bead-beaten solubilized fraction of C. burnetii termed CBUSF was prepared as a positive control antigen. Briefly, formaldehyde-inactivated C. burnetii bacteria were frozen in liquid nitrogen for 1 min, thawed at 37°C for 1 min, and then disrupted by using FastProtein Blue Matrix in a FastPrep instrument (Q-Biogene, Irvine, CA) according to the manufacturer's instructions. This cycle was repeated three times. The final pellet was resuspended at 2% (wt/vol) in N-lauroylsarcosine (Sigma, St. Louis, MO) in distilled H2O and mixed for 1 h. The supernatant was collected by centrifugation at 13,000 × g for 5 min and then dialyzed against distilled H2O. The final product was quantified by using a Micro-BCA protein assay kit (Pierce).
ELISA.
Microplates (96 well; Fisher Scientific, Pittsburgh, PA) were coated overnight at 4°C with 100 μl of a 2-μg/ml antigen solution. Plates were then blocked with 200 μl of 0.5% nonfat milk for 2 h at 37°C. Then, 50 μl of a 1:50 dilution of human serum was added to each well, and the plates were incubated for 1 h at 37°C. Plates were washed three times with PBST, and then 100 μl of a 1:5,000 dilution of biotin-labeled goat anti-human IgG gamma chain-specific antibody (Sigma) was added to each well. Plates were incubated for 1 h at 37°C, wells were washed three times with PBST, and then 100 μl of ABC solution from a Vectastain Elite PK-6100 kit (Vector Laboratories, Inc., Burlingame, CA) was added to each well. After a 1-h incubation at room temperature, the plates were washed with PBST, and the peroxidase activity was detected with o-phenylenediamine dihydrochloride (Sigma). Substrate reactions were developed for 15 min and then stopped with 100 μl of 1 M H2SO4. Reactions were measured at 490 nm by using a Spectra Max M2 (Molecular Devices, Sunnyvale, CA), and data were analyzed by GraphPad Prism (San Diego, CA). A positive cutoff for a given antigen was defined as the mean absorbance of IFA-negative samples plus two standard deviations. The percent sensitivity for a given antigen was calculated as the number of ELISA-positive sera from the IFA-positive pool divided by the number of IFA-positive sera. The percent specificity for a given antigen was calculated as the number of ELISA-negative sera from the IFA-negative pool divided by the number of IFA-negative sera.
Bioinformatics.
Potentially secreted C. burnetii antigens were identified by using a list of predicted C. burnetii secreted proteins compiled by the PathoSystems Resource Integration Center (PATRIC) (http://patric.vbi.vt.edu/special_projects/?amode=view&spId=8). The subcellular location of immunogenic proteins was predicted by using PSORTb.
RESULTS AND DISCUSSION
Synthesis and characterization of TAP products.
A total of 1,988 ORFs representing 97.2% of C. burnetii's 2,046 coding sequences were successfully amplified into TAP products (Fig. 1). Protein was synthesized from TAP products by IVTT and dot blots performed to ascertain the number of reactions that produced full-length recombinant protein. Based on a positive reaction between the Penta-His monoclonal antibody and the C-terminal His6 tag, full-length recombinant protein was produced from 1,492 TAP products representing 72.9% of the predicted C. burnetii proteome (data not shown). Based on signal intensities, there was obvious variation in the amount of protein produced by IVTT from individual TAP products. There were deficiencies in the production of the previously described C. burnetii antigens Hsp60 (CBU1718) (32), which was not amplified, and Com1 (CBU1910) (12), which was amplified into a TAP product but poorly expressed by IVTT.
Immunoproteome characterization by protein microarray.
Protein microarrays were generated by spotting His6 tag-positive and -negative IVTT reactions onto nitrocellulose-coated glass slides. Antigen-specific antibody responses to C. burnetii infection or vaccination were then assessed by individually probing microarrays with sera derived from five Q-Vax vaccinees, five Q fever patients (both early and late serum samples), and six naive individuals.
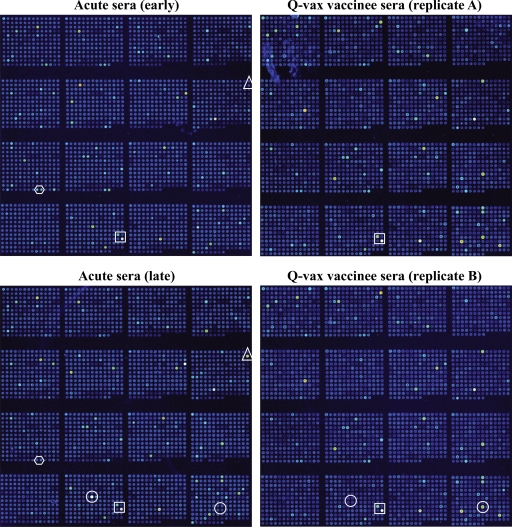
Representative fluorescence signals of probed microarrays are depicted in Fig. 2. Arrays probed with naive serum showed few immunoreactive proteins (data not shown). Conversely, arrays probed with early (20 days after the onset of clinical symptoms) and late (96 days after the onset of clinical symptoms) acute Q fever serum showed multiple immunoreactive proteins with stronger signal intensities for individual proteins generally associated with arrays probed with late immune sera. A subset of proteins was selectively recognized by early or late sera. Temporal development of antigen-specific antibody responses has been previously described in a guinea pig model of Q fever (6). Early antigens would likely be optimal as Q fever serodiagnostic antigens, while late antigens, with their more prominent role in development of protective immunity, may prove more efficacious in subunit vaccines. Although there was obvious overlap between proteins recognized by Q fever convalescent and Q-Vax vaccinee sera, each serum type also recognized a unique subset of proteins. Proteins selectively recognized by patient sera may represent secreted proteins that are present at low levels in killed whole-cell vaccine preparations (discussed in more detail below). Arrays probed in duplicate showed nearly identical fluorescence patterns, demonstrating the reproducibility of the microarray procedure. Some immunoreactive proteins were His tag negative, indicating IVTT can incompletely synthesize proteins to result in C-terminal truncations (data not shown).
FIG. 2.
Protein microarray analysis of the human humoral response to C. burnetii infection or vaccination. Microarrays were probed with paired “early” and “late” acute Q fever patient sera that was obtained at 20 and 96 days, respectively, after the onset of clinical Q fever (left panels) or in duplicate with serum from a human Q-Vax vaccinee (right panels). A laser confocal scanner was used to visualize reactive proteins with bound PBXL-3-conjugated streptavidin. Proteins within boxes are recognized by both Q fever patient and vaccinee sera. Proteins within circles are differentially recognized by Q fever patient and vaccinee sera. Proteins within hexagons and triangles are differentially recognized by early and late Q fever patient sera, respectively.
Comparison of TAP product and plasmid-based arrays.
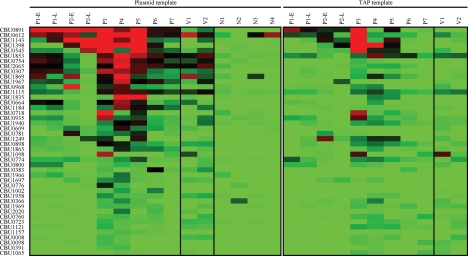
Results in our laboratory suggested that protein is more efficiently synthesized by IVTT from expression plasmids than from TAP fragments. Therefore, to directly compare the two methods, we cloned ORFs encoding the 44 most immunoreactive proteins identified above from the TAP product proteome array into a T7 expression plasmid. Proteins were then generated by IVTT from both plasmid and TAP templates and printed as single replicates onto smaller arrays. Arrays were then probed with 9 sera from acute Q fever patients (including two pairs of early and late sera), two Q-Vax vaccinee sera, and 16 naive sera. As depicted in Fig. 3 and detailed in Table 1, patient and vaccinee sera generally recognized the same proteins on plasmid- and TAP fragment-based arrays. However, the fluorescence intensity of individual proteins was generally greater on plasmid-based arrays, indicating more efficient IVTT protein production from plasmid templates.
FIG. 3.
Fluorescence intensity plots of plasmid- and TAP fragment-based protein microarrays probed with human sera. Plasmid- and TAP fragment-based protein microarrays were probed individually with sera from two Q-Vax vaccinees (V1 and V2) and seven acute Q fever patients (P1 to P7) that included paired early (E) and late (L) serum from patients 1 and 2. Arrays were also probed with sera from 16 C. burnetii-naive individuals (N). The average fluorescence intensity values were determined for each protein, with the intensity plot showing the most reactive proteins in red. Proteins are ordered from top (most reactive) to bottom (least reactive) according to the fluorescence intensity values of plasmid-based arrays. Four representative intensity plots of plasmid-based arrays probed with naive serum samples are shown, while only the intensity plots of TAP-fragment-based arrays probed with patient sera are shown.
TABLE 1.
Forty-four immunoreactive C. burnetii proteins identified by protein microarraya
| ORF no. | Protein function | ORF name | Signal peptideb | Predicted subcellular locationc | Avg array signal intensityd
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasmid
|
TAP
|
|||||||||
| P | V | N | P | V | N | |||||
| CBU0891 | Hypothetical exported membrane-associated protein | + | Unknown | 35,248 | 3,618 | 124 | 12,752 | 111 | - | |
| CBU0612 | Outer membrane protein OmpH | ompH | + | Outer membrane | 24,825 | 20,955 | 2,881 | 9,277 | 3,540 | 762 |
| CBU1143 | Preprotein translocase, YajC subunit | yajC | + | Noncytoplasmic | 23,756 | 3,222 | 665 | 9,927 | 521 | 10 |
| CBU1398 | 2-Oxoglutarate dehydrogenase | sucB | Cytoplasmic | 21,849 | -e | 224 | 11,471 | - | 31 | |
| CBU0545 | LemA protein | lemA | Unknown | 15,922 | 4,348 | 248 | 7,689 | 1,393 | - | |
| CBU1853 | GtrA family protein | Cytoplasmic membrane | 15,594 | 14,574 | 563 | 6,644 | 6,529 | 443 | ||
| CBU0754 | Efflux transporter, RND family, MFP subunit | + | Cytoplasmic membrane | 13,898 | 8,861 | 297 | 374 | - | - | |
| CBU2065 | Hypothetical exported protein | Cytoplasmic membrane | 13,577 | 15,646 | 440 | 1,276 | - | 35 | ||
| CBU0307 | OmpA-like transmembrane domain protein | + | Outer membrane | 12,930 | 5,479 | 60 | 643 | 219 | - | |
| CBU1869 | Hypothetical exported protein | + | Noncytoplasmic | 12,281 | 15,781 | 778 | 2,309 | 342 | 247 | |
| CBU1967 | Drug resistance transporter, Bcr/CflA family | + | Cytoplasmic membrane | 10,013 | 6,071 | 46 | 812 | 322 | - | |
| CBU0968 | Phospholipase D | + | Cytoplasmic membrane | 9,850 | 634 | 265 | 1,213 | 1,360 | 18 | |
| CBU1115 | Hypothetical protein | Unknown | 9,085 | 10,329 | 498 | 3,435 | 5,584 | 227 | ||
| CBU1835 | Protoporphyrinogen oxidase | Cytoplasmic | 8,558 | 427 | 28 | 1,148 | - | - | ||
| CBU0664 | Transposase, ISAs1 family | Cytoplasmic | 8,422 | 3,334 | 712 | 740 | - | 73 | ||
| CBU1184 | Acyltransferase family protein | Unknown | 7,432 | 2,943 | 49 | 296 | 495 | - | ||
| CBU0718 | Hypothetical membrane-associated protein | Unknown | 6,335 | - | 199 | 3,461 | - | - | ||
| CBU0935 | RNA-binding protein | + | Unknown | 5,997 | 3,258 | 304 | 3,619 | 1,182 | 311 | |
| CBU1940 | ATP synthase F0, C subunit | Cytoplasmic membrane | 4,539 | 3,128 | 401 | 1,931 | 1,364 | 76 | ||
| CBU0609 | Mevalonate kinase | Unknown | 4,234 | 1,206 | 73 | 54 | 793 | - | ||
| CBU0781 | Ankyrin repeat protein | ankG | Secreted | 4,186 | - | 420 | 561 | 152 | 52 | |
| CBU1249 | DNA-binding protein | Unknown | 4,017 | 467 | 324 | 5,260 | 1,002 | 95 | ||
| CBU0898 | Thyroglobulin type 1 repeat domain protein | Unknown | 3,914 | 3,834 | 135 | 2,427 | 900 | 58 | ||
| CBU1865 | Hypothetical membrane-associated protein | Cytoplasmic membrane | 3,225 | 758 | 246 | 688 | 444 | 85 | ||
| CBU1098 | Hypothetical cytosolic protein | Cytoplasmic | 3,142 | 4,049 | 71 | 2,468 | 9,747 | 19 | ||
| CBU0774 | Stress-responsive transcriptional regulator PspC | pspC | Unknown | 2,983 | 835 | 335 | 2,984 | 2,145 | 237 | |
| CBU0800 | Hypothetical protein | Unknown | 1,817 | 1,064 | 31 | 1,051 | 945 | 27 | ||
| CBU0383 | DNA-3-methyladenine glycosidase I | tag | Unknown | 1,790 | 3,220 | 58 | 285 | - | 152 | |
| CBU1966 | Glutamyl-tRNA reductase | hemA | Cytoplasmic | 1,624 | - | 172 | 23 | - | - | |
| CBU1697 | Endonuclease III | nth | Unknown | 1,620 | 1,961 | 22 | 669 | - | 72 | |
| CBU0776 | ABC transporter, ATP-binding protein | Cytoplasmic membrane | 1,448 | - | 94 | 2 | - | 9 | ||
| CBU1002 | Biotin operon repressor/biotin synthetase | birA | Unknown | 1,254 | - | 8 | 3 | 163 | - | |
| CBU1958 | Hypothetical ATPase | Unknown | 1,248 | 3,259 | 119 | 394 | 225 | 46 | ||
| CBU0366 | Phosphate regulon sensor protein PhoR | phoR | Cytoplasmic membrane | 1,010 | - | 456 | 944 | 1,009 | 400 | |
| CBU1969 | DnaK suppressor protein | dksA | Cytoplasmic | 1,007 | 765 | 81 | 793 | 1,732 | 26 | |
| CBU2020 | Glutamate/gamma-aminobutyrate antiporter | Cytoplasmic membrane | 1,004 | 601 | - | 78 | - | 57 | ||
| CBU0760 | Sensor protein GacS | Unknown | 749 | 1,423 | 37 | 1,036 | 640 | 23 | ||
| CBU0723 | Hypothetical protein | Unknown | 704 | 2,520 | 126 | 683 | 920 | 172 | ||
| CBU1121 | Hypothetical protein | Unknown | 672 | 1,724 | 79 | 1,295 | 778 | 90 | ||
| CBU1157 | Hypothetical exported lipoprotein | Unknown | 522 | - | 26 | 21 | - | 23 | ||
| CBU0008 | Hypothetical protein | Unknown | 513 | 2,408 | 109 | 494 | 1,377 | 89 | ||
| CBU0098 | Nicotinate-nucleotide pyrophosphorylase | nadC | Cytoplasmic | 468 | - | 11 | 504 | 1,010 | 5 | |
| CBU0391 | Riboflavin biosynthesis protein RibF | ribF | Cytoplasmic | 453 | - | 6 | 96 | 588 | 22 | |
| CBU1065 | 2′-5′ RNA ligase | Cytoplasmic | 158 | 323 | 45 | 643 | 726 | 2 | ||
Immunoreactive proteins ordered from top (most reactive) to bottom (least reactive) based on average fluorescence intensity values of plasmid-based arrays probed Q fever patient sera.
Predicted signal peptide as determined by SignalP.
Predicted subcellular location as determined by Psort.
P, Q fever patient sera; V, Q-Vax vaccinee sera; N, naive sera.
-, signal absent with all sera.
Features of immunoreactive proteins.
Thirteen of the forty-four immunogenic proteins are annotated as hypothetical proteins (22) (Table 1). Based on predicted signal peptides, 9 immunoreactive proteins among the top 44 (20.5%) were predicted to be secreted into the periplasm or beyond. Eight of these proteins were among the top fifteen (53.3%), a finding which contrasts markedly with the percentage of predicted secreted proteins within the entire proteome (10.2%). A similar bias in signal sequence-containing proteins was observed in a proteome microarray screen of F. tularensis antigens (10, 25). In addition to a type II secretory pathway, C. burnetii encodes a Dot/Icm type IV secretion system that translocates effector molecules into the host cell cytosol (19). Immunoreactive CBU0781 (AnkG), an ankyrin repeat domain-containing protein, has been recently shown to be secreted in a Dot/Icm-dependent manner (19). Secreted immunogens that disassociate with C. burnetii are likely present in very low amounts in the Q-Vax vaccine. Consistent with this idea, AnkG's patient to vaccinee average fluorescence array signal ratio was 489 to 1. This contrasts with a ratio of 2.1 to 1 for the most reactive protein CBU0891, a hypothetical membrane associated protein (Table 1).
A few C. burnetii antigens have been identified by immunoscreening a phage expression library (32) and proteins separated by two-dimensional gel electrophoresis (6). Among the 16 proteins showing the strongest average fluorescence signals by microarray, CBU0891, CBU1398 (2-oxglutarate dehydrogenase, SucB), and CBU0664 (ISAs1 family transposase) were previously identified as immunogens by Zhang et al. (32), indicating reproducibility between microarray and conventional antigen screening procedures. Antibody recognition of SucB indicates cytosolic housekeeping enzymes are exposed to the host's immune system. Indeed, in this report and elsewhere (6, 32), additional C. burnetii housekeeping enzymes, such as isoleucyl-tRNA synthetase and isocitrate dehydrogenase, have been identified in antigen screens. Moreover, metabolic enzymes of other bacterial pathogens are known to trigger B-cell responses (26, 30).
ELISA validation of protein microarray data.
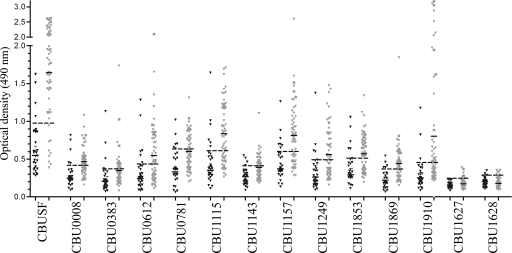
To validate protein microarray results using another serological method, 10 reactive proteins showing a range of average fluorescence intensities were purified as recombinant His6-tagged protein and tested by ELISA. The 10 proteins chosen for testing were: CBU0008 (hypothetical protein), CBU0381 (DNA-3-methyladenine glycosidase), CBU0612 (outer membrane protein OmpH), CBU0781 (AnkG), CBU1115 (hypothetical protein), CBU1143 (preprotein translocase YajC), CBU1157 (hypothetical exported lipoprotein) CBU1249 (putative DNA-binding protein), CBU1853 (GtrA family protein), and CBU1869 (hypothetical exported protein). CBU0891 was not tested because of difficulty in obtaining recombinant protein. Recombinant His6-tagged Com1 (CBU1910), a characterized C. burnetii protein antigen (12), and C. burnetii cell extracts (CBUSF), were included as positive controls. As negative controls, recombinant IcmE and IcmK, two array-negative proteins that are part of the organism's type IV secretion system (29), were included.
ELISA was conducted using 55 and 5 serum samples from human acute and chronic Q fever patients, respectively, that tested positive for C. burnetii antibodies by immunofluorescence assay (see Materials and Methods). ELISA was also conducted using 32 IFA-negative human sera, including the 16 naive sera used in microarray screens. Seven of ten recombinant proteins were positive by ELISA when tested with sera from acute Q fever patients. When tested with sera from endocarditis patients, all recombinant proteins were positive (Fig. 4 and Table 2). Consistent with microarray results, the negative control proteins IcmE and IcmK were negative by ELISA. We did not observe a clear correlation between IFA titer and responses by ELISA to specific recombinant proteins. However, convalescent human sera with high IFA titers consistently demonstrated positive reactivity against all recombinant proteins, whereas several convalescent-phase sera with low IFA titers did not react strongly with any recombinant protein (data not shown). Sensitivity and specificity for the 10 recombinant proteins ranged from 31.6 to 61.6% to 78.1 to 90.0%, respectively. CBUSF gave the highest sensitivity at 85%. While none of the individual recombinant proteins provided complete coverage for IFA-positive samples, six out ten proteins showed greater sensitivities than the positive control CBU1910 (Com1). Combining several recombinant proteins significantly increased sensitivity but also moderately reduced specificity (data not shown). While a final recombinant protein mixture equal to or superior to CBUSF was not defined, these studies did support the concept that a limited multiplex assay can be developed to replace whole-cell reagents in a new Q fever diagnostic.
FIG. 4.
ELISA validation of immunogenic proteins identified by microarray. Recombinant proteins corresponding to 10 antigens identified by protein microarray were generated and tested by ELISA for IgG reactivity of human sera. A sampling of proteins that ranged from strongly (e.g., CBU1143) to weakly (e.g., CBU0008) reactive were tested. Recombinant Com1 (CBU1910), a characterized C. burnetii protein antigen, was included as a positive control. Recombinant IcmE and IcmK, which tested negative by protein microarray, were included as controls. ELISA was conducted on 55 and 5 serum samples from human acute (gray triangles) and chronic Q fever patients (boxes), respectively, that tested positive for C. burnetii antibodies by indirect immunofluorescence assay (see Materials and Methods). ELISA was also conducted on 32 IFA-negative sera (black triangles). Black lines denote the mean reactivity of positive and negative sera. Dashed black lines denote the positive cutoff, which was calculated as the mean ELISA reactivity of IFA-negative sera samples plus two times the standard deviation. CBUSF, C. burnetii soluble fraction.
TABLE 2.
ELISA reactivity of human sera to recombinant C. burnetii proteins
| ORF | Protein function | %
|
Ratioc (positive/negative) | |
|---|---|---|---|---|
| Specificitya | Sensitivityb | |||
| CBUSFd | 87.5 | 85 | 2.9 | |
| CBU0008 | Hypothetical protein | 84.0 | 60.0 | 1.8 |
| CBU0383 | DNA-3-methyladenine glycosylase | 87.5 | 31.6 | 1.7 |
| CBU0612 | Outer membrane protein OmpH | 81.2 | 51.6 | 2.2 |
| CBU0781 | Ankyrin repeat protein | 81.3 | 40.0 | 1.8 |
| CBU1115 | Hypothetical protein | 81.2 | 56.6 | 2.3 |
| CBU1143 | Protein translocase subunit YajC | 90.6 | 33.3 | 1.6 |
| CBU1157 | Hypothetical exported lipoprotein | 78.1 | 61.6 | 2.2 |
| CBU1249 | DNA-binding protein | 87.5 | 45.0 | 2.1 |
| CBU1853 | GtrA family protein | 84.3 | 56.6 | 1.8 |
| CBU1869 | Hypothetical exported protein | 90.0 | 55.0 | 2 |
| CBU1910 | Outer membrane protein Com1 | 90.0 | 50.0 | 3.1 |
| CBU1627 | IcmE | 96.9 | 11.6 | 1.1 |
| CBU1628 | IcmK | 90.6 | 8.3 | 0.9 |
The percent specificity was calculated as the number of ELISA-negative sera from the IFA-negative pool divided by the number of IFA-negative sera.
The percent sensitivity was calculated as the number of ELISA-positive sera from the IFA-positive pool divided by the number of IFA-positive sera.
The positive/negative ratio was calculated as the mean OD490 of proteins probed with IFA-positive sera.
That is, the C. burnetii soluble fraction.
While the primary goal of the present study was to identify C. burnetii proteins with serodiagnostic potential, some of the identified immunogens may be candidates for a Q fever subunit vaccine. Both cell-mediated and humoral immune responses are important for protection against Q fever (33), and accumulating evidence suggests that an efficacious vaccine based on recombinant antigen is feasible (27, 31, 34). Indeed, several purified proteins including CBU0781 (AnkG), CBU1157 (lipoprotein), and CBU1143 (YajC) induce strong gamma interferon recall responses in purified CD4+ T cells of vaccinated or infected mice (J. E. Samuel, unpublished data). Interestingly, YajC, an inner membrane protein involved in Sec-dependent secretion (9), is both a B-cell and a T-cell antigen of Brucella abortus (28). Testing of microarray-identified antigens for T-cell antigenicity does not require purified recombinant protein as IVTT-produced antigen, either coupled to latex beads by an affinity tag or contained within in crude IVTT lysates, both effectively stimulate T cells in proliferation assays (13, 14).
In summary, we have used a high-throughout proteome microarray screening method to identify C. burnetii proteins recognized by the human humoral immune response to C. burnetii infection and vaccination. This screening method allows profiling of antibody responses of large cohorts of infected and/or vaccinated animals and humans to rapidly identify consensus immunodominant antigens on a whole-proteome basis. Identification of immunodominant antigens of C. burnetii will aid in the development of safe and effective recombinant protein-based vaccines and reliable serodiagnostic tests that do not require biosafety level 3 facilities for antigen production. We are currently developing a second-generation C. burnetii IVTT-based microarray that includes proteins unique to two chronic Q fever endocarditis isolates (G and K) (3). Comparisons of the immunoproteome of acute and chronic disease isolates may identify unique serodiagnostic antigens that allow differentiation of acute and chronic Q fever.
Acknowledgments
We thank Harlan Caldwell for critical review of the manuscript, Chad Burk, and Siddiqua Hirst for microarray probing and analysis, and Shahed Toosi for assistance in molecular cloning.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R.A.H.) and by the Public Heath Service grants U54 AI057156, R01 AI057768, U54 AI065359, and U01 AI061363 from the National Institute of Allergy and Infectious Diseases (J.E.S. and P.L.F.).
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., A. Jasinskas, M. A. Kayala, D. H. Davies, A. C. Steere, P. Baldi, and P. L. Felgner. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect. Immun. 76:3374-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beare, P. A., S. F. Porcella, R. Seshadri, J. E. Samuel, and R. A. Heinzen. 2005. Preliminary assessment of genome differences between the reference Nine Mile Isolate and two human endocarditis Isolates of Coxiella burnetii. Ann. N. Y. Acad. Sci. 1063:64-67. [DOI] [PubMed] [Google Scholar]
- 4.Brinkman, M. B., M. McKevitt, M. McLoughlin, C. Perez, J. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 44:888-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockrell, D. C., P. A. Beare, E. R. Fischer, D. Howe, and R. A. Heinzen. 2008. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J. Microbiol. Methods 72:321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, S. A., E. R. Fischer, D. C. Cockrell, D. E. Voth, D. Howe, D. J. Mead, J. E. Samuel, and R. A. Heinzen. 2007. Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect. Immun. 75:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. H., X. Liang, J. E. Hernandez, A. Randall, S. Hirst, Y. Mu, K. M. Romero, T. T. Nguyen, M. Kalantari-Dehaghi, S. Crotty, P. Baldi, L. P. Villarreal, and P. L. Felgner. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 102:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. H., M. M. McCausland, C. Valdez, D. Huynh, J. E. Hernandez, Y. Mu, S. Hirst, L. Villarreal, P. L. Felgner, and S. Crotty. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell. Mol. Life Sci. 60:2034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyles, J. E., B. Unal, M. G. Hartley, S. L. Newstead, H. Flick-Smith, J. L. Prior, P. C. Oyston, A. Randall, Y. Mu, S. Hirst, D. M. Molina, D. H. Davies, T. Milne, K. F. Griffin, P. Baldi, R. W. Titball, and P. L. Felgner. 2007. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 7:2172-2183. [DOI] [PubMed] [Google Scholar]
- 11.Gat, O., H. Grosfeld, N. Ariel, I. Inbar, G. Zaide, Y. Broder, A. Zvi, T. Chitlaru, Z. Altboum, D. Stein, S. Cohen, and A. Shafferman. 2006. Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect. Immun. 74:3987-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrix, L. R., L. P. Mallavia, and J. E. Samuel. 1993. Cloning and sequencing of Coxiella burnetii outer membrane protein gene com1. Infect. Immun. 61:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jing, L., D. H. Davies, T. M. Chong, S. Chun, C. L. McClurkan, J. Huang, B. T. Story, D. M. Molina, S. Hirst, P. L. Felgner, and D. M. Koelle. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 82:7120-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez, J. E., P. A. Beare, R. A. Heinzen, J. Norimine, K. K. Lahmers, G. H. Palmer, and W. C. Brown. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 332:129-141. [DOI] [PubMed] [Google Scholar]
- 15.Marmion, B. 2007. Q fever: the long journey to control by vaccination. Med. J. Aust. 186:164-166. [DOI] [PubMed] [Google Scholar]
- 16.Marmion, B. P., R. A. Ormsbee, M. Kyrkou, J. Wright, D. A. Worswick, A. A. Izzo, A. Esterman, B. Feery, and R. A. Shapiro. 1990. Vaccine prophylaxis of abattoir-associated Q fever: eight years’ experience in Australian abattoirs. Epidemiol. Infect. 104:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mora, M., C. Donati, D. Medini, A. Covacci, and R. Rappuoli. 2006. Microbial genomes and vaccine design: refinements to the classical reverse vaccinology approach. Curr. Opin. Microbiol. 9:532-536. [DOI] [PubMed] [Google Scholar]
- 19.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regis, D. P., C. Dobano, P. Quinones-Olson, X. Liang, N. L. Graber, M. E. Stefaniak, J. J. Campo, D. J. Carucci, D. A. Roth, H. He, P. L. Felgner, and D. L. Doolan. 2008. Transcriptionally active PCR for antigen identification and vaccine development: in vitro genome-wide screening and in vivo immunogenicity. Mol. Biochem. Parasitol. 158:32-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scola, B. L. 2002. Current laboratory diagnosis of Q fever. Semin. Pediatr. Infect. Dis. 13:257-262. [DOI] [PubMed] [Google Scholar]
- 22.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon, J. G., and R. A. Heinzen. 2007. Infection of human monocyte-derived macrophages with Coxiella burnetii. Methods Mol. Biol. 431:189-200. [DOI] [PubMed] [Google Scholar]
- 24.Sharma, J., Y. Zhong, F. Dong, J. M. Piper, G. Wang, and G. Zhong. 2006. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect. Immun. 74:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundaresh, S., A. Randall, B. Unal, J. M. Petersen, J. T. Belisle, M. G. Hartley, M. Duffield, R. W. Titball, D. H. Davies, P. L. Felgner, and P. Baldi. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23:i508-i518. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira-Gomes, A. P., A. Cloeckaert, G. Bezard, R. A. Bowden, G. Dubray, and M. S. Zygmunt. 1997. Identification and characterization of Brucella ovis immunogenic proteins using two-dimensional electrophoresis and immunoblotting. Electrophoresis 18:1491-1497. [DOI] [PubMed] [Google Scholar]
- 27.Varghees, S., K. Kiss, G. Frans, O. Braha, and J. E. Samuel. 2002. Cloning and porin activity of the major outer membrane protein P1 from Coxiella burnetii. Infect. Immun. 70:6741-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vemulapalli, R., A. J. Duncan, S. M. Boyle, N. Sriranganathan, T. E. Toth, and G. G. Schurig. 1998. Cloning and sequencing of yajC and secD homologs of Brucella abortus and demonstration of immune responses to YajC in mice vaccinated with B. abortus RB51. Infect. Immun. 66:5684-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel, J. P. 2004. Turning a tiger into a house cat: using Legionella pneumophila to study Coxiella burnetii. Trends Microbiol. 12:103-105. [DOI] [PubMed] [Google Scholar]
- 30.Voland, P., D. L. Weeks, D. Vaira, C. Prinz, and G. Sachs. 2002. Specific identification of three low molecular weight membrane-associated antigens of Helicobacter pylori. Aliment. Pharmacol. Ther. 16:533-544. [DOI] [PubMed] [Google Scholar]
- 31.Williams, J. C., T. A. Hoover, D. M. Waag, N. Banerjee-Bhatnagar, C. R. Bolt, and G. H. Scott. 1990. Antigenic structure of Coxiella burnetii: a comparison of lipopolysaccharide and protein antigens as vaccines against Q fever. Ann. N. Y. Acad. Sci. 590:370-380. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, G., K. Kiss, R. Seshadri, L. R. Hendrix, and J. E. Samuel. 2004. Identification and cloning of immunodominant antigens of Coxiella burnetii. Infect. Immun. 72:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, G., and J. E. Samuel. 2004. Vaccines against Coxiella infection. Expert Rev. Vaccines 3:577-584. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y. X., N. Zhi, S. R. Yu, Q. J. Li, G. Q. Yu, and X. Zhang. 1994. Protective immunity induced by 67 K outer membrane protein of phase I Coxiella burnetii in mice and guinea pigs. Acta Virol. 38:327-332. [PubMed] [Google Scholar]