Abstract
The genome of the basidiomycete pathogenic yeast Cryptococcus neoformans carries two UDP-glucose epimerase genes (UGE1 and UGE2). UGE2 maps within a galactose cluster composed of a galactokinase homologue gene and a galactose-1-phosphate uridylyltransferase. This clustered organization of the GAL genes is similar to that in most of the hemiascomycete yeast genomes and in Schizosaccharomyces pombe but is otherwise not generally conserved in the fungal kingdom. UGE1 has been identified as necessary for galactoxylomannan biosynthesis and virulence. Here, we show that UGE2 is necessary for C. neoformans cells to utilize galactose as a carbon source at 30°C but is not required for virulence. In contrast, deletion of UGE1 does not affect cell growth on galactose at this temperature. At 37°C, a uge2Δ mutant grows on galactose in a UGE1-dependent manner. This compensation by UGE1 of UGE2 mutation for growth on galactose at 37°C was not associated with upregulation of UGE1 transcription or with an increase of the affinity of the enzyme for UDP-galactose at this temperature. We studied the subcellular localization of the two enzymes. Whereas at 30°C, Uge1p is at least partially associated with intracellular vesicles and Uge2p is on the plasma membrane, in cells growing on galactose at 37°C, Uge1p colocalizes with Uge2p to the plasma membrane, suggesting that its activity is regulated through subcellular localization.
Cryptococcus neoformans is an environmental microbe responsible for severe diseases in immunocompromised individuals (4). It is found in soils, decaying vegetation, and bird droppings. C. neoformans is thought to be acquired early in life and to stay in dormancy until an immune defect occurs (11). Then, C. neoformans multiplies and disseminates to various organs, including the central nervous system, where it causes meningoencephalitis that is fatal if untreated. Its main virulence factor is the polysaccharide capsule, which is essential for virulence. This capsule is composed mainly of two polysaccharides, a large molecule called glucuronoxylomann (made of an α-mannose chain with glucuronic acid, xylose, and O-acetyl residues) and a smaller molecule called galactoxylomannan (GalXM) (made of a galactose chain with mannose and xylose residues) (for reviews, see references 21 and 3).
We have been conducting a program of systematic deletion of all genes potentially involved in capsule polysaccharide biosynthesis. Analysis of virulence and organ dissemination profiles of the resulting mutant strains led to the identification of a gene named UGE1 as necessary for GalXM biosynthesis and virulence (28). UGE1 encodes a putative UDP-glucose epimerase, an enzyme involved in galactose metabolism. Confirming the central role of galactose metabolism in C. neoformans virulence, we have also described a second gene, named UGT1, encoding a putative UDP-galactose transporter and similarly necessary for GalXM biosynthesis and virulence (28).
In species ranging from Escherichia coli to mammals, galactose is metabolized via a series of reactions collectively known as the Leloir pathway (reviewed in reference 17). A set of four enzymes (galactose mutarotase, galactokinase, galactose-1-phosphate uridylyltransferase, and UDP-galactose epimerase) converts β-d-galactose into glucose-1-phosphate. In Saccharomyces cerevisiae, GAL1 and GAL7 encode the galactokinase and galactose-1-phosphate uridylyltransferase, respectively; the mutarotase and UDP-galactose epimerase activities are catalyzed by a single protein encoded by the gene GAL10 (7). Mutation of the epimerase-mutarotase gene or the galactose-1-phosphate uridyl transferase gene results in galactose sensitivity: these mutant strains stop growing in response to even trace quantities of galactose even in the presence of an alternative carbon source (glycerol/ethanol) (33). Similarly, in humans, mutation in one these genes results the inherited metabolic disorder named galactosemia (31). Chinese hamster ovary cells lacking UDP-galactose epimerase activities exhibit impaired growth when exposed to galactose at concentrations of above 0.125 mM, with this phenotype being bypassed by the addition of uridine to the culture medium (36).
Surprisingly, in C. neoformans, the deletion of UGE1 is not associated with galactose sensitivity. Thus, a uge1Δ mutant can grow on galactose at 30°C, and the addition of galactose to the medium bypasses the thermosensitive phenotype associated with this gene deletion (28). Analysis of the C. neoformans genome sequence identified a UGE1 gene paralogue which we named UGE2. Here, we report an investigation of how UGE1 and UGE2 contribute to galactose metabolism, GalXM biosynthesis, and C. neoformans virulence by regulating the equilibrium between UDP-galactose and UDP-glucose.
MATERIALS AND METHODS
Strains and culture conditions.
The C. neoformans strains used in this study all originated from the serotype A strain KN99α (30) and are listed in Table SA in the supplemental material. The strains were routinely cultured on yeast extract-peptone-dextrose (YPD) medium at 30°C (39). Synthetic dextrose was prepared as described previously (39). The capsule sizes were estimated after 24 h of growth in capsule-inducing medium at 30°C as previously described (22). Production of melanin and urease was assessed after spotting 105 cells of each strain on l-Dopa or Christensen agar medium, respectively (32, 44); the plates were read after 48 h of incubation at 30°C. The bacterial strain Escherichia coli XL1-Blue (Stratagene, La Jolla, CA) was used for the propagation of all plasmids.
RNA extraction and Northern blot analysis.
Cells were grown in YPD liquid culture to a density of 5 × 107 cells/ml. RNA was extracted with Trizol reagent (Invitrogen) following the manufacturer's instructions. Total RNA (10 μg) was separated by denaturing agarose gel electrophoresis, transferred onto a Hybond-N+ membrane (Amersham), and probed with [32P]dCTP-radiolabeled DNA fragments. The banding pattern was quantified with a Typhoon 9200 imager and Image Quantifier 5.2 software (Molecular Dynamics).
Strain construction.
The genes described in this report were deleted by transforming the KN99α strain with a disruption cassette constructed by overlapping PCR as previously described (27). The primer sequences used are given in Table SB in the supplemental material. The transformants were then screened for homologous integration, first by PCR and then by Southern blotting, as previously described (29). The tagged plasmids, pNATSTM and pHYGSMT, used to amplify the selective marker were kindly provided by Jennifer Lodge (St. Louis University School of Medicine). The uge2Δ strain was reconstituted by inserting a 3.9-kbp PCR-amplified fragment (using the primers UGE2-5′5 and UGE2-3′3) between the KspI and SpeI sites of a plasmid containing a nourseothricin resistance cassette (18). The resulting plasmid, pNE394, was digested with SpeI and used to transform strain NE369 (MATα uge2Δ) by biolistic DNA delivery. Transformants were selected on YPD medium containing 100 μg/ml of nourseothricin (Werner BioAgents). Three nourseothricin-resistant strains were obtained; all grew on galactose. Two were stored at −80°C for further studies.
Recombinant protein production.
UGE1 and UGE2 cDNAs were amplified by PCR and inserted into the pQ-30 E. coli expression vector (Qiagen). The E. coli BL21 transformant strains were grown in 50 ml of yeast extract tryptone medium containing ampicillin (50 μg/ml) and kanamycin (30 μg/ml) to an optical density at 600 nm of 0.5; gene expression was induced by addition of 50 μM of IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation overnight at room temperature. The cells were then disrupted by sonication and centrifuged at 3,000 × g. The supernatant was recovered, and the recombinant proteins were purified by affinity chromatography on an Ni-nitrilotriacetic acid column (Qiagen) following the manufacturer's procedures. The protein solution was adjusted to 20% (wt/vol) glycerol (final concentration, 700 μg/ml) and stored in aliquots at −80°C.
UDP-galactose epimerase assay.
Epimerase activity was assayed using an NADH-coupled assay developed by Wilson and Hogness (46) with some minor modifications. The 1-ml assay mixture consisted of 100 mM glycine buffer (pH 8.7), 1 mM β-NAD+ (Sigma), and 0.8 mM UDP-galactose (Sigma). The reaction was started by adding 10 μl of epimerase (140 μg/ml) in 50 mM Tris HCl (pH 7.6)-1% bovine serum albumin-1 mM dithiothreitol-1 mM EDTA-1 mM β-NAD+ and stopped by incubation for 10 min at 100°C. The UDP-glucose produced was determined by addition of 0.04 unit of bovine UDP-glucose dehydrogenase (Sigma) and incubation for 10 min at 30°C; the increase in absorbance due to the formation of NADH was then measured at 340 nm. Km values were determined by varying the UDP-galactose concentration between 0.4 mM and 3.2 mM. The experiment was conducted in triplicate.
Subcellular localization with fluorescent protein fusions.
To localize the Uge proteins, the UGE1 and UGE2 genes under the control of their own promoters were joined in frame to the sequences encoding the green fluorescent protein or the DsRed protein at their C-terminal ends. Primers used for amplification are listed in Table SB in the supplemental material. The uge1Δ and uge2Δ strains and the uge1Δ uge2Δ double mutant were transformed with plasmids containing the Hyg or the Neo selectable marker and the Uge-fluorescent protein fusion by biolistic delivery (42). Tagged fluorescent UGE1 versions were integrated in the genome using the Hyg marker. It should be noted that the UGE1 deletion is associated with hypersensitivity to hygromycin such that the uge1Δ::NAT1 uge2Δ::HYG1 double mutants were sensitive to hygromycin (data not shown), and this allowed the reutilization of this marker for the reintroduction of this gene. Tagged fluorescent UGE2 versions were integrated in the genome using the Neo selectable marker. Transformants were analyzed for fluorescence under various growth conditions.
Expression of UGE1 and UGE2 in S. cerevisiae.
UGE1 and UGE2 cDNAs were amplified by PCR and inserted into the vector pCM190 (12). The Euroscarf S. cerevisiae strain BY4742 (Matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gal10::kanMX4) was transformed using a lithium acetate procedure (13) and tested on 1% galactose medium.
Analysis of the sugar composition.
Cells were grown at 30°C or 37°C for 3 days in capsule induction medium. After elimination of the cells by centrifugation and filtration, the total polysaccharide in the culture supernatant was precipitated with ethanol, resuspended in distilled water, filtered, and lyophilized. The polysaccharide compositions were analyzed by gas-liquid chromatography after acid hydrolysis with trifluoroacetic acid as previously described (35).
Virulence assays.
Six-week-old male BALB/c mice (Charles River Laboratories, France) were used for all in vivo experiments. Strains were grown on YPD medium at 30°C overnight. For the dissemination assay, 105 C. neoformans cells were injected into the tail vein of each mouse. On day 1, the mice were killed and the brain, spleen, and lungs were recovered. Each organ was homogenized, and adapted dilutions were plated on Sabouraud-chloramphenicol plates. The experiments were repeated independently three times.
The relative virulence of the strains was evaluated by intravenously inoculating groups of seven mice with 105 cells of each cryptococcal strain. Animals were observed daily, and any deaths were recorded.
Gene identification.
The Cryptococcus neoformans var. grubii Uge2p sequence we used as a query in BLASTP searches against the proteomes of 36 fungi (Ashbya gossypii, Aspergillus fumigatus, Aspergillus nidulans, Aspergillus terreus, Batrachochytrium dendrobatidis, Botrytis cinerea, Candida albicans, Candida glabrata, Candida guillermondii, Candida lusitaniae, Candida tropicalis, Chaetomium globosum, Coccidioides immitis, Coprinus cinereus, Cryptococcus neoformans var. neoformans, Debaryomyces hansenii, Fusarium graminearum, Fusarium oxysporum, Fusarium verticillioides, Histoplasma capsulatum, Kluyveromyces lactis, Lodderomyces elongisporus, Magnaporthe grisea, Neosartorya fischeri, Neurospora crassa, Phanerochaete chrysosporium, Pichia stipitis, Puccinia graminis, Rhizopus oryzae, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Sclerotinia sclerotiorum, Stagonospora nodorum, Uncinocarpus reesei, Ustilago maydis, and Yarrowia lipolytica). For each query, the top hits were retained provided that their E values were less than 1e−20. Proteins sharing at least 25% sequence identity with Cryptococcus neoformans var. grubii Uge2p as analyzed with ClustalW were selected (see Table SC in the supplemental material). Phylogenetic analysis was performed via the server http://www.phylogeny.fr/, using MUSCLE (9) for the multiple alignment, Gblocks (6) to remove the poorly aligned positions, and PHYML (14) for tree reconstruction. Branch support was calculated using the approximate likelihood ratio test described previously (1). The substitution model used was WAG plus gamma distribution with an estimated alpha parameter and an estimated proportion of invariable sites.
RESULTS
C. neoformans possesses two putative UDP-glucose epimerase genes in its genome.
The gene UGE1 is necessary for GalXM biosynthesis and virulence in C. neoformans (28). BLAST analysis revealed the presence of a paralogue of UGE1, called UGE2, in the C. neoformans var. grubii genome. Uge1p and Uge2p share 65% amino acid sequence identity (Fig. 1A). Hydropathy analysis indicates that both proteins have a potential N-terminal transmembrane domain (Fig. 1B); both sequences include an epimerase motif, but neither has a mutaronase domain similar to that in the Gal10 protein of S. cerevisiae (25). In fact, we were unable to identify any putative maturonase gene in the C. neoformans genome (24). The presence of two UDP-glucose epimerases has not previously been described for organisms in which this enzyme has been mostly studied (S. cerevisiae, E. coli, and human), where only one UDP-glucose epimerase equilibrates the UDP-glucose/UDP-galactose ratio (17). We looked for putative UDP-glucose epimerase genes in the large number of fungal genomes now sequenced and annotated (Table 1). We identified at least one GAL10 orthologue in most fungi but not in Candida glabrata and Ashbya gossypii (8, 16). The list of fungi covered most of the evolutionary tree of fungi, and although not statistically representative because some genera are clearly overrepresented, it is clear that the presence of more than one UDP-glucose epimerase is not unusual.
FIG. 1.
(A) Amino acid alignment of the two C. neoformans Uge sequences. (B) Hydropathy profiles of the Uge proteins determined by TMHMM1.0 analysis.
TABLE 1.
Putative GAL10 paralogues in fungi
| Fungus | UDP-glucose epimerase-homologous gene(s) | Clustering of GAL1 with one GAL10 paralogues |
|---|---|---|
| Ashbya gossypii | None | |
| Aspergillus fumigatus | Afu3g07910, Afu4g14090, Afu5g10780 | No |
| Aspergillus nidulans | AN2951, AN4727 | No |
| Aspergillus terreus | ATEG 07631, ATEG 01678 | No |
| Batrachochytrium dendrobatidis | BDEG07987 | No |
| Botrytis cinerea | BC1G 0445, BC1G11467 | No |
| Candida albicans | GAL10 | Yes |
| Candida glabrata | None | |
| Candida guillermondii | PCGUG 05863 | Yes |
| Candida lusitaniae | CLUG02291 | Yes |
| Candida tropicalis | CTRG04618 | Yes |
| Chaetomium globosum | CHGG 04003 | No |
| Coccidioides immitis | CIMG 00721 | No |
| Coprinus cinereus | CC1G_00553 | No |
| Cryptococcus neoformans var. grubii | UGE1, UGE2 | Yes |
| Cryptococcus neoformans var. neoformans | UGE1, UGE2 | Yes |
| Debaryomyces hansenii | DEHA0C02838g | Yes |
| Fusarium graminearum | FG05689, FG03048 | No |
| Fusarium oxysporum | FOXG09188, FOXG02479 | No |
| Fusarium verticillioides | FVEG06791, FVEG05667 | No |
| Histoplasma capsulatum | HCAG00955 | No |
| Kluyveromyces lactis | GAL10 | Yes |
| Lodderomyces elongisporus | LELG_01648 | Yes |
| Magnaporthe grisea | MGG8012 | No |
| Neosartorya fischeri | NFIA076450, NFIA069260, NFIA102220 | No |
| Neurospora crassa | NCU4442 and NCU5133 | No |
| Phanerochaete chrysosporium | PC160.1 | No |
| Pichia stipitis | GALK | Yes |
| Puccinia graminis | PGTG06253, PGTG14524 | No |
| Rhizopus oryzae | RO3G12257, RO3G01911, RO3G05148 | No |
| Saccharomyces cerevisiae | GAL10 | Yes |
| Schizosaccharomyces pombe | SPBPB2BE-12c, SPB23G5-14c | Yes |
| Sclerotinia sclerotiorum | SS1G08326, SS1G04477 | No |
| Stagonospora nodorum | SNOG14014, SNOG 14228 | No |
| Uncinocarpus reesei | UREG 00714 | No |
| Ustilago maydis | UM06057 | No |
| Yarrowia lipolytica | YALI0E26829g | No |
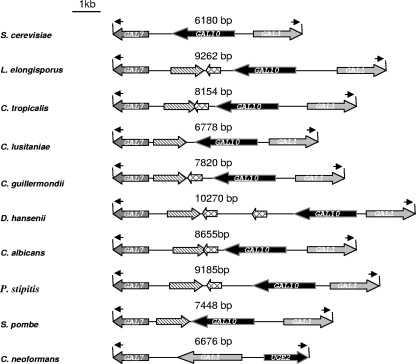
UGE2 belongs to a cluster of genes involved in galactose assimilation, whereas UGE1 is not clustered with any other GAL genes. UGE2 is localized immediately upstream from a close homologue of S. cerevisiae GAL1 in the opposite orientation, and a close homologue of S. cerevisiae GAL7 is downstream from GAL1 on the same strand (Fig. 2). This galactose gene cluster organization is very similar to those in S. cerevisiae and Kluyveromyces lactis (45) (Fig. 2) except that the GAL7 gene in C. neoformans is on the other side of the GAL10-GAL1 cluster. We manually reannotated all the regions surrounding the GAL10 genes in the different fungal genomes. We found that GAL10 and GAL1 orthologues are generally not clustered. Indeed, this particular clustered organization of the galactose genes is restricted to the hemisacomycete genomes, with Yarrowia lipolytica being the only member of this class of fungi to not have any GAL cluster. As shown in Fig. 2, the GAL cluster can be restricted to the three GAL genes, like in S. cerevisiae and K. lactis. For all other members, the GAL cluster includes two additional putative open reading frames (ORFs). An ORF encoding a protein sharing similarities with dTDP glucose-6-dehydratases is present upstream from the GAL7 gene on the opposite strand (26); this ORF is also present in S. pombe. Another small ORF, encoding a putative sybindin-like protein, is present downstream (10). This ORF is not present in C. lusitaniae and in S. pombe but is duplicated in D. hansenii. It overlaps the dTDP glucose-6-dehydratase gene in C. albicans and C. tropicalis.
FIG. 2.
Organization of the galactose gene cluster in fungi. A galactose cluster is found in only some hemiascomycte yeasts and in S. pombe and C. neoformans. In ascomycetes, GAL10 encodes a double enzyme (UDP-glucose epimerase-mutaronase), whereas UGE2 and UGE1 in C. neoformans and the paralogue of GAL10 in S. pombe (SBPB23G5-14c), which is located outside of the galactose cluster, are shorter and encode only putative UDP-glucose epimerase activity., encodes a putative dTDP glucose-6-dehydratase., encodes a sybindin-like protein.
Outside the hemiascomycetes, the clustered organization of the GAL genes is conserved only in S. pombe and C. neoformans; C. neoformans is the only organism outside the ascomycete phylum to have a GAL gene cluster.
Both UGE1 and UGE2 can complement a gal10 mutation in S. cerevisiae.
The cDNAs of UGE1 and UGE2 were amplified by PCR and inserted into an S. cerevisiae expression vector under the control of a Tet-off promoter. The resulting plasmids were used to transform an S. cerevisiae gal10 mutant strain. In the absence of doxycycline, the transformants were able to grow on 1% galactose, whereas the strain transformed with the control plasmid was not (Fig. 3). These results indicate that both UGE genes encode UDP-glucose epimerases and that both enzymes are able to catalyze the transformation of UDP-galactose to UDP-glucose.
FIG. 3.
Both UGE1 and UGE2 can complement an S. cerevisiae gal10 mutation. An S. cerevisiae gal10 mutant strain was transformed with plasmids containing UGE1 or UGE2 cDNAs under the control of a Tet-off promoter, and 105 cells were spotted onto each glucose and galactose medium. Cells were grown at 30°C for 3 days and photographed. As a control, the S. cerevisiae gal10 mutant strain was transformed with the same plasmid containing no cDNA.
UGE2 is not necessary for C. neoformans dissemination and virulence.
We deleted the UGE2 gene using the hygromycin marker (see Materials and Methods). The uge2Δ strains displayed a normal colonial morphology and a wild-type growth rate on glucose, and production of capsule, melanin, and urease was not affected (data not shown). The polysaccharide secreted by these mutants at 30°C and 37°C was the same as that secreted by the wild type; thus, GalXM production was not affected by deletion of this gene (data not shown).We studied the dissemination of uge2Δ and wild-type strains to various target organs (Fig. 4). In contrast to the case for the uge1Δ strain (28), the dissemination of the uge2Δ strain was indistinguishable from that of the wild type. Moreover, the virulence of the uge2Δ and reconstructed strains did not differ significantly from that of the wild-type. Thus, unlike UGE1, UGE2 is clearly inessential for C. neoformans virulence.
FIG. 4.
Virulence of the uge2Δ strains. (A) Dissemination of uge2Δ (black bars) and wild-type (white bars) cells after 1 day of infection. The various organs (brain [B], lungs [L], and spleen [S]) were homogenized and plated, and the numbers of CFU per organ were recorded after 3 to 5 days of incubation at 30°C. Three mice were used for each strain at each time point, and the results reported are mean values and standard deviations. (B) Survival of mice after infection with strains KN99α (wild type [WT]), NE369 (uge2Δ), and NE447 (uge2Δ + UGE2).
UGE2 is necessary for C. neoformans growth on galactose at 30°C but not at 37°C.
Like S. cerevisiae gal10 mutants, the uge2Δ strains were not able to grow on galactose (1%) at 30°C (Fig. 5) and displayed galactose sensitivity (defective growth on ethanol-galactose and glycerol-galactose media at 30°C [data not shown]). This galactose sensitivity was not relieved by the addition of uracil or uridine to the medium (data not shown). Reintroduction of the UGE2 gene into the mutant strains restored growth on galactose. In contrast, uge1Δ strains grew on galactose at 30°C, suggesting that uge1 is not necessary for galactose assimilation/detoxification at this temperature.
FIG. 5.
Growth defects associated with UGE1 and UGE2 disruptions. C. neoformans cells were grown in liquid YPD medium overnight and washed with sterile water, and serial dilutions (5 × 105, 5 × 104, 5 × 103, and 5 × 102 cells) of each strain were spotted onto various media and observed after incubation for 3 days.
Surprisingly, at 37°C, the uge2Δ strains were able to grow on galactose (Fig. 5) suggesting that another enzyme assimilated/detoxified galactose at this temperature. To test whether the enzyme was Uge1p, we deleted the UGE1 gene from a uge2Δ background using a nourseothricin marker. The double mutant strain cumulated all the phenotypes associated with each single deletion: it did not grow on glucose at 37°C, displayed a larger-than-wild-type capsule, did not produce GalXM, had a growth defect on glucose at 30°C, and did not grow on galactose at 30°C. However, the double mutant did not grow on galactose at 37°C, whereas both single mutants did, suggesting that at this temperature and on this medium, UGE1 compensates for the absence of UGE2.
UGE2 expression, but not UGE1 expression, is induced by the presence of galactose.
The fact that UGE1 is able to compensate for the absence of UGE2 on galactose at 37°C but not at 30°C (see above) suggests that Uge1p is regulated in a temperature-dependent manner on galactose. The easiest hypothesis was that UGE1 transcription would be upregulated at 37°C on galactose in the absence of UGE2 and thus compensate for the absence of its closest paralogue. We thus studied the expression of the two UGE genes in strains grown on various carbon sources by Northern blotting. As shown in Fig. 6, transcription of UGE2 was undetectable in cells grown on glucose and was very strongly induced in the presence of galactose at 30°C and 37°C. This regulation is similar to that of the S. cerevisiae GAL10 gene. In contrast, UGE1 expression was not affected by the presence of galactose in the medium; the expression was similar under all condition tested in the absence or the presence of UGE2.
FIG. 6.
Expression of UGE genes under various growth conditions. Cells were cultured on YPD overnight and subcultured on various media and at various temperatures up to a cell density of 5 × 107 cells/ml. Total RNA was extracted and separated under denaturing condition and probed with UGE1, UGE2, and ACT1 gene-specific probes.
The enzymatic properties of Uge1p are not affected by the temperature.
Our second hypothesis was that Uge1p has different affinities for UDP-galactose or UDP-glucose at different temperatures. We made various constructions to produce both Uge1 and Uge2 recombinant enzymes in E. coli. For each, we made two constructs including or excluding the putative transmembrane domain described above. For Uge2p, although denaturing gel electrophoresis unambiguously showed production of a recombinant protein, the protein obtained was always in the insoluble cell extract at all temperatures (4°C, 25°C, and 37°C) and IPTG concentrations (1 mM and 50 μM) tested (data not shown). For Uge1p, the truncated gene similarly gave an insoluble protein, but the vector with the complete coding sequence produced a soluble protein of the correct size, which was purified on an affinity column (Fig. 7A). We tested the activity of the recombinant enzyme at 30°C and 37°C on UDP-galactose (Fig. 7B). The recombinant Uge1p converted UDP-galactose to UDP-glucose, but the affinity of the enzyme was largely unaffected by a change in the temperature; the affinity of the enzyme for UDP-galactose was, if anything, lower at 37°C (apparent Km = 610 ± 75 μM) than at 30°C (apparent Km = 282 ± 34 μM).
FIG. 7.
Uge1p can catalyze the epimerization of UDP-galactose in UDP-glucose at 30°C and 37°C. (A) Purification of recombinant Uge1p in E. coli, showing 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining of total E. coli lysate (lane 1), soluble supernatant (lane 2), and the purified protein after affinity column purification (lane 3). (B) Lineweaver-Burk plots of purified recombinant Uge1p UDP-galactose epimerase activity at 30°C (▪) and at 37°C (□).
Uge1p and Uge2p subcellular localization.
Our third hypothesis was that Uge1p has different subcellular localizations at 30 and 37°C: at one location Uge1p may be involved exclusively in GalXM biosynthesis, transforming UDP-glucose to UDP-galactose, whereas at the other it may participate in galactose assimilation, transforming UDP-galactose to UDP-glucose. We constructed fluorescently tagged versions of Uge1p and Uge2p. These protein fusions were completely functional (data not shown). Strains producing these proteins were cultivated at 30°C on galactose and searched for the fluorescent tags: Uge1p fluorescence appeared at least partly in a punctate, dotted pattern, indicating that the protein was partly associated with vesicles, while Uge2p mainly had a plasma membrane-like localization, although there was some cytoplasmic labeling (Fig. 8). We checked that the fluorescence observed was indeed specific for the Uge proteins (Fig. 8). We also checked that the localization of Uge1 was not affected by the deletion of UGE2 (data not shown). We then constructed a strain expressing the fluorescent versions of both Uge1p and Uge2p. When the cells were cultivated at 30°C on galactose, the localizations of the proteins were similar to those observed using the strains expressing only one fluorescent protein (Fig. 9). Overlaying the pictures on each other showed that the two proteins did not colocalize to any great extent at this temperature (Fig. 9). When the temperature was shifted to 37°C and the cells grown on galactose, the subcellular localization of Uge1p changed such that it was also found at the plasma membrane, like Uge2p. Overlaying the pictures on each other showed at least partial colocalization of the two proteins (Fig. 9). Thus, the carbon source and the temperature regulate the subcellular localization of Uge1p. These results strongly suggest that the subcellular localization of Uge1p determines, at least in part, its function in C. neoformans.
FIG. 8.
Subcellular localization of Uge proteins. Cells expressing the fluorescently tagged versions of the Uge proteins were grown on galactose at 30°C and examined by epifluorescence or by bright-field microscopy. It should be noted that the level of autofluorescence is negligible when cells are grown under such conditions. RFP, red fluorescent protein; GFP, green fluorescent protein.
FIG. 9.
Temperature-dependent colocalization of Uge proteins. Cells producing the fluorescence-tagged Uge proteins (strain NE537) were grown on galactose at 30°C and 37°C and examined by epifluorescence or by bright-field (BF) microscopy.
DISCUSSION
Origin of the GAL gene cluster.
UGE2 is located within a GAL cluster very similar in organization to those in most of the hemiascomycete yeasts and in the archeascomycete yeast S. pombe. The presence of this similar genomic organization in these evolutionarily very distant organisms is surprising. These clusters may have originated from horizontal transfer from a bacterium. Indeed, the E. coli gal operon, containing the four Leloir pathway genes (galM, galK, galT, and galE), has a similar organization. Were this the case, there must have been either multiple independent gene transfers or one transfer to an ancestral organism followed by the loss of this gene organization in most fungi. Constructions of phylogenetic trees provide no evidence for specific clustering of any bacterial protein sequences with those of any fungal phylum containing the GAL cluster (see Fig. SA and Table SC in the supplemental material), as would be expected if there had been multiple horizontal gene transfers. Alternatively, the GAL clusters may have been formed by genomic rearrangements leading to a similar organization in the three different classes representing two phyla. Indeed, there is now evidence that the gene order in eukaryotic genomes is not random (19). According to current models of the dynamics of gene orders in eukaryotes, clustering of genes is selected to keep particular combinations of alleles in linkage disequilibrium (19). In the present case, the galactose sensitivity associated with the loss of the GAL10 or GAL7 gene can be compensated for by the loss of GAL1. In other words, it is better to lose the complete pathway than only part of it. Analysis of the galactose assimilation pathway in hemiascomycete yeast has recently illustrated this phenomenon. When one of the four Leloir genes is lost or mutated, the three others are also lost or mutated (16). Thus, in this case, linkage disequilibrium might be the driving force for natural selection to cluster these genes. The same has recently been suggested for the DAL gene cluster, which is the only other cluster of at least three genes in the S. cerevisiae genome whose products catalyze successive enzymatic steps (47). Also, a study has reported evidence for the independent origin of the maltase gene cluster in two Drosophila species, D. viridis and D. melanogaster (43).
The absence of the GAL gene cluster from some filamentous ascomycetes is associated with the absence of a GAL4 orthologue and a different type of GAL gene regulation (15, 37), suggesting that this class uses another metabolic and gene regulation strategy to assimilate this alternative carbon source. The absence of coregulation of the GAL genes might also explain why the GAL clusters, whether formed or acquired, have not been maintained in these fungi. Indeed, natural selection favors conservation of clusters of coregulated genes (20). The presence of the galactose gene cluster in diverse fungi may thus be a unique example of evolutionary convergence leading to the formation or maintenance of gene clustering.
Uge1p delocalization and Uge2p transcription regulate their functions.
In the absence of galactose, UGE2 is not transcribed and thus does not participate in the anabolism of GalXM. This result by itself explains why a uge1Δ mutant strain is GalXM negative even in the presence of a functional UGE2 gene. However, at 37°C, the uge2Δ mutant grows on galactose but the uge1Δ uge2Δ double mutant does not: therefore, Uge1p participates in the assimilation of galactose at 37°C. Neither UGE1 transcription nor the activity of the protein in vitro toward UDP-galactose is higher at this temperature than at 30°C. The only pertinent difference we observed was the subcellular localization of the protein. Uge1p was clearly associated, at least partially, with Golgi-like vesicles at 30°C on glucose whereas at 37°C on galactose it localized at least partly to the plasma membrane. We have no definitive proof of a causal relationship between the differential localization of the protein and its metabolic function. However, the colocalization of Uge1p and Uge2p at 37°C on galactose at the plasma membrane suggests strongly that under these conditions, Uge1p delocalizes from the Golgi-like vesicles to the plasma membrane to back up Uge2p to transform UDP-galactose to UDP-glucose. Neither the galactose nor the temperature is by itself sufficient to modify the function of Uge1p. Indeed, the uge2Δ mutant strain cannot grow on galactose at 30°C; thus, the presence of galactose is not sufficient to modify the function of Uge1. Similarly, the uge2Δ mutant strain grown at 37°C on glucose does not display any GalXM defect, as demonstrated by the composition of the secreted polysaccharide. This suggests that at this temperature, Uge1 still fully participates in the transformation of UDP-glucose to UDP-galactose. Thus, both a temperature of 37°C and the presence of galactose seem to be necessary to signal a modification of Uge1p function.
The presence of two UDP-glucose epimerases in C. neoformans is reminiscent of the situation in plants, in which five paralogous proteins are present (2). However, unlike in C. neoformans, all these proteins have particular functions in various carbohydrate biosynthesis pathways, and galactose tolerance parallels the total UDP-galactose-epimerase activity rather than being determined by the presence or absence of a particular UGE isoform (34). On the other hand, recent cytological data and transcriptional pattern analysis suggest that, like for the Uge proteins in C. neoformans, the function of each of the proteins might be dependent on the subcellular localization of the protein and on its transcriptional regulation during plant development (2, 34, 38).
Is this metabolic mechanism an adaptation for C. neoformans to live in the environment or in the host?
In the host, C. neoformans may be subjected to the two signals sufficient to drive the translocation of Uge1p (i.e., a temperature of 37°C and the presence of galactose at the surface of the host cells), and therefore this metabolic regulation may be an adaptation to the host. Some recent observations seem to contradict this view. First, Candida glabrata, which is not present in the environment, has completely lost its galactose assimilation pathway (8) but can nevertheless survive and multiply in the host. Second, the deletion of the Candida albicans GAL10 homologue renders the cells galactose sensitive but does not significantly reduce virulence as assessed by mouse studies (40). These findings suggest that other carbon sources are available in the host and that the concentration of free galactose is very low.
In the environment, plant polysaccharides can be galactose rich and C. neoformans probably uses this source of carbon, but the temperature is rarely close to 37°C. However, 37°C is not the optimal growth temperature for C. neoformans. Indeed, this yeast grows much better at lower temperatures (unlike fungal pathogens not acquired from the environment, such as Candida species), and the pattern of gene regulation at 37°C seems to indicate that this temperature is stressful for C. neoformans (23). Thus, the signal that changes the function of Uge1p in the presence of galactose might not be specifically temperature related but might be associated with extreme environmental conditions. We looked at the effects of various other stresses (heavy metals, high osmolarity, and presence of oxidants) on the capacity of a uge2Δ strain to grow on galactose but could not identify any in vitro growth conditions that mimic the effect of temperature (data not shown). However, environmental conditions are most certainly much more complex than those in a petri dish in the laboratory, and C. neoformans has to cope with potential predators such as amoebae and worms and also with fluctuations of the temperature, humidity, and pH. It also has to find nutrients and adapt its metabolism to be able to use them (5). Thus, the environment should be considered to be a very stressful situation in which C. neoformans has to maintain both functional virulence factors (i.e., capsule and melanin) (41) and also the plasticity of its metabolism to be able to exploit as many carbon and nitrogen sources as possible.
Supplementary Material
Acknowledgments
We thank Jenny Lodge for signature-tagged markers. Gene identification was possible thanks to the following genome sequencing project websites: http://www.broad.mit.edu/annotation/fungi/fgi/, http://www.tigr.org/tdb/e2k1/cna1/, http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=search&term, and http://genolist.pasteur.fr/CandidaDB/. We thank Estelle Mogensen and Christophe d'Enfert for critical reading of the manuscript.
This work was supported by a grant from ANR to G.J. (Erapathogenomics Program).
Footnotes
Published ahead of print on 26 September 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55539-552. [DOI] [PubMed] [Google Scholar]
- 2.Barber, C., J. Rösti, A. Rawat, K. Findlay, K. Roberts, and G. J. Seifert. 2006. Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana. J. Biol. Chem. 28117276-17285. [DOI] [PubMed] [Google Scholar]
- 3.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 5.Casadevall, A., and L. A. Pirofski. 2007. Accidental virulence, cryptic pathogenesis, Martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot. Cell 62119-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castresana, J., R. Guigó, and M. M. Albà. 2004. Clustering of genes coding for DNA binding proteins in a region of atypical evolution of the human genome. J. Mol. Evol. 5972-79. [DOI] [PubMed] [Google Scholar]
- 7.Douglas, H. C., and D. C. Hawthorne. 1964. Enzymatic expression and genetic linkage of genes controlling galactose utilization in Saccharomyces. Genetics 49837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dujon, B., D. Sherman, G. Fisher, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. B. Barbe, S., S. Blanchin, J. M. Beckerich, Beyne, E., C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 43035-44. [DOI] [PubMed] [Google Scholar]
- 9.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 191792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ethell, I. M., K. Hagihara, Y. Miura, F. Irie, and Y. Yamaguchi. 2000. Synbindin, a novel syndecan-2-binding protein in neuronal dendritic spines. J. Cell Biol. 15153-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Hermoso, D., G. Janbon, and F. Dromer. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 373204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13837-848. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, R. D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 2201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 15.Hartl, L., C. P. Kubicek, and B. Seiboth. 2007. Induction of the gal pathway and cellulase gene involves no transcriptional inducer function of the galackinase in Hypocrea jecorina. J. Biol. Chem. 28218654-18659. [DOI] [PubMed] [Google Scholar]
- 16.Hittinger, C. T., A. Rokas, and S. B. Carroll. 2004. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl. Acad. Sci. USA 10114144-14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holden, H. M., I. Rayment, and J. B. Thoden. 2003. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 27843885-43888. [DOI] [PubMed] [Google Scholar]
- 18.Hua, J. H., J. D. Meyer, and J. K. Lodge. 2000. Development of positive markers for the fungal pathogen, Cryptococcus neoformans. Clin. Diagn. Lab Immunol. 7125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst, L. D., C. Pal, and M. J. Lercher. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5299-310. [DOI] [PubMed] [Google Scholar]
- 20.Hurst, L. D., E. J. B. Williams, and C. Pal. 2002. Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet. 18604-606. [DOI] [PubMed] [Google Scholar]
- 21.Janbon, G. 2004. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 4765-771. [DOI] [PubMed] [Google Scholar]
- 22.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42453-469. [DOI] [PubMed] [Google Scholar]
- 23.Kraus, P. R., M. J. Boily, S. S. G. Giles, J. E. Stajich, A. Allen, G. M. Cox, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic microarray. Eukaryot. Cell 31249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftus, B., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. Anderson, J. A. Fraser, J. Allen, I. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. Haas, J. C. Huang, G. Janbon, S. Jones, M. I. Krzywinski, K. J. Kwon-Chung, K. B. Lengeler, R. Maiti, M. Marra, R. E. Marra, C. Mathewson, T. G. Mitchell, M. Pertea, F. Riggs, S. L. Salzberg, J. Schein, A. Shvartsbeyn, H. Shin, C. Specht, B. Suh, A. Tenney, T. Utterback, B. L. Wickes, N. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome and transcriptome of Cryptococcus neoformans, a basidiomycetous fungal pathogen of humans. Science 3071321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar, S., J. Ghatak, S. Mukkerji, H. Bhattacharjee, and A. Bhaduri. 2004. UDPgalactose 4-epimerase from Saccharomyces cerevisiae. Eur. J. Biochem. 271753-759. [DOI] [PubMed] [Google Scholar]
- 26.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 1775539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moyrand, F., Y. C. Chang, U. Himmelreich, K. J. Kwon-Chung, and G. Janbon. 2004. Cas3p belongs to a seven member family of capsule structure designer proteins. Eukaryot. Cell 31513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyrand, F., T. Fontaine, and G. Janbon. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 64771-781. [DOI] [PubMed] [Google Scholar]
- 29.Moyrand, F., and G. Janbon. 2004. UGD1 encoding the Cryptococcus neoformans UDP-glucose dehydrogenase is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 31601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, K., G. Cox, P. Wang, D. Toffaletti, J. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 714831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novelli, G., and J. K. V. Reichardt. 2000. Molecular basis of disorders of human galactose metabolism: past, present, and future. Mol. Genet. Metab. 7162-65. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, G. D., C. D. Horstmeier, G. A. Land, and J. H. Foxworth. 1978. Rapid urea broth test for yeasts. J. Clin. Microbiol. 7584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross, K. L., C. N. Davis, and J. L. Fridovich-Keil. 2004. Differential roles of the Leloir pathway enzymes and metabolites in defining galactose sensitivity in yeast. Mol. Genet. Metab. 83103-116. [DOI] [PubMed] [Google Scholar]
- 34.Rösti, J., C. J. Barton, S. Albrecht, P. Dupree, M. Pauly, K. Findlay, K. Roberts, and G. J. Seifert. 2007. UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 191565-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawardeker, J. S., J. H. Sloneker, and A. Jeanes. 1965. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Biochem. 371602-1604. [Google Scholar]
- 36.Schulz, J. M., K. L. Ross, K. Malmstrom, M. Krieger, and J. L. Fridovich-Keil. 2005. Mediators of galactose sensitivity in UDP-galactose 4′-epimerase-impaired mammalian cells. J. Biol. Chem. 28013493-13502. [DOI] [PubMed] [Google Scholar]
- 37.Seiboth, B., L. Karaffa, E. Sandor, and C. P. Kubicek. 2002. The Hypocrea jeronica gal10 (uridine 5′-diphosphate-glucose 4-epimerase-encoding) gene differs from yeast homologues in structure, genomic organization and expression. Gene 295143-149. [DOI] [PubMed] [Google Scholar]
- 38.Seifert, G. J., C. Barber, B. Wells, L. Dolan, and K. Roberts. 2002. Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channelling from UDP-d-galactose into cell wall polymers. Curr. Biol. 121840-1845. [DOI] [PubMed] [Google Scholar]
- 39.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 40.Singh, V., S. V. Satheesh, M. L. Raghavendra, and P. P. Sadhale. 2007. The key enzyme in galactose metabolism, UDP-galactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet. Biol. 44563-574. [DOI] [PubMed] [Google Scholar]
- 41.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 9815245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1751405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vieira, C. P., J. Viera, and D. L. Hartl. 1997. The evolution of small gene clusters: evidence for an independent origin of the maltase gene cluster in Drosophila viridis and Drosophila melanogaster. Mol. Biol. Evol. 14985-993. [DOI] [PubMed] [Google Scholar]
- 44.Walton, F. J., A. Idnurm, and J. Heitman. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 571381-1396. [DOI] [PubMed] [Google Scholar]
- 45.Webster, T. D., and R. C. Dickson. 1988. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 258011-8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, D. B., and D. S. Hogness. 1964. The enzymes of the galactose operon in Escherichia coli. I. Purification and characterisation of the uridine diphosphogaclactose 4-epimerase. J. Biol. Chem. 2392469-2481. [PubMed] [Google Scholar]
- 47.Wong, S., and K. H. Wolfe. 2005. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 37777-782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.