Abstract
Thioredoxins usually perform a role as a thiol-disulfide oxidoreductase using their active-site cysteines. The fission yeast Schizosaccharomyces pombe contains two thioredoxins: Trx1 for general stress protection and Trx2 for mitochondrial functions. The Δtrx2 mutant grows as well as the wild type on complex media containing glucose. However, on nonfermentable carbon source such as glycerol, the mutant did not grow, indicating a defect in mitochondrial function. The mutant also exhibited auxotrophy for arginine and cysteine on minimal medium. In order to find the reason for the unexpected arginine auxotrophy, we searched for multicopy suppressors and found that the arg3+ gene encoding ornithine carbamoyltransferase (OCTase) in the urea cycle of the arginine biosynthetic pathway rescued the arginine auxotrophy. The levels of arg3+ transcript, Arg3 protein, and OCTase activity were all decreased in Δtrx2. Through immunocoprecipitation, we observed a direct interaction between Trx2 and Arg3 in cell extracts. The mutant forms of Trx2 lacking either one or both of the active site cysteines through substitution to serines also rescued the arginine auxotrophy and restored the decreased OCTase activity. They also rescued the growth defect of Δtrx2 on glycerol medium. This contrasts with the thiol-dependent action of overproduced Trx2 in complementing glutathione reductase. Therefore, Trx2 serves multiple functions in mitochondria, protecting mitochondrial components against thiol-oxidative damage as a thiol-disulfide oxidoreductase, and supporting urea cycle and respiration in mitochondria in a manner independent of active site thiols.
In various organisms, glutathione (GSH) and peptide thiols in thioredoxin (Trx) and glutaredoxin (Grx) provides antioxidative environment by reducing disulfide bonds (13, 30, 55). Thioredoxins, initially isolated as a hydrogen donor for ribonucleotide reductase (15), are small proteins with two conserved active cysteines. They efficiently reduce disulfide bonds in a wide variety of proteins and are reduced by thioredoxin reductase using NADPH (14, 30, 55). Thioredoxins function as an antioxidative agent not only by reducing disulfide bonds in oxidized substrates but also by providing electrons to thioredoxin-dependent peroxidases. They also serve as electron donors for several enzymes such as methionine sulfoxide reductase and 3′-phosphoadenosyl-5′-phosphosulfate (PAPS) reductase (30). The eukaryotic signal transduction pathway is modulated by thioredoxins, as observed in regulating the activities of NF-κB (13, 36) and AP-1 family transcription factors (8, 19, 23) and in antiapoptotic regulation (40).
In addition to its redox reaction, thioredoxin is also known to play a key role in promoting growth and assembly of viruses in Escherichia coli such as M13 and T7 (16, 29, 38). In T7 it participates as an accessory protein of the phage-encoded DNA polymerase complex (18, 46). Trx stabilizes the complex between T7 DNA polymerase and DNA and confers processivity on the polymerizing reaction. Surprisingly, the oxidoreductase activity is not required for the function, and substitution of both cysteines in Trx did not significantly affect the maximum polymerase activity (17).
Mitochondria are well known as the primary energy-generating system in eukaryotic cells. Besides its central task of ATP generation, mitochondria play multiple roles to support biochemical pathways for carbon and nitrogen metabolism. Various amino acid biosynthetic enzymes and metabolic pathways are localized in mitochondria, and the tricarboxylic acid cycle links both carbon and nitrogen metabolism by oxidizing organic acids from glycolysis and providing α-ketoglutarate as a carbon skeleton for amino acid synthesis (48). In addition, mitochondria participate in iron-sulfur cluster assembly, fatty acid oxidation, calcium signaling, and apoptosis (3, 6, 27).
As by-products of aerobic respiration, reactive oxygen species (ROS) are generated in mitochondria and cause damages in various components (5, 45, 50). Mitochondrial defense system against ROS includes a number of antioxidant enzymes such as Mn-superoxide dismutase, glutathione peroxidase, and thioredoxin peroxidase (34, 50). Trx, Grx, and GSH maintain the redox homeostasis not only in the cytosol but also in mitochondria. It has been reported that the mitochondrial Trx gene, TRX2, is an essential gene in chicken and that Trx2-deficient cells undergo apoptosis with accumulation of intracellular ROS (47). Overexpression of human mitochondrial Trx causes increased membrane potential (7) and inhibits mitochondrial ASK1-mediated apoptosis (57). Lack of mitochondrial Trx2 also causes early embryonic lethality in mice (35). In contrast to the critical requirement of mitochondrial thioredoxin in avian and mammalian cell survival, its counterpart in the yeast Saccharomyces cerevisiae (Trx3) is dispensable for cell survival in complex, minimal, or respiratory media and even for survival under oxidative stress conditions (37, 49).
The fission yeast S. pombe relies heavily on mitochondria for growth under most laboratory conditions as partly reflected by its petit-negative physiology (41). In this respect, it can serve as a good model system to study genes that affect various aspects of mitochondrial function. Maintenance of oxidation-labile iron-sulfur cluster assembly system in mitochondria, for example, is critical for aerobic growth of S. pombe even under nutrient-rich conditions. Glutathione reductase (GR) supports this function (25), and we previously found that thioredoxin Trx2, when overproduced, can replace GR (43). In the present study we examine the role of mitochondrial thioredoxin in further detail and report an unexpected novel finding that it is required for the urea cycle of arginine biosynthesis as well for efficient energy generation on glycerol, both of which do not involve the thiol-dependent oxidoreductase activity of thioredoxin.
MATERIALS AND METHODS
Yeast strains and culture media.
S. pombe strains used in the present study are ED668 (h+ ade6-M216 leu1-32 ura4-D18), ED665 (h− ade6-M210 leu1-32 ura4-D18), JL38 (ura4+ in ED665 background), JL36 (ura4+ Δpgr1 nmt-pgr1 in ED665), and JY31b (trx2::ura4+ in ED668). Growth and maintenance of all of the strains were done as described previously (2, 33). Cells were grown in YES (0.5% yeast extract, 3% dextrose) medium or Edinburgh minimal medium (EMM) with appropriate supplements as described by Alfa et al. (2). For respiratory growth, glycerol medium (0.5% yeast extract, 0.1% dextrose, 3% glycerol, and 250 mg of supplements/liter) was used.
Construction of Δtrx2 mutants.
To disrupt the trx2+ gene, the BglII fragment of the trx2 open reading frame cloned in pTZ18R vector was replaced with 1.8-kb ura4+ gene cassette. The 3.2-kb HindIII fragment containing the recombinant construct was introduced to ED665/ED668 diploid cells for direct homologous recombination at the trx2+ locus. The transformants were selected by the ura+ marker, and the expected disruption was confirmed by both colony PCR and genomic Southern hybridization. A haploid Δtrx2 strain was isolated through tetrad analysis.
Construction of mutants and recombinants.
Construction of pREP1-trx2+ and pREP42-trx2-EGFP-C was described previously (43). Substitution mutagenesis of active-site cysteines to serines in Trx2 was done by using mutagenic primers; T2-mut1 (5′ GCGGTCCTTCGAAATACCTCAAACC 3′, the BstBI site is underlined) and T2-mut2 (antisense of T2-mut1) for Cys50 substitution or T2-mut3 (5′ GACTGGTCCGGACC TTCGAAATACCTC 3′, the BspEI and BstBI sites are underlined) and T2-mut4 (antisense of T2-mut3) for substitution of both Cys47 and Cys50. The arg3+ gene fragment was amplified with the primer pair ARG3-F (5′ GATTTACTGCAGTTGGTAGAAGGC 3′, the PstI site is underlined) and ARG3-R (5′ GCATTGTTTGCAGGATCCCCAGAT 3′, the BamHI site is underlined). The 2.3-kb amplified DNA was subcloned into PstI/BamHI site of pREP1. For C-terminal Myc tagging, the 9× His-human rhinovirus 3C protease-9xMyc (HPM) cassette originated from pJS-HPM53H (a modified TAP-tagging vector kindly provided by J.-H. Seol, Seoul National University) was cloned into the SmaI site of pREP41, generating pREP41-HPM vector. To construct chimeric Arg3-Myc protein with C-terminally fused 9× Myc tag, PCR products with the primers Arg3-N (5′ CAGTTTGCAATTGCATATGTCTTT C 3′, the NdeI site is underlined) and Arg3-HPM (5′ ATTAAGGATTCCCGGGTAGGCTGAG 3′, the SmaI site is underlined) were generated, cut with NdeI and SmaI, and cloned into pREP41-HPM. To construct pREP41-Gld-Myc, a 1.4-kb DNA fragment containing the open reading frame of a putative glycerol dehydrogenase gene (gld1; SPAC13F5.03c) was cloned in a similar way.
Northern hybridization.
RNAs from exponentially grown cells in YES medium were separated on an agarose gel containing formaldehyde, transferred onto a Hybond-N+ membrane (Amersham), and fixed by UV-cross-linker (43). Hybridization was performed in Rapid-Hyb buffer (Amersham) with radioactively labeled arg3+ probes generated by PCR as recommended by the manufacturer. The signal was visualized by exposing the membrane to X-ray film, and the radioactivity was quantified with a PhosphoImager (BAS-5000) and a MultiGauge (Fuji).
OCTase assay.
Ornithine carbamoyltransferase (OCTase) activity was measured as described by Lee and Nussbaum (24) with some modification. Crude cell extracts were added to 700 μl of reaction mixture composed of 5 mM ornithine, 15 mM carbamoyl phosphate, and 270 mM triethanolamine (pH 7.7), followed by incubation at 37°C for 30 min. The reaction was stopped by adding 250 μl of 3:1 phosphoric acid-sulfuric acid (vol/vol). Citrulline production was determined by measuring the absorbance at 490 nm, after the addition of 50 μl of 3% 2,3-butanedione monoxime and further incubation in the dark at 95 to 100°C for 15 min.
Western immunoblot analysis.
Crude cell extracts (100 μg of total protein) prepared from cells grown to an optical density at 595 nm (OD595) of ∼1.5 in YES medium were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and subjected to Western analysis using anti-Arg3 polyclonal antibody raised in mice. The secondary antibody (goat anti-mouse IgGAM; Cappell) conjugated with horseradish peroxidase was used at 10−5 dilution, and the light development was detected by using an ECL system (Amersham). The signal was visualized by the LAS-3000 imaging system (Fuji) and quantified with a MultiGauge (Fuji).
Coimmunoprecipitation.
Crude cell extracts containing 1 mg of total protein were mixed with 10 μl of agarose beads conjugated with monoclonal antibodies against c-Myc or green fluorescent protein (GFP; Santa Cruz Biotech). Each sample was incubated at 4°C with rocking for 2 h, followed by washing with buffer A (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, and 100 mg of sodium azide/liter). Pellets were resuspended in 1× SDS sample buffer and separated by SDS-PAGE. They were subjected to Western analysis with monoclonal antibodies against GFP or c-Myc (Santa Cruz Biotech).
RESULTS
The Δtrx2 mutant shows auxotrophy for arginine and cysteine.
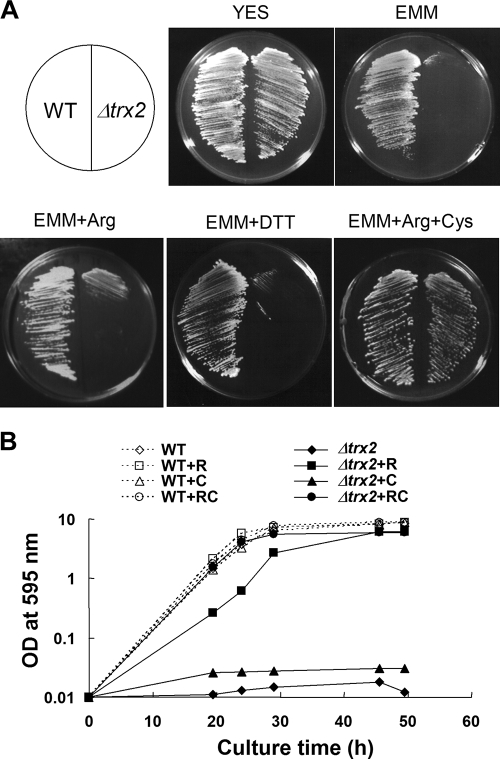
The S. pombe mutant devoid of mitochondrial thioredoxin gene (Δtrx2) was constructed and examined for growth on different media. On complex YES medium, the Δtrx2 mutant grew as well as the wild type. However, on a minimal medium plate (EMM) it did not grow unless supplemented with Casamino Acids (Fig. 1A). Among amino acids, arginine alone partially restored growth, whereas additional supplement with cysteine resumed full growth. In contrast to the Δtrx1 (cytosolic thioredoxin) mutant that shows cysteine auxotrophy (44), the growth defect of Δtrx2 was hardly reversed by supplementing with cysteine alone, nor by dithiothreitol. In liquid minimal medium, the growth stimulatory effect of arginine was more pronounced in the mutant, reaching an OD595 near 6.0, albeit with a slightly slower growth rate than that of the wild type (Fig. 1B). Supplementation with both arginine and cysteine completely restored the fast growth of Δtrx2 in liquid minimal medium. The cysteine synthesis has been related to thioredoxin, since one of its biosynthetic enzymes (PAPS reductase) is known to require thioredoxin for its activity. The arginine auxotrophy, however, has not been associated with thioredoxin in previous studies.
FIG. 1.
Requirement of arginine and cysteine for the growth of Δtrx2. (A) Growth on plates. Wild-type (JL38) and Δtrx2 (JY31b) cells were streaked on complex (YES) and minimal (EMM) media either not supplemented or supplemented with 1 mM dithiothreitol, arginine, or arginine plus cysteine. The photos were obtained after 4 days of incubation at 30°C. (B) Growth in liquid media. The wild-type (open symbols connected with dotted lines) and Δtrx2 (filled symbols connected with solid lines) cells were inoculated from overnight seed culture to an OD595 of 0.01 in EMM (diamonds), EMM plus arginine (R, squares), EMM plus cysteine (C, triangles), and EMM plus R and C (circles). Cell growth was monitored by measuring the OD595. WT, wild type.
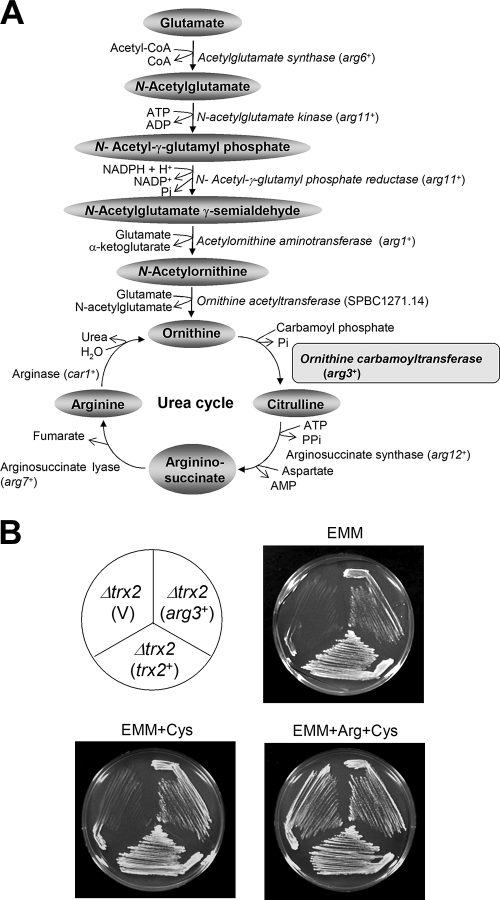
The gene for OCTase (arg3+) in the urea cycle of arginine biosynthetic pathway suppresses Δtrx2 phenotype.
Arginine is synthesized from glutamate via ornithine and the urea cycle, as summarized in Fig. 2A. Among the enzymes in the arginine biosynthetic pathway, six (acetylglutamate synthase, N-acetylglutamate kinase, N-acetyl-γ-glutamyl phosphate reductase, acetylornithine aminotransferase, ornithine acetyltransferase, and OCTase; presented in italics in Fig. 2A) were reported to reside in mitochondria (31, 54). We suspected that one or more of these mitochondrial enzymes could have been compromised by the loss of Trx2. To test this hypothesis, we cloned each of these genes with its own promoter on multicopy vector (pREP1) and introduced it to Δtrx2 mutant. The transformants that grew on minimal EMM plates were selected. We found that the arg3+ gene encoding OCTase partially and fully recovered growth on EMM and EMM+cysteine plates, respectively (Fig. 2B). This suggests that OCTase, which converts ornithine to citrulline through carbamoylation in the urea cycle, could be the target affected by trx2 disruption. In contrast to arginine biosynthetic enzymes, most cysteine biosynthetic enzymes are predicted to reside in the cytosol, with the exception of mitochondrial cysteine synthase encoded by cys 11+ and cyc12+ (31). Overexpression of Cys12, however, did not alleviate the cysteine requirement of the trx2 disruptant (data not shown).
FIG. 2.
Introduction of arg3+ gene encoding OCTase overcomes arginine auxotrophy of the Δtrx2 mutant. (A) Biosynthetic pathway for arginine from glutamate. The enzymes that are reported to be mitochondrial proteins are presented in italics. The standard names for encoding genes as they appear in S. pombe database (www.genedb.org/pombe) are indicated in parentheses (arg6+, SPBC725.14; arg11+, SPAC4G9.09c; arg1+, SPCC777.09c; arg3+, and SPAC4G9.10; arg12+, SPBC428.05c; arg7+, SPBC1773.14; car1+, SPBP26C9.02c). (B) Complementation by the arg3+ gene. The Δtrx2 (JY31b) cells were introduced with cloned trx2+ or arg3+ genes containing their own promoters in pREP1-based vector (V) and incubated on EMM plates with supplements at 30°C for 4 days.
Loss of Trx2 causes a decrease in OCTase.
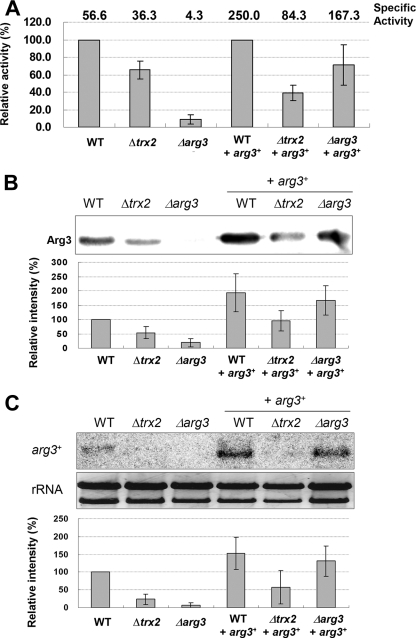
To confirm the relationship between Trx2 and Arg3, OCTase activity was measured in the wild-type, Δtrx2, and Δarg3 cells. In exponentially growing cells, the OCTase activity was reduced in the Δtrx2 mutant to ca. 60% of the wild-type level (Fig. 3A). OCTase activity in Δarg3 mutant was ca. 8% of the wild-type level, confirming that Arg3 provides the major activity for OCTase. When the same set of cells were introduced with arg3+ gene with its own promoter on pREP-based vector, the specific enzyme activities all increased, confirming the OCTase activity of the arg3+ gene product. However, even with the provision of multicopy arg3+ genes, the OCTase activity was much lower in Δtrx2 cells than in the wild-type and in Δarg3 mutant backgrounds, indicating that Trx2 is necessary for proper level of Arg3 enzyme activity.
FIG. 3.
Levels of OCTase, Arg3 protein, and arg3+ transcripts in Δtrx2. The wild-type (ED668), Δtrx2 (JY31b), and Δarg3 cells with or without pREP-arg3+ plasmid were harvested at exponential phase (OD595 ∼1.5 in YES medium) to prepare cell crude extract and RNA. The amount of OCTase and Arg3 protein in cell extracts was measured by determining the enzyme activity (A) and by Western blotting (B). (C) The arg3+ transcripts were detected by Northern analysis with rRNAs as loading controls. The relative amounts of OCTase, Arg3 protein, and arg3+-specific RNA compared to the wild-type level (set as 100) are also presented. In panel A, the relative enzyme activities in strains with pREP-arg3+ plasmid are presented compared to the wild-type background. The specific enzyme activities are shown at the top. Average values with standard deviations from four (A and C) or three (B) independent experiments are given. WT, wild type.
To unravel how the loss of Trx2 caused a decrease in OCTase activity, we examined the expression level of the arg3+ gene. The Western blot against the Arg3 protein demonstrated that in the exponentially growing Δtrx2 cells, Arg3 protein level was reduced to ca. 50% of its wild-type level (Fig. 3B). This result coincides with the decrease in OCTase activity in Δtrx2 cells. When the level of arg3+ mRNA was examined by Northern analysis, we found that it was also lowered to ca. 30% level in Δtrx2 (Fig. 3C). Reductions in the level of protein and transcripts were consistently observed in Δtrx2 cells provided with pREP-arg3+ plasmids compared to the wild-type and Δarg3 mutant backgrounds. We observed that the stability of arg3+ mRNA did not change in Δtrx2 (data not shown). The decreased mRNA level was restored by introducing trx2+ gene on pREP-based plasmid (data not shown). When the protein half-life of Arg3 was examined by Western blotting after cycloheximide treatment, no significant difference was observed between wild-type and Δtrx2 cells (data not shown). Therefore, it seems most likely that Trx2 affects arg3+ gene expression at the level of transcription, which in turn affects the protein level, and hence the activity.
The active-site cysteines of Trx2 is not required to maintain OCTase activity.
Trx2 shares many conserved residues with other known thioredoxins, including the highly conserved active-site sequence of Trp-Cys-Gly-Pro-Cys, which is essential for oxidoreductase activity. To investigate whether the thiol-dependent oxidoreductase activity of Trx2 is necessary to maintain OCTase, one (C50) or both (C47 and C50) of cysteine residues in the active site were mutated to serines, resulting in T2-CS and T2-SS mutants, respectively. The wild-type and mutant trx2 genes were introduced into Δtrx2 cells on pREP-based multicopy plasmid retaining their own promoters. To our surprise, both the T2-CS and the T2-SS variants allowed Δtrx2 cells to overcome arginine auxotrophy (Fig. 4A). The OCTase activity also increased in cell extracts expressing either wild-type or mutant Trxs, a finding consistent with growth promotion on minimal medium (Fig. 4B).
FIG. 4.
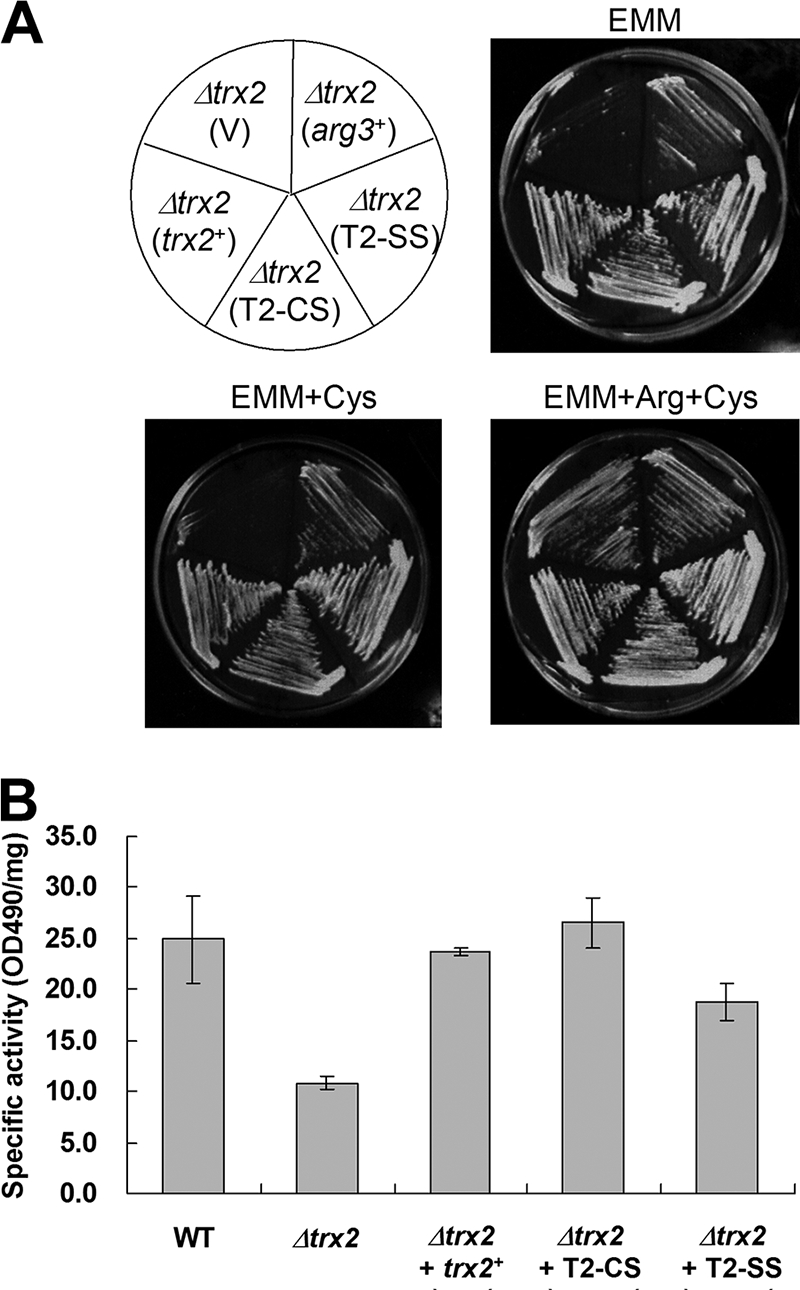
Thiol-independent complementation of arginine auxotrophy by mutant Trx2. (A) The Δtrx2 (JY31b) mutant was transformed with pREP1 (V) or pREP-based recombinant plasmids with cloned arg3+, trx2+, or mutated trx2 with C50S (T2-CS) or C47/50S (T2-SS) substitutions. The cloned genes contained their own promoters to allow expression. Cells were streaked onto EMM plates supplemented with cysteine or with cysteine plus arginine, followed by incubation at 30°C for 4 days. (B) OCTase activity of wild-type (ED668) or Δtrx2 cells complemented with various trx2 constructs. Freshly grown cells at the early exponential phase (OD595 ∼0.4) were used to prepare crude cell extracts. Error bars indicate the standard deviations from three independent experiments. WT, wild type.
Direct interaction of Trx2 with Arg3 protein.
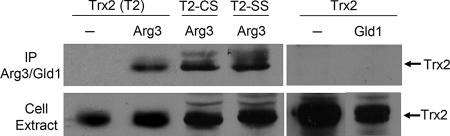
We examined whether Trx2 directly interacts with Arg3 protein in vivo by coimmunoprecipitation analysis. Arg3 and Trx2 were C-terminally tagged with 9×Myc and GFP, respectively, and introduced into the wild-type cells through pREP41- and pREP42-based expression system, respectively. Cell extracts were immunoprecipitated with anti-Myc antibody against Arg3-Myc, followed by Western blotting with anti-GFP antibody against Trx2-GFP. The results in Fig. 5 demonstrate that Trx2 was coprecipitated with Arg3. The specificity of coprecipitation was examined by probing interaction of Trx2 with another mitochondrial enzyme (a putative glycerol dehydrogenase; Gld1) that is similarly tagged at C terminus with Myc. The negative result with Gld1 demonstrates that the interaction between Trx2 and Arg3 is specific. Interaction with Trx2 mutants was examined likewise, revealing that both mutant forms were able to interact with Arg3 as the wild type. When immunocoprecipitation was performed with anti-GFP antibody against Trx2, followed by Western with anti-Myc against Arg3, Arg3 was detected only in cells with Trx2 or their variants (data not shown). Therefore, it is evident that Trx2 interacts with Arg3 protein in vivo, and this interaction is not affected by the loss of active-site cysteines.
FIG. 5.
Interaction of Trx2 with Arg3 monitored by coimmunoprecipitation. Cell extracts prepared from ED665 containing pREP41-Arg3-Myc and pREP42-Trx2-GFP were subjected to immunoprecipitation with anti-Myc antibodies conjugated on agarose beads. Total crude extract and the immunoprecipitates were resolved on SDS-PAGE, followed by Western blotting with anti-GFP antibodies. As a negative control, parallel immunoprecipitation was done with cells expressing Trx2-GFP alone and cells containing pREP41-Gld1 (glycerol dehydrogenase)-Myc instead of pREP-Arg3-Myc. Trx2 variants (T2-CS and T2-SS) were also fused with GFP tag and examined in the same way.
Trx2 and its cysteine mutants support growth on glycerol, a nonfermentable carbon source.
On complex media with glycerol as a carbon source, S. pombe cells rely heavily on aerobic respiration for growth. We found that Δtrx2 cells were unable to grow on glycerol medium (Fig. 6A). The respiration rate of Δtrx2 was drastically reduced in both YES and glycerol media, suggesting a role for Trx2 in maintaining proper mitochondrial function including respiration (data not shown). Introduction of trx2+ gene on pREP-based vector restored growth on glycerol (Fig. 6A). Interestingly, the T2-CS and T2-SS variants of Trx2 also restored growth on glycerol plate. Considering that YES or glycerol medium does not lack arginine, it is evident that the role of Trx2 to maintain proper mitochondrial function goes beyond amino acid biosynthesis and does not require the thiol-dependent oxidoreductase activity of Trx2.
FIG. 6.
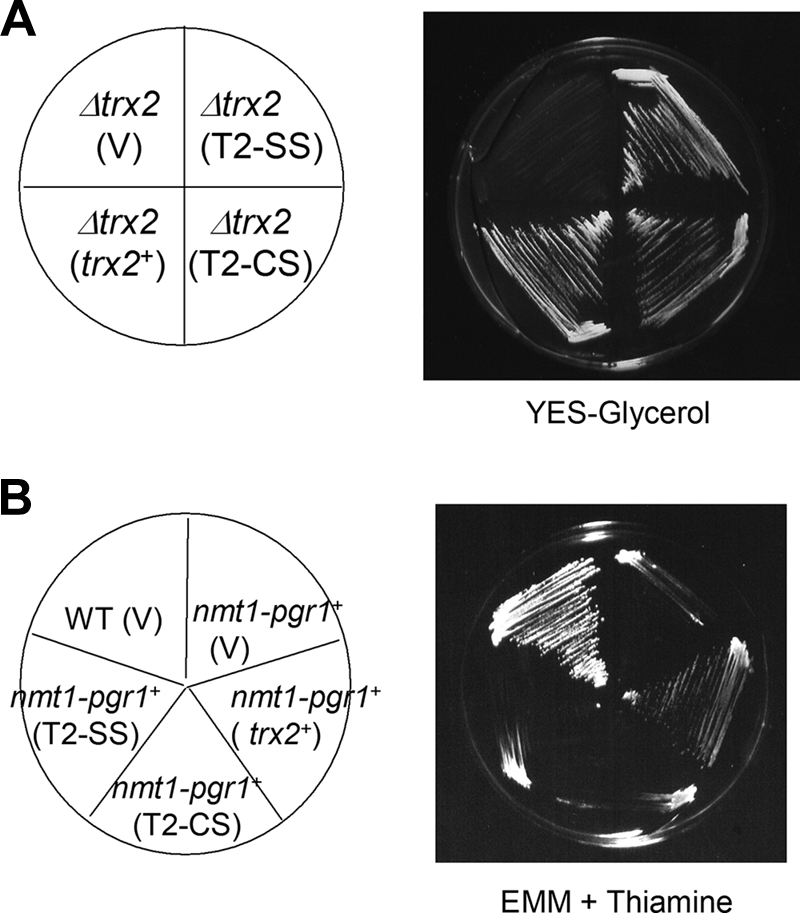
Thiol-independent function of Trx2 for growth on glycerol versus thiol-dependent function for substituting GR. (A) Growth on a modified YES medium that contain 3% glycerol instead of glucose. The Δtrx2 mutant was transformed with pREP1 (V), or pREP-trx2+, pREP-T2-CS, or pREP-T2-SS and monitored for growth on plates containing glycerol as a carbon source. (B) Complementation of GR deficiency. A pgr1 mutant strain (JL36 [25]) that contain thiamine-repressible pgr1+ gene (Pnmt1-pgr1) in the chromosome was transformed with pREP1 (V), pREP-trx2+, pREP-T2-CS, and pREP-T2-SS. Thiamine represses expression from Pnmt-pgr1+ without affecting trx2 gene expression from its own promoter. The wild type (JL38) with control vector (V) was also examined in parallel. The growth was monitored on EMM plates containing 10 μM thiamine after incubation at 30°C for 4 days. WT, wild type.
Suppression of GR deficiency requires active site cysteines in Trx2.
In our previous work, the trx2+ gene was found as a multicopy suppressor of GR deficiency, which hinders cell growth under all conditions examined. The restoration of growth by overproduced Trx2 coincided with the restoration of oxidant-labile Fe-S enzymes that were compromised in GR-deficient mutants. We examined whether the role of overproduced Trx2 as a substitute for GR is also independent of the thiol oxidoreductase activity. Using a thiamine-suppressible nmt1-pgr1+ strain that lacks GR and therefore cannot grow on plates containing thiamine (25), we examined the compensatory activity of Trx2 variants. The results in Fig. 6B demonstrate that the compensating effect of Trx2 for GR is dependent on its thiol-oxidoreductase activity, since active site cysteine mutants did not restore growth. Therefore, the compensatory role of Trx2 to replace GR is mediated through providing thiol-disulfide redox activity, whereas other roles to support arginine biosynthesis and mitochondrial respiratory function are independent of thiol-redox function.
DISCUSSION
S. pombe possesses two thioredoxins: Trx1 for general stress protection (44) and Trx2 for mitochondrial functions (43). A recently annotated thioredoxinlike protein (Txl1 or Trx3) contains an N-terminal thioredoxinlike domain and C-terminal domain of unknown function, but its function is not well characterized except for defense against alkylhydropeorxide (20) or other oxidants (21). We present evidence here that Trx2, the mitochondrial thioredoxin in S. pombe, contributes to maintain proper mitochondrial functions that include aerobic respiration and biosynthesis of arginine. Trx2 served to maintain proper level of OCTase (Arg3) in particular by regulating its synthesis. It even interacted directly with it. Arginine is one of the most versatile amino acids that serve as a precursor for making not only proteins but also nitric oxide, urea, polyamine, proline, glutamate, creatine, and agmatine (10). It is dispensable for healthy adult humans but is essential for young, growing animals. Interestingly, most of the enzymes in the arginine biosynthetic pathway (from glutamate to citrulline) are localized in mitochondria. It has been reported that some of these mitochondrial arginine biosynthetic enzymes are associated with mitochondrial nucleoids in S. cerevisiae (Arg5/6 [11, 22]), as well as in humans (Arg4; carbamoyl phosphate synthetase [56]). In S. cerevisiae Arg5/6 has been shown to associate with specific nuclear loci and regulate nuclear gene expression. Since the arginine pathway is tightly interlinked with carbon and nitrogen metabolism, it seems logical that this pathway could be at a crossroad to connect nutrient signals with mitochondrial function, as well as with nuclear gene expression.
The fact that loss of Trx2 contributed to decreasing arg3+ transcription could be interpreted as a phenomenon reflecting communication between mitochondria and the nucleus. Recent findings on mitochondrial stress signaling accumulate to indicate that mitochondrial dysfunction due to mitochondrial DNA loss or altered membrane potential triggers retrograde response that involves multiple factors in the signaling pathway to regulate nuclear target gene expression (28, 39). Concentration changes in metabolites originating from mitochondria such as oxaloacetate, α-ketoglutarate, glutamate, ammonia, and [Ca2+] are known as signaling molecules to trigger this pathway. Although the RTG-dependent pathway in S. cerevisiae has been investigated in most detail, the composition and the mechanism of the signaling pathway appear to be diverse among organisms (4). It is tempting to speculate from our study that a kind of retrograde signaling that involves Trx2 may exist in S. pombe to connect mitochondrial status with nuclear gene expression. In this case, mitochondrial dysfunction resulting from the lack of Trx2 somehow downregulates gene expression for Arg3, shutting off arginine synthesis and the ornithine cycle. There is no available information on retrograde mitochondrial signaling in S. pombe, and it appears highly intriguing to unravel the mechanism by which Trx2 affects nuclear gene expression.
The observation that Trx2 supports efficient respiration and Arg3 (OCTase) production independently of thiol oxidoreductase activity implies that these functions are mediated through protein-protein interaction without involving thiol-disulfide redox reaction. This hypothesis has been partly confirmed by direct interaction between Arg3 and Trx2. How the interaction of Trx2 with Arg3 in mitochondria is related to arg3+ gene expression in the nucleus is very intriguing. Considering previous reports that thioredoxins can stabilize proteins, enhance enzyme activities, and assist proper folding, it is conceivable that it can somehow ensure optimal OCTase activity, maintaining a proper balance of metabolites, which in turn signal proper gene regulation. According to this scenario, the loss of Trx2 would disturb this balance and decrease arg3+ expression. The validity of this scenario requires further systematic analysis of Trx2 function as well as search for additional interaction partners. In our hands, Arg3 enzyme activity itself was not affected by Trx2 in vitro (data not shown), suggesting that Trx2 may not modulate OCTase enzyme activity in vivo. How Trx2 is related to the expression of arg3+ gene is an interesting question for future investigations.
The observation that Δtrx2 mutation in S. pombe caused a severe decrease in mitochondrial functions contrasts the absence of such phenotype in S. cerevisiae mutant (ΔTRX3) that lack mitochondrial thioredoxin (37, 49). Although OCTases are present from bacteria to higher eukaryotes, the regulation of its synthesis could be different. In contrast to S. pombe and other higher eukaryotes, OCTase in S. cerevisiae is reported to be a cytosolic enzyme (53, 54) and is regulated by protein-protein interaction with arginase (1). In higher eukaryotes, mitochondrial TRX2 is an essential gene as reported for cell viability in chickens and for embryonic development in mice (35, 47). Previously, the reason for the essentiality of mitochondrial Trx for mammalian cell survival was proposed to be due to the inhibition of apoptosis and cell death (35, 47, 57). On the basis of present study, we present another possible reason for Trx2 requirement for cell survival, namely, to maintain OCTase activity and to ensure proper mitochondrial function. In mammals, OCTase deficiency causes urea cycle disorder, resulting in the accumulation of ammonia and excess or deficiency of other metabolites that lead to hyperammonemia, encephalopathy, and respiratory alkalosis (32). In humans, inherited OCTase deficiency leads to an increase in blood ammonia and glutamate/glutamine level and a decrease in citrulline and arginine concentration. Clinically severe symptoms start at a younger age even within several days after birth (9). Therefore, it is worthwhile to examine whether mitochondrial Trx regulates OCTase in other eukaryotes including humans.
Our study in the S. pombe system has demonstrated that mitochondrial thioredoxin as a thiol oxidoreductase can replace the function of GR when overproduced (43) and serve as an electron donor for other oxidoreductases such as Gpx1, a thioredoxin peroxidase (26). We presented additional novel roles of mitochondrial thioredoxin to support optimal mitochondrial function and to ensure the proper level of Arg3, both of which do not involve thiol-disulfide redox function. The present findings imply that Trx2 participates in communication between mitochondria and the nucleus to adjust nuclear gene expression in response to changes in mitochondrial function.
Acknowledgments
This study was supported by a National Research Laboratory grant (2004-02397) from the Ministry of Science and Technology to J.-H.R. J.-Y.S. was supported by the second stage of BK21 program for Biological Sciences at Seoul National University.
Footnotes
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Alami, M., E. Dubois, Y. Oudjama, C. Tricot, J. Wouters, V. Stalon, and F. Messenguy. 2003. Yeast epiarginase regulation, an enzyme-enzyme activity control. J. Biol. Chem. 27821550-21558. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, C., P. Fantes, J. Hyams, M. Mcleod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 3.Borutaite, V., and G. Brown. 2003. Mitochondria in apoptosis of ischemic heart. FEBS Lett. 241-5. [DOI] [PubMed] [Google Scholar]
- 4.Butow, R., and N. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 141-15. [DOI] [PubMed] [Google Scholar]
- 5.Cahill, A., X. Wang, and J. B. Hoek. 1997. Increased oxidative damage to mitochondrial DNA following chronic ethanol consumption. Biochem. Biophys. Res. Commun. 235286-290. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 1251241-1252. [DOI] [PubMed] [Google Scholar]
- 7.Damdimopoulos, A. E., A. Miranda-Vizuete, M. Pelto-Huikko, J.-Å. Gustafsson, and G. Spyrou. 2002. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J. Biol. Chem. 27733249-33257. [DOI] [PubMed] [Google Scholar]
- 8.Delaunay, A., A. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 195157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endo, F., T. Matsuura, K. Yanagita, and I. Matsuda. 2004. Clinical manifestations of inborn errors of the urea cycle and related metabolic disorders during childhood. J. Nutr. 134(Suppl. 6)1605S-1609S. [DOI] [PubMed] [Google Scholar]
- 10.Guoyao, W. U., and S. M. Morris. 1998. Arginine metabolism: nitric oxide and beyond. Biochem. J. 3361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, D. A., H. Zhu, X. Zhu, T. Royce, M. Gerstein, and M. Snyder. 2004. Regulation of gene expression by a metabolic enzyme. Science 306482-484. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Herrero, E., and J. Ros. 2002. Glutaredoxins and oxidative stress defense in yeast. Methods Enzymol. 348136-146. [DOI] [PubMed] [Google Scholar]
- 14.Hirota, K., M. Murata, Y. Sachi, H. Nakamura, J. Takeuchi, K. Mori, and J. Yodoi. 1999. Distinct roles of thioredoxin in the cytoplasm and in the nucleus: a two-sept mechanism of redox regulation of transcription factor NF-κB. J. Biol. Chem. 27427891-27897. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren, A. 1984. Enzymatic reduction-oxidation of protein disulfides by thioredoxin. Methods Enzymol. 107295-300. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54237-271. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 26413963-13966. [PubMed] [Google Scholar]
- 18.Huber, H. E., M. Russel, P. Model, and C. C. Richardson. 1986. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. J. Biol. Chem. 26115006-15012. [PubMed] [Google Scholar]
- 19.Huber, H. E., S. Tabor, and C. C. Richardson. 1987. Escherichia coli thioredoxin stabilizes complex of bacteriophage T7 DNA polymerase and primed template. J. Biol. Chem. 26216224-16232. [PubMed] [Google Scholar]
- 20.Izawa, S., K. Maeda, K. Sugiyama, J. Mano, and Y. Inoue. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 27428459-28465. [DOI] [PubMed] [Google Scholar]
- 21.Jiménez, A., L. Mateos, J. R. Pedrajas, A. Miranda-Vizuete, and J. L. Revuelta. 2007. The txl1+ gene from Schizosaccharomyces pombe encodes a new thioredoxin-like 1 protein that participates in the antioxidant defense against tert-butyl hydroperoxide. Yeast 24481-490. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. J., E. M. Jung, H. J. Jung, Y. S. Song, E. H. Park, and C. J. Lim. 2007. Cellular functions and transcriptional regulation of a third thioredoxin from Schizosaccharomyces pombe. Can. J. Microbiol. 53775-783. [DOI] [PubMed] [Google Scholar]
- 23.Kucej, M., and R. A. Butow. 2007. Evolutionary tinkering with mitochondrial nucleoids. Trends. Cell. Biol. 17586-592. [DOI] [PubMed] [Google Scholar]
- 24.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 216139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. T., and R. L. Nussbaum. 1989. An arginine to glutamine mutation in residue 109 of human ornithine transcarbamylase completely abolishes enzymatic activity in Cos1 cells. J. Clin. Investig. 841762-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., I. Dawes, and J. H. Roe. 1997. Isolation, expression, and regulation of the pgr1+ gene encoding glutathione reductase absolutely required for the aerobic growth of Schizosaccharomyces pombe. J. Biol. Chem. 27223042-23049. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S.-Y., J.-Y. Song, E.-S. Kwon, and J.-H. Roe. 2008. Gpx1 is a stationary phase-specific thioredoxin peroxidase in fission yeast. Biochem. Biophys. Res. Commun. 36767-71. [DOI] [PubMed] [Google Scholar]
- 28.Lill, R., and U. Mühlenhoff. 2006. Iron-sulfur protein biogenesis in eukaryote: components and mechanisms. Annu. Rev. Cell Dev. Biol. 22457-486. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z., and R. Butow. 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40159-185. [DOI] [PubMed] [Google Scholar]
- 30.Mark, D. F., and C. C. Richardson. 1976. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 73780-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masutani, H., and J. Yodoi.2002. Thioredoxin. Methods Enzymol. 347279-286. [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, S. Horinouchi, and M. Yoshida. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24841-847. [DOI] [PubMed] [Google Scholar]
- 33.Mian, A., and B. Lee. 2002. Urea-cycle disorders as a paradigm for inborn errors of hepatocyte metabolism. Trends Mol. Med. 8583-589. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 35.Netto, L. E. S., A. J. Kowaltowski, R. F. Castilho, and A. E. Vercesi. 2002. Thiol enzymes protecting mitochondria against oxidative damage. Methods Enzymol. 348260-270. [DOI] [PubMed] [Google Scholar]
- 36.Nonn, L., R. R. Williams, R. P. Erickson, and G. Powis. 2003. The Absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 23916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto, T., K. Asamitsu, and T. Tetsuka. 2002. Thioredoxin and mechanism of inflammatory response. Methods Enzymol. 347349-360. [DOI] [PubMed] [Google Scholar]
- 38.Pedrajas, J. R., E. Kosmidou, A. Miranda-Vizuete, J. A. Gustafsson, A. P. Wright, and G. Spyrou. 1999. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem. 2746366-6373. [DOI] [PubMed] [Google Scholar]
- 39.Russel, M., and P. Model. 1986. The role of thioredoxin in filamentous phage assembly. J. Biol. Chem. 26114997-15005. [PubMed] [Google Scholar]
- 40.Ryan, M. T., and N. J. Hoogenraad. 2007. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76701-722. [DOI] [PubMed] [Google Scholar]
- 41.Saitoh, M., H. Nishitoh, M. Fujii, K. Takeda, K. Tobiume, Y. Sawada, M. Kawabata, K. Miyazono, and H. Ichijo. 1998. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 172596-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schäfer, B. 2003. Genetic conservation versus variability in mitochondria: the architecture of the mitochondrial genome in the petite-negative yeast Schizosaccharomyces pombe. Curr. Genet. 43311-326. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, J.-Y., J. Cha, J. Lee, and J.-H. Roe. 2006. Glutathione reductase and a mitochondrial thioredoxin play an overlapping role for maintaining iron-sulfur enzymes in fission yeast. Eukaryot. Cell 51857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song, J.-Y., and J.-H. Roe. 2008. The role and regulation of Trx1, a cytosolic thioredoxin in Schizosaccharomyces pombe. J. Microbiol. 46408-414. [DOI] [PubMed] [Google Scholar]
- 46.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62797-821. [DOI] [PubMed] [Google Scholar]
- 47.Tabor, S., H. E. Huber, and C. C. Richardson. 1987. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 26216212-16223. [PubMed] [Google Scholar]
- 48.Tanaka, T., F. Hosoi, Y. Yamaguchi-Iwai, H. Nakamura, H. Masutani, S. Ueda, A. Nishiyama, S. Takeda, H. Wada, G. Spyrou, and J. Yodoi. 2002. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 211695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, N. L., D. A. Day, and A. H. Millar. 2004. Targets of stress-induced oxidative damage in plant mitochondria and their impact on cell carbon/nitrogen metabolism. J. Exp. Bot. 551-10. [DOI] [PubMed] [Google Scholar]
- 50.Trotter, E. W., and C. M. Grant. 2005. Overlapping roles of the cytoplasmic and mitochondrial redox regulatory systems in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 4392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turrens, J. F. 2003. Mitochondrial formation of reactive oxygen species. J. Physiol. 552335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Urrestarazu, L., S. Vissers, and J. Wiame. 1977. Change in location of ornithine carbamoyltransferase and carbamoylphosphate synthetase among yeasts in relation to the arginase/ornithine carbamoyltransferase regulatory complex and the energy status of the cells. Eur. J. Biochem. 79473-481. [DOI] [PubMed] [Google Scholar]
- 54.Van Huffel, C., E. Dubois, and F. Messenguy. 1992. Cloning and sequencing of arg3 and arg11 genes of Schizosaccharomyces pombe on a 10-kb DNA fragment: heterologous expression and mitochondrial targeting of their translation products. Eur. J. Biochem. 20533-43. [DOI] [PubMed] [Google Scholar]
- 55.Vlamis-Gardikas, A., and A. Holmgren. 2002. Thioredoxin and glutaredoxin isoforms. Methods Enzymol. 347286-296. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., and D. F. Bogenhagen. 2006. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 28125791-25802. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, R., R. Al-Lamki, L. Bai, J. W. Streb, J. M. Miano, J. Bradley, and W. Min. 2004. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ. Res. 941483-1491. [DOI] [PubMed] [Google Scholar]