Abstract
Bacterial chemoreceptors form ternary signaling complexes with the histidine kinase CheA through the coupling protein CheW. Receptor complexes in turn cluster into cellular arrays that produce highly sensitive responses to chemical stimuli. In Escherichia coli, receptors of different types form mixed trimer-of-dimers signaling teams through the tips of their highly conserved cytoplasmic domains. To explore the possibility that the hairpin loop at the tip of the trimer contact region might promote interactions with CheA or CheW, we constructed and characterized mutant receptors with amino acid replacements at the two nearly invariant hairpin charged residues of Tsr: R388, the most tip-proximal trimer contact residue, and E391, the apex residue of the hairpin turn. Mutant receptors were subjected to in vivo tests for the assembly and function of trimers, ternary complexes, and clusters. All R388 replacements impaired or destroyed Tsr function, apparently through changes in trimer stability or geometry. Large-residue replacements locked R388 mutant ternary complexes in the kinase-off (F, H) or kinase-on (W, Y) signaling state, suggesting that R388 contributes to signaling-related conformational changes in the trimer. In contrast, most E391 mutants retained function and all formed ternary signaling complexes efficiently. Hydrophobic replacements of any size (G, A, P, V, I, L, F, W) caused a novel phenotype in which the mutant receptors produced rapid switching between kinase-on and -off states, indicating that hairpin tip flexibility plays an important role in signal state transitions. These findings demonstrate that the receptor determinants for CheA and CheW binding probably lie outside the hairpin tip of the receptor signaling domain.
Motile bacteria sense attractant and repellent gradients by means of a signaling complex composed of chemoreceptors (methyl-accepting chemotaxis proteins [MCPs]), a histidine kinase (CheA), and a coupling protein (CheW) (see reference 17 for a recent review). MCPs are homodimeric transmembrane molecules defined by a highly conserved cytoplasmic signaling domain (Fig. 1A). Escherichia coli contains four MCPs, Tsr (serine), Tar (aspartate and maltose), Trg (ribose and galactose), and Tap (dipeptides and pyrimidines), as well as Aer, an MCP-like aerosensor (7, 16, 28, 30, 36, 42). Tar and Tsr are the most abundant chemoreceptor molecules in E. coli and the most studied (17). Amino acid ligands bind directly to their periplasmic domains to modulate, through CheW, the activity of their associated CheA molecules (17).
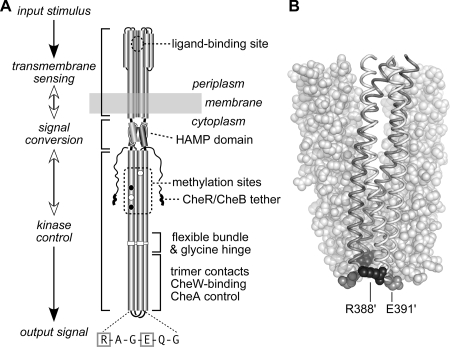
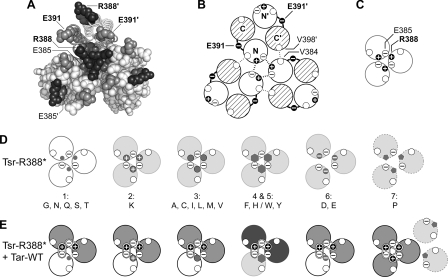
FIG. 1.
(A) Functional architecture of Tsr and other MCP molecules. Native Tsr is a homodimer; each subunit is 551 residues in length. Most of the secondary-structure elements are alpha helices (shown as thickened cylinders). Structural elements of the transmembrane-sensing, HAMP, and kinase control domains are shown in their native arrangement. The kinase control domain is organized as a four-helix bundle of coiled coils. The Tsr residues in the hairpin loop at the cytoplasmic tip of the coiled-coil bundle are shown: the boxed residues (R388 and E391) are the subjects of the present study. Several of the adaptation (methylation) sites are initially synthesized as glutamate residues (white circles); others are synthesized as glutamine residues (black circles) and subsequently converted to glutamates by an irreversible CheB-mediated deamidation. The C terminus of each subunit ends in a pentapeptide (NWETF) to which the adaptation enzymes, CheR and CheB, can bind. An ∼30-residue segment, thought to serve as a flexible linker, joins this tethering site to the kinase control domain. (B) Structure of the Tsr trimer of dimers. This view corresponds to the flexible-bundle and trimer contact portion of the dimer schematic in panel A, with the membrane-distal tip at the bottom. Two dimers are shown space filled (white); the third is shown as a backbone trace (gray and white subunits) with key residues in space fill mode: R388 (black) and E391 (gray). Note that R388 in the gray subunit lies (mostly hidden) at the trimer interface, whereas its counterpart in the white subunit lies at the trimer perimeter. The convention used throughout this report is to designate residues in the nonaxial subunits of the trimer with primes, e.g., R388′ and E391′.
In the absence of attractant ligands, E. coli chemoreceptors stimulate CheA autophosphorylation (17). Phospho-CheA subsequently donates its phosphoryl groups to the CheY response regulator to enhance clockwise (CW) rotation of the flagellar motors, producing random changes in swimming direction (“tumbles”). Attractant ligands suppress CheA activity, lowering phosphorylated-CheY levels and promoting counterclockwise (CCW) rotation, the default motor state that produces forward swimming movements (“runs”) (17). Chemoreceptor complexes sense chemoeffector gradients temporally, using reversible methylation as a memory store of recent chemical conditions. The CheR enzyme transfers methyl groups to four to six specific glutamic acid residues in the MCP cytoplasmic domain; the CheB enzyme demethylates those sites. The interplay between CheR and CheB activities regulates the steady-state MCP methylation level. Chemical stimuli shift the relative enzyme activities, through changes in the substrate properties of the signaling MCP molecules and through phosphorylation control of CheB, to update the methylation record, culminating in sensory adaptation to the new chemical environment (17).
Chemoreceptor signaling complexes are exquisitely sensitive to chemical stimuli and, through sensory adaptation, maintain high sensitivity over a wide range of chemoeffector concentrations (19, 37, 41). The high-gain signaling properties of chemoreceptors appear to arise from cooperative interactions within and between receptor signaling teams in macromolecular arrays (39). Receptors of different detection specificities form commingled clusters, typically at the cell poles (20, 29, 49). The receptor network generates large flagellum-controlling output signals in response to small chemical changes detected by a subset of its component receptors. Although the mechanisms of CheA activation and deactivation are not well understood, receptors appear to form signaling teams and higher-order networks through a trimer-of-dimers organization (see reference 34 for a review). Trimer-based signaling teams could produce signal amplification by allowing team members to respond cooperatively to ligand-induced conformational changes. Additional amplification could arise through conformational interactions between signaling teams in the receptor lattice, for example, through shared contacts with the CheA kinase (17).
The key to understanding how the receptor network operates is to elucidate the structures of receptor signaling teams. The receptor residues that promote trimer formation—in a crystal structure of the Tsr signaling domain (22) and between MCP molecules in vivo (4, 43)—are localized near the tip of the cytoplasmic domain (Fig. 1A). The principal trimer contact residues are nearly invariant in all MCPs (1, 24, 50), which enables receptors with different detection specificities to form mixed-trimer signaling teams (2, 4, 43, 44). The trimer contact region of the receptor also contains the binding surfaces for CheW and CheA that lead to ternary-complex formation and function (5, 14, 31), but the actual docking determinants have not been identified.
A variety of structural models have been proposed for the ternary signaling complex, all of which have CheA and CheW binding to the receptor cytoplasmic tip (14, 25, 31, 32, 38). The hairpin loop at the tip of the receptor signaling domain is highly conserved in MCPs (consensus sequence, R-A-G-E/D-X-G) (1), implying that loop residues have important signaling functions. However, these receptor residues have received scant experimental attention. Coleman et al. (13) created alanine and cysteine replacements at the two hairpin glycine residues in the aspartate receptor, Tar, and found that they prevented kinase activation in vitro. Ames et al. (4) created alanine, proline, and tryptophan replacements at the arginine residue, which is the most tip-proximal of the trimer contact sites, in the serine receptor, Tsr, and found a unique combination of signaling defects in each mutant. The present study reports a comprehensive mutational survey of the two conserved, charged tip residues in Tsr: R388 and E391. In the X-ray structure of the Tsr trimer of dimers, R388 residues at the trimer interface form salt bridges to the interfacial E385 residues in neighboring dimers, contributing to trimer stability (22). The R388 residues in the subunits on the perimeter of the trimer could conceivably play different functional roles (see Fig. 1B). In contrast, E391 residues in the receptor dimers occupy roughly similar positions in the trimer of dimers and do not contribute to the trimer interaction. We found that both of these charged residues played important signaling roles in the receptor, but neither appears to be a critical binding determinant for ternary-complex assembly. Rather, R388 probably influences receptor function through its role as a trimer contact residue. E391, although less critical, appears to regulate conformational transitions between the kinase-activating and kinase-deactivating receptor signaling states.
MATERIALS AND METHODS
Bacterial strains.
All strains used in this study were derivatives of E. coli K-12 strain RP437 (35) and contained the following markers relevant to this study: strains UU1250 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (4), UU1535 [Δaer-1 Δ(tar-cheB)2234 Δtsr-7028 Δtrg-100] (8), UU1581 [Δ(flhD-flhB)4 Δtsr-7028 Δtrg-100] (43), UU1623 [Δ(tsr)7028 Δtap-3654 Δtrg-100] (6), UU1624 [Δ(tsr)7028 Δtap-3654 Δtrg-100 Δaer-1] (15), UU2377 [tsr-R69E Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (6), UU2378 [tsr-T156K Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (6), RP8607 [Δtsr-7028 Δ(cheW-tap)2217 Δtrg-100] (P. Ames and J. S. Parkinson, unpublished data), and RP9352 [Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 ΔcheZ-6725] (27).
Plasmids.
The plasmids used were as follows: pRR48, a derivative of pBR322 (9) that confers ampicillin resistance and has an expression/cloning site with a tac promoter and an ideal (perfectly palindromic) lac operator under the control of a plasmid-encoded lacI repressor inducible by IPTG (isopropyl-β-d-thiogalactopyranoside) (44); pCS53, a derivative of pRR48 that expresses functionally wild-type Tsr-S366C under IPTG control (44); pVS49, a derivative of pACYC184 (12) that confers chloramphenicol resistance and makes a functional yellow fluorescent protein (YFP)-CheZ fusion protein under inducible arabinose control (V. Sourjik, personal communication); pVS102, a relative of pVS49 that makes a functional YFP-CheR fusion protein under inducible arabinose control (21); and pPA801 a relative of pVS49 that makes a functional YFP-CheW fusion protein under inducible arabinose control (Ames and Parkinson, unpublished).
Directed mutagenesis.
Plasmid mutations were generated by QuickChange PCR mutagenesis, as previously described (4). For all-codon mutagenesis, we used oligonucleotide primer pools that were fully degenerate (i.e., that had equal frequencies of the four bases [A, C, G, and T] at all three base positions of the targeted codon). Mutations were verified by sequencing the entire tsr coding region in the mutant plasmid.
Chemotaxis assays.
Host strains carrying mutant derivatives of pCS53 were assessed for chemotactic ability on tryptone soft-agar plates (33) containing 50 μg/ml ampicillin and various concentrations of IPTG. The plates were incubated for 7 to 10 h at 32.5°C.
Swimming behavior.
Cells carrying pCS53 derivatives were grown at 30°C in tryptone broth plus 50 μg/ml ampicillin, induced with 100 μM IPTG after 1 h, and grown for an additional 3 to 4 h to mid-log phase. Cells in culture medium were placed on a glass slide and observed with a phase-contrast microscope. Swimming patterns were classified by eye as “smooth” (mostly running with few or no tumbling episodes), “random” (alternating runs and tumbles), or “tumbly” (frequent tumbling with few episodes of running).
Flagellar-rotation assays.
The flagellar-rotation patterns of strains carrying pCS53 derivatives and induced with 100 μM IPTG were analyzed by antibody tethering as described previously (27). The overall percentage of time spent in CW rotation was calculated from the percentage of cells in each of five rotational categories (exclusively CCW, CCW-reversing, balanced CCW-CW, CW-reversing, and exclusively CW), as described previously (4).
Expression levels of mutant Tsr proteins.
Tsr expression from pCS53 derivatives was analyzed in strain UU1535 to avoid multiple modification states. Cells were grown, and lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (43). Tsr proteins were visualized by Western blotting with a polyclonal rabbit antiserum directed against the highly conserved portion of the Tsr signaling domain (3) and a fluorescent secondary antibody (Cy5-goat anti-rabbit; Amersham Biosciences, Pittsburgh, PA) and quantified with a Molecular Dynamics Typhoon instrument.
Trimer cross-linking.
The abilities of mutant Tsr proteins containing an S366C reporter to form trimers of dimers were assessed by in vivo cross-linking with tris-2-maleimidoethyl-amide (TMEA) (Pierce) in host strain UU1535 as described previously (43, 44).
Receptor clustering.
Mutant pCS53 derivatives were transformed into host strains carrying a compatible plasmid for a YFP-tagged reporter: pVS49/RP9352, pVS102/UU1535 or pVS102/UU1581, and pPA801/RP8607. Cells were grown at 30°C in tryptone broth with 12.5 μg/ml chloramphenicol, 50 μg/ml ampicillin, 100 μM IPTG, and a reporter-specific concentration of l(+)-arabinose: 0.005% for pVS49, 0.01% for pVS102, and 0.004% for pPA801. Cells in mid-log phase were examined by fluorescence microscopy and analyzed as previously described (4, 40). In brief, at least 100 cell images were scored for the presence of one or more fluorescent foci (clusters). In this study, no attempt was made to distinguish between polar and lateral clusters. The fraction of cells with clusters was normalized to the corresponding value for cells containing the wild-type receptor and the same fluorescent reporter.
Protein structural display.
Structure images were prepared with MacPyMOL software (http://www.pymol.org).
RESULTS
Construction of Tsr-R388 and Tsr-E391 mutants.
To survey the effects of single amino acid replacements at Tsr residues R388 and E391, we constructed mutations at the corresponding codons of a plasmid-borne tsr gene, using degenerate PCR primers to randomize all three base positions of the target codon. The parental plasmid, pCS53, expresses a full-function variant of wild-type Tsr that has a reporter site (S366C) for detecting trimer-of-dimer formation by TMEA cross-linking (43). Hereafter, we refer to pCS53 as the parental wild-type plasmid and to Tsr-S366C as wild-type Tsr. Candidate mutant plasmids were identified by DNA sequencing; we obtained a complete set of amino acid replacements at both of the targeted residues. To facilitate discussion, we will use the designations R388* and E391* to refer in general to mutant plasmids and mutant Tsr proteins.
We subjected R388* and E391* mutants to various in vivo assays that measured aspects of Tsr function. Those tests and example results (Fig. 2, 3, and 4) are described below, with all test results summarized in Fig. 5. Based on those findings, the R388* and E391* mutants were arranged into groups of amino acid replacements that produced similar functional properties: seven groups of R388 mutants and five groups of E391 mutants.
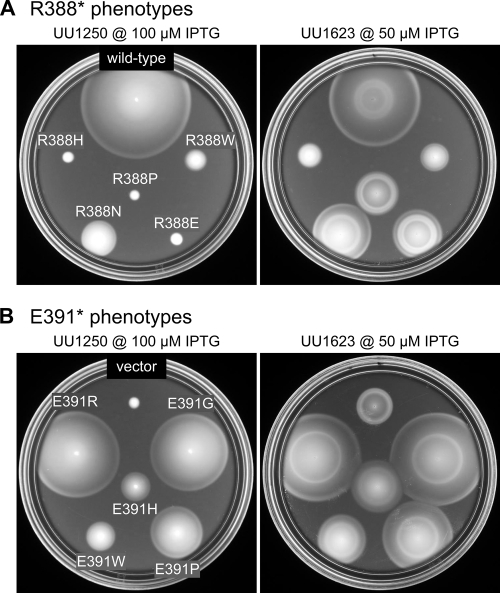
FIG. 2.
Examples of Tsr-R388 and Tsr-E391 mutant phenotypes. In the left-hand images, mutant receptor plasmids were tested in UU1250 (receptorless host) on soft-agar plates containing 100 μM IPTG and incubated for 8.5 h at 32.5°C. In the right-hand images, the same mutant plasmids were tested in UU1623, a host containing wild-type aspartate (Tar) and oxygen (Aer) receptors, on soft-agar plates containing 50 μM IPTG and incubated for 7 h at 32.5°C. (A) R388 mutants. The wild-type control plasmid was pCS53. In UU1250, R388N shows slight Tsr function while the other mutants show none. In UU1623, R388H and R388W jam Tar function and are not functionally rescued, R388N and R388E do not jam and are functionally rescued, and R388P shows neither jamming ability nor functional rescue (compare to vector control in panel B). (B) E391 mutants. The vector control plasmid was pRR48. In UU1250, full Tsr function (E391R and E391G), moderate Tsr function (E391P), and minimal Tsr function (E391W and E391H) are shown. In UU1623, mutants with partial Tsr function show functional rescue by wild-type Tar and do not jam Tar function.
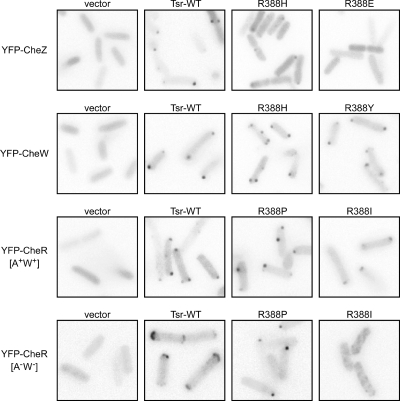
FIG. 3.
Examples of clustering phenotypes of Tsr-R388* and Tsr-E391* mutants. Each image shows an inverted gray-scale portion of the original fluorescence image. A few panels are composites containing images from several areas of the same micrograph. Cells appear as light-gray shapes due to background cytoplasmic fluorescence; the dark spots correspond to receptor clusters. Fluorescent reporters are listed on the left; Tsr mutants or controls are listed above each panel. The host strains (from top to bottom) were RP9352, RP8607, UU1535, and UU1581.
FIG. 4.
Examples of flagellar-rotation patterns of Tsr-R388* and Tsr-E391* mutants. Each panel summarizes the rotational behavior of 100 tethered cells of the receptorless strain UU1250 carrying the indicated pCS53 mutant derivative (see Materials and Methods). The cross-hatched bars indicate the fast-switching phenotype (see the text). WT, wild type.
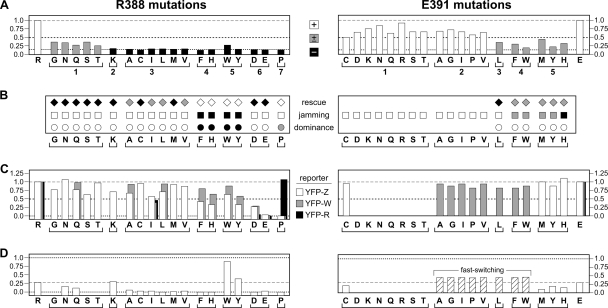
FIG. 5.
Functional properties of Tsr-R388* and Tsr-E391* mutants. For comparison purposes, dashed gray lines show wild-type (WT) values, dotted black lines show the limit value(s) for a particular test, and dashed black lines show 50% values used as the threshold criteria for functional defects. (A) Tsr function relative to WT. Mutant derivatives of pCS53 (IPTG-inducible tsr) were transferred to receptorless strain UU1250 and tested for serine chemotaxis on tryptone soft-agar plates containing 100 μM IPTG. Colony sizes and morphologies were recorded after 8 h of incubation at 32.5°C. The wild-type controls are at the left and right ends of the panels. Mutants, indicated by single-letter amino acid replacements, are grouped below according to the results of additional function tests. The dashed gray lines (at 1.0), dashed black lines (at 0.5), and dotted black lines (at 0.15), show the wild-type, 50%, and vector control values as described above. The white bars indicate wild-type serine ring morphology, the gray bars indicate a ring with aberrant morphology, and the black bars denote no ring. (B) Complementation behavior. White symbols indicate absence of an effect, gray symbols indicate a partial effect, and black symbols indicate a full effect. The tests are described in Materials and Methods and Results. (C) Clustering proficiency relative to WT. The normalized fraction of cells that show one or more clusters with the indicated fluorescent reporters (YFP-Z, YFP-CheZ; YFP-W, YFP-CheW; YFP-R, YFP-CheR). Note that the scale for R388* mutants begins below 0.0 to emphasize mutants with near-zero values. (D) CW rotation. CW time was calculated from flagellar-rotation patterns in the receptorless strain UU1250, as described in Materials and Methods. Note that the scale for R388* mutants begins below 0.0 to emphasize mutants with near-zero values. E391* mutants in groups 2, 3, and 4 exhibited fast-switching behavior (see the text).
Tsr function of R388* and E391* mutants.
Mutant plasmids were tested for Tsr function in UU1250, a host strain that lacks all five MCP family transducers (4), by assessing the sizes and chemotactic ring morphologies of transformant colonies on tryptone soft-agar plates over a range of inducer (IPTG) concentrations (Fig. 5A). At 100 μM IPTG, an optimal concentration for wild-type Tsr function, all amino acid replacements at E391 supported serine chemotaxis to some extent; 13/19 had wild-type serine ring morphology (Tsr+), and 6/19 had altered ring morphology (Tsr±). In contrast, most R388* mutants were completely defective in Tsr function; 14/19 mutants were Tsr− and 5/19 were Tsr±. The levels of the Tsr* proteins under these same growth and inducer conditions ranged from 0.6 to 1.3 of wild type (data not shown), indicating that the mutant phenotypes were most likely due to altered Tsr function rather than to aberrant expression or stability of the mutant proteins. Examples of the mutant colony morphologies are shown in Fig. 2, left.
Jamming and rescue tests.
Receptors of different types form mixed signaling teams based on a trimer-of-dimers organization (4, 34, 43). In such mixed teams, wild-type aspartate (Tar) receptors can functionally rescue some Tsr defects and some mutant Tsr receptors can abrogate or jam the functions of their wild-type Tar partners (2, 4). The jamming and rescue assays provide functional tests for trimer formation. To look for jamming and/or rescue effects of R388* and E391* proteins, mutant plasmids were transferred into UU1624, a strain lacking all chemoreceptors but Tar, or strain UU1623, which lacks all chemoreceptors but Tar and the aerotaxis transducer Aer. The jamming and rescue behaviors of Tsr* mutants were identical in both strains and are summarized in Fig. 5B. Examples of the soft-agar plate assays used to assess these effects are shown in Fig. 2, right.
In these tests, cells expressing wild-type Tsr form colonies with an outer ring indicative of serine chemotaxis and an inner ring indicative of aspartate chemotaxis (Fig. 2A). Based on their colony morphologies, R388* mutants showed three rescue and jamming patterns. R388* mutants in groups 4 (F, H) and 5 (W, Y) jammed Tar function, indicating formation of mixed, but defective, trimers of dimers (Fig. 5B and R388H and R388W in Fig. 2A). These two mutant groups exhibited no rescue of Tsr function by Tar at any expression level tested (i.e., IPTG concentrations of 50, 100, and 150 μM). Rescue was noted in nonjamming mutant groups 1, 2, 3, and 6 (R388N and R388E in Fig. 2A). The rescuable mutant proteins must be capable of forming mixed, and functional, trimers of dimers. Only R388P (group 7) was neither rescuable nor jamming (Fig. 2A), which could mean that this Tsr* protein cannot form stable mixed trimers of dimers with Tar.
Tsr+ E391* mutant groups 1 and 2 had considerable Tsr function and could not be assessed for functional rescue. Not surprisingly, these mutant proteins did not jam wild-type Tar function (Fig. 5B and E391R, E391G, and E391P in Fig. 2B). One other mutant, E391L (group 3), was also nonjamming, but alone it was sufficiently defective in Tsr function (Tsr±) to see functional rescue in the coexpression tests with Tar. The remaining Tsr± E391* mutants (groups 4 and 5) were functionally rescued at low induction levels (see E391W and E391H in Fig. 2B) but jammed Tar function at higher induction levels (not shown in Fig. 2). These expression-dependent behaviors probably mean that mixed trimers with one Tsr* member can function, whereas those with two Tsr* members cannot.
Heterodimer tests.
Receptor subunits in trimers of dimers can occupy two distinct spatial environments, at the trimer interface or at the trimer periphery. Residues that lie at the trimer interface in one subunit of a dimer occupy the peripheral position in the other subunit of the dimer (Fig. 6). Some receptor residues could conceivably play different functional roles in each location. To determine whether R388 or E391 might play such multiple roles, we asked whether heterodimeric molecules containing a mutant residue in one subunit and its wild-type counterpart in the other could support Tsr function, as assessed by soft-agar chemotaxis assays. These tests were done by expressing R388* or E391* subunits in host strains (UU2377 and UU2378) with other receptors (Tar, Tap, Trg, and Aer) deleted and whose chromosomal tsr genes carried a recessive serine-binding lesion (R69E or T156K, respectively) (6). Tsr molecules have two symmetric serine-binding sites at the dimer interface, but one functional site suffices for serine sensing (18, 26, 48). Thus, in these tests, neither mutant homodimer can mediate serine chemotaxis, whereas heterodimers will have one functional serine-binding site. Those heterodimers will support Tsr function if the R388* or E391* subunit carries a recessive defect.
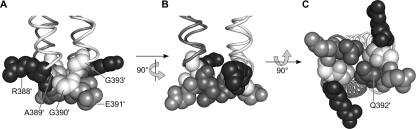
FIG. 6.
Structural features of the tip region in receptor trimers of dimers. (A) End-on view of the trimer in Fig. 1B showing the cytoplasmic tip and the relative positions of R388 (black), E391 (gray), and E385 (dark-gray) residues in the trimer of dimers. (Symmetry-related residues in the trimer are not explicitly labeled.) Note that the two E391 residues in a dimer occupy roughly equivalent positions in the trimer, whereas the E385 and R388 residues do not. At the trimer interface, R388 forms a salt bridge with E385 in the adjacent dimer, which serves to stabilize the trimer arrangement. The corresponding residues (R388′ and E385′) in the nonaxial subunits of the trimer do not engage in interactions between dimers. (B) Schematic of a trimer containing three Tsr-WT (wild-type) dimers. Helices at the N-terminal side of the tip are shown as open, and those at the C-terminal side of the tip are shown with cross-hatching. The E391 residues lie in the loops connecting the N- and C-terminal helices of each subunit. In the trimer, each dimer contributes residue contacts from two helices (N and C′) to the trimer interface, with the majority of interactions, including those between R388 and E385 shown here (thick dashed lines), involving residues in the N-terminal helices. A few additional trimer contacts involve interactions (thin dotted lines) between residues in the N and C′ helices, for example, V384 (N) and V398′ (C′). (C) Schematics of the major (N-terminal) trimer contact helices in trimers of dimers from Fig. 7B showing the R388 and E385 interactions. The unfilled circles represent V384. (D) Schematics of the major (N-terminal) trimer contact helices in Tsr-R388* mutant trimers of dimers. The white helices depict mutants with residual Tsr function, and the light-gray helices depict mutants with no Tsr function. The small dark-gray symbols depict the mutant amino acid replacements. All mutants except R388D, R388E, and R388P probably make trimers with reduced stability (group 1, G, N, Q, S, T; group 2, K; group 3, (A, C, I, L, M, V) or altered geometry (group 4, F, H; group 5, W, Y). The D, E (group 6) replacements introduce acidic residues that probably prevent stable trimer formation through charge repulsion with E385. The P (group 7) replacement probably destabilizes the N helices, disrupting the major trimer contact interface (dashed outline). (E) Schematics as in panel D showing the probable composition and function of mixed trimers of dimers in R388* jamming and rescue tests with Tar-WT. Tsr* helices, functional, white, and nonfunctional, light gray; Tar helices, functional, gray, and nonfunctional, dark gray. Mixed trimers containing a group 1, 2, 3, or 6 Tsr* dimer exhibit both Tar and Tsr functions because they have most of the correct residue contacts at the trimer interface. However, mixed trimers containing group 4 or 5 Tsr* subunits (F, H/W, Y) have neither Tsr nor Tar function, perhaps owing to an altered trimer geometry or dynamic behavior caused by the Tsr* member. The R388P receptor (group 7), which is neither rescuable nor jamming, probably fails to form stable mixed trimers with Tar.
The Tsr+ mutations in E391* groups 1 and 2 were not tested because the mutant homodimers are fully functional. However, the Tsr± mutations in E391* groups 3 to 5 were sufficiently defective and proved recessive in these heterodimer tests (Fig. 5B). All R388* mutations were sufficiently defective for the heterodimer test, and most of those lesions also proved to be recessive. However, R388* mutations in groups 4 (F, H) and 5 (W, Y), which have the largest side chain volumes, were fully dominant, whereas R388P (group 7) was partially dominant (Fig. 5B).
Cross-linking test for trimer-of-dimer formation.
TMEA, a trifunctional thiol-reactive cross-linker, readily enters cells and captures Tsr-S366C molecules in two- and three-subunit products that are thought to derive from the interfacial subunits in trimers of dimers (43). Assuming that all receptor molecules in a cell join trimers of dimers and that trimer-of-dimer associations are stable in the presence of CheW and CheA (44), TMEA should cross-link a maximum of 50% of the receptor subunits. We surveyed UU1535 cells carrying R388* and E391* plasmids for trimer formation with TMEA cross-linking treatments of 15 s and 15 min. (UU1535 lacks all receptors and also the MCP-modifying enzymes CheR and CheB to simplify Tsr band patterns on sodium dodecyl sulfate-polyacrylamide gel electrophoresis.) Cross-linking levels of wild-type Tsr increased somewhat over this period, reaching 52% cross-linking at 15 min (data not shown). However, the 15-s reaction also yielded a substantial signal (33%), consistent with previous observations that TMEA reacts rapidly and provides a “snapshot” of the receptor population (data not shown) (44). Overall, the R388* receptors had cross-linking values that averaged 36% ± 6% and 45% ± 8% at the two time points, with no significant differences between the various replacement groups (all mutants but R388V were tested) (data not shown). The E391* receptors showed similar cross-linking averages of 36% ± 7% and 58% ± 6% (all E391* were tested) (data not shown). Thus, none of the R388* or E391* mutants showed significant differences from the wild-type cross-linking values, and we conclude that the mutant proteins are capable of making normal levels of trimers of dimers detectable by the TMEA assay.
Cluster formation tests.
Wild-type receptors form signaling clusters, typically at the cell poles, that interact with the six cytoplasmic Che components of the signaling system through direct and indirect binding interactions (reviewed in reference 20). We assessed cluster formation by R388* and E391* receptors with fluorescently tagged Che reporter proteins: YFP-CheW binds directly to the receptor trimer contact region at the cytoplasmic tip (10), YFP-CheR binds directly to the NWETF pentapeptide at the C termini of receptor subunits (20, 47), and YFP-CheZ binds to CheAS, an alternative form of CheA that lacks a phosphorylation domain (11). In turn, CheAS and CheAL, the phosphorylation-competent form of CheA, bind to CheW and receptors to form ternary signaling complexes. Examples of receptor clustering phenotypes using these three YFP-Che reporters are shown in Fig. 3, with the results of all tests summarized in Fig. 5C.
We first tested the R388* mutants with the YFP-CheZ reporter to assess their abilities to form ternary signaling complexes. With this reporter in a strain lacking CheZ, wild-type Tsr produced one or more discernible clusters in 73% of cells (data not shown and Fig. 3). The partially functional group 1 R388* mutants and the nonfunctional (but rescuable) group 2 and 3 mutants exhibited clustering signals above 50% of wild type (Fig. 5C). Group 4 and 5 R388* mutants (Tsr−; nonrescuable; strong jamming and dominance) showed less clustering, with three of four mutants yielding signals below 50% of wild type (Fig. 5C). Moreover, those cells with clusters had noticeably higher cytoplasmic backgrounds and smaller clusters, indicating that many of the reporter molecules had not been recruited to the cluster (R388H in Fig. 3). Mutants in groups 6 and 7 had substantially reduced clustering signals. R388E (Fig. 3) and R388P (not shown) produced no evident clusters with the YFP-CheZ reporter. The R388* receptors with low clustering values (groups 4 to 7) could be defective in clustering per se, in CheW binding, or in CheAS binding, as all of these functions underlie cluster detection with the YFP-CheZ reporter.
To distinguish these possibilities, we next asked whether the YFP-CheW reporter could detect cluster formation by any of the group 4 to 7 R388* mutant receptors. With this reporter in a strain lacking CheW, wild-type Tsr produced 84% cluster-containing cells (data not shown and Fig. 3), and one of the partially functional group 1 mutants (R388Q), checked as a positive control, exhibited a comparable signal (Fig. 5C). Two of the group 3 mutants (R388A and R388L) had essentially wild-type clustering signals with this reporter, indicating that the mutant proteins clustered and bound CheW with normal efficiency. Thus, their reduced signal with the YFP-CheZ reporter most likely reflected a defect in ternary-complex assembly. Similarly, the group 4 and 5 mutants exhibited more robust signals with the CheW clustering reporter than they did with the CheZ reporter, implying that these mutant proteins are also defective in ternary-complex assembly (R388H and R388Y in Fig. 3). However, one group 3 mutant (R388I) exhibited less than half the wild-type signal and the group 6 and 7 mutants produced few or no clusters with the YFP-CheW reporter, consistent with defects in CheW binding and/or clustering (Fig. 5C).
To distinguish CheW-binding defects from clustering defects, we examined the R388I, R388D, R388E, and R388P mutant receptors with a YFP-CheR reporter, which binds directly to the C termini of receptor molecules (20, 47). In a host strain lacking the CheR/CheB proteins, wild-type Tsr produced clusters in about 67% of the cells with the CheR reporter (data not shown and Fig. 3). The clustering signal of R388I was about 50% of wild type, similar to those seen for other reporters with this mutant, indicating a somewhat reduced propensity for clustering (Fig. 5C). R388D and R388E showed essentially no clustering with the CheR reporter, indicating a severe defect in cluster formation. R388D exhibited less extreme clustering defects with the CheZ and CheW reporters, suggesting that ternary-complex components may assist cluster formation by this mutant receptor. In contrast, the R388P protein exhibited a normal clustering frequency with the YFP-CheR reporter, even though it showed no clustering with other reporters (Fig. 5C).
To determine whether the YFP-CheR reporter clustering patterns of R388I and R388P receptors were dependent on CheA/CheW, we also examined them in a host strain (UU1581) with all chemotaxis-related genes deleted. In this strain, wild-type receptors form broad polar clusters (“caps”) detectable with the CheR reporter (Fig. 3). The R388I receptor failed to form polar clusters or caps in UU1581 (Fig. 3), indicating that ternary-complex components contribute substantially to the R388I clusters seen with the CheW and CheZ reporters. However, R388P molecules formed tight clusters in UU1581 (Fig. 3), suggesting that these mutant receptors might be associating by a different mechanism.
The E391* mutants, which exhibited more Tsr function than their R388* counterparts, showed normal behavior in selected clustering tests (Fig. 5C). One representative of the Tsr+ group 1 mutants (E391C) had a wild-type clustering signal with the YFP-CheZ reporter, as did the Tsr± group 5 (E391M, E391Y, and E391H) mutants (Fig. 5C). Mutants in groups 2 to 4 produced normal clustering signals with the YFP-CheW reporter.
Swimming and flagellar-rotation tests.
A strain's swimming behavior and underlying pattern of flagellar rotation reflect its steady-state level of phospho-CheY, which in turn depends on the autophosphorylation activity of CheA. To assess the abilities of R388* and E391* receptors to form CheA-activating ternary complexes, we first asked whether the mutant receptors could produce tumbling episodes during swimming, which are indicative of CW flagellar rotation and CheA activation. To examine any CW output signals they might produce, we surveyed mutant plasmids in strain UU1535, which lacks chemoreceptors and the CheR/CheB adaptation enzymes but has wild-type levels of CheW and CheA. In this strain, wild-type Tsr elicits nearly incessant tumbling behavior, reflecting high CheA activity in ternary signaling complexes (6). Consistent with their demonstrable Tsr function, all E391* plasmids produced tumbling behavior in UU1535 (data not shown), indicating some ability to activate CheA. In contrast, the R388* mutants exhibited a range of behaviors in UU1535 swimming tests: group 1 and 2 mutants were mainly tumbly, and group 3 mutants exhibited occasional tumbling, indicating some ability to activate CheA (data not shown). The group 4 to 7 mutants (with one exception) swam smoothly, characteristic of the CCW default flagellar rotation that prevails in the absence of CheA activation. The exceptional mutant, R388W, exhibited substantial tumbling behavior. Note that this mutant receptor also showed higher levels of CheW binding and ternary-complex formation in clustering tests than did the other group 5 mutant, R388Y (Fig. 5C).
To obtain a more quantitative measure of CheA activation by R388* and E391* receptors, we examined the flagellar-rotation patterns of tethered UU1250 cells carrying mutant plasmids. The results are summarized in Fig. 5D with examples of the patterns observed shown in Fig. 4. UU1250 cells carrying the wild-type Tsr plasmid spent ∼28% of their time rotating in the CW direction when induced with 100 μM IPTG. Most R388* mutants exhibited little or no CW behavior, indicating substantial defects in CheA activation (R388H and R388E in Fig. 4). This pattern was the case for all mutants in groups 3, 4, 6, and 7 (Fig. 5D). The group 2 mutant (R388K) had a normal rotation pattern (Fig. 4). Two of the partially functional group 1 mutants (R388N and R388Q) also exhibited some CW output but were more CCW biased than the wild type (e.g., R388N in Fig. 4). In contrast, group 5 mutants produced CW-biased rotation patterns (Fig. 5D). R388W caused extreme CW output (Fig. 4), whereas R388Y produced cells with variable extents of CW bias, as well as some with exclusively CCW behavior (Fig. 4).
All E391* receptors tested by flagellar rotation produced detectable levels of CW rotation in UU1250 (Fig. 5D), consistent with the UU1535 swimming tests. The Tsr± group 5 mutants (E391H, E91M, and E391Y) exhibited CCW-biased output patterns (E391H in Fig. 4) with 9 to 19% CW output (Fig. 5D). One representative of the Tsr+ group 1 mutants was examined and had a relatively normal rotation pattern in UU1250 (E391C in Fig. 4). In contrast, the Tsr+ group 2 E391* mutants displayed a novel rotation pattern characterized by reversal rates approximately fivefold higher than in wild-type cells (data not shown). Overall, these mutants spent nearly 50% of their time in the CW rotational mode (E391G in Fig. 4). The E391* mutants in groups 3 and 4, which had less Tsr function than those in group 2, displayed similar fast-switching rotation patterns (E391W in Fig. 4). Evidently, receptors with fast-switching properties can function with near wild-type efficiency (e.g., E391G in Fig. 3). Interestingly, all fast-switching receptors contained hydrophobic or nonpolar amino acid replacements (G, P, A, V, I, L, F, and W).
DISCUSSION
Structural role of R388 in the Tsr trimer of dimers.
The R388 residues in a Tsr dimer occupy two distinctly different structural environments in the trimer of dimers (Fig. 6A). At the trimer interface, R388 promotes dimer-dimer interaction by forming a salt bridge and hydrogen bonds through water to the E385 residue in the neighboring Tsr dimer (22). The guanidinium groups of those axial R388 side chains also pack against one another, which may contribute further to the stability of the trimer arrangement (Fig. 6A). In contrast, the R388 residues in the nonaxial dimer subunits are solvent exposed on the periphery of the trimer tip. The guanidinium groups of those R388 side chains pack against the carboxyl group of the E385 residue in the same subunit, which might contribute to helix stability in the dimer tip.
The trimer-of-dimers arrangement buries nearly 1,000 Å2 of surface area near the receptor tips (22). Two of the four helices in each dimer, the N-terminal helix from one subunit and the C-terminal helix from the other, contribute trimer-stabilizing determinants (shown schematically in Fig. 6B). The axial N-terminal helix bears R388 and most of the other trimer-stabilizing residues (V384, N381, L380, L378, I377, N376, and F373). Several of those residues (V384 and N376) promote trimer formation through hydrophobic or polar interactions with residues (V398 and R409) in the C-terminal helices of the other subunits (22) (a V384-V398 pair is shown in Fig. 6B). In all, five residue pairs contribute to interdimer interactions in the trimer; another (N381) forms a hydrogen-bonded network with its axial counterparts to stabilize the trimer interface (22). Thus, the R388-E385 pair is one of a number of residue interactions that contribute to trimer stability (Fig. 6B).
Altered trimer shape or stability can account for R388* functional defects.
The tip residues involved in trimer-stabilizing contacts are highly conserved throughout the MCP superfamily (1). We found that every amino acid replacement at R388 drastically impaired Tsr function and that most replacements destroyed function entirely (Fig. 5A). We propose that the functional defects of R388* mutants are due to the structural role of this residue in the trimer-of-dimers arrangement. Loss of the ionic interactions between R388 and E385 at the trimer interface should reduce trimer stability. Moreover, some amino acid replacements at R388 might create steric clashes or vacant spaces that additionally perturb trimer geometry. The resultant changes in trimer stability are probably rather modest ones, as the additional stabilizing pairs involved in trimer formation have not been altered. Therefore, it is not surprising that TMEA cross-linking efficiencies were unaltered in any R388* mutants. This assay for trimer formation may not be sufficiently sensitive to detect subtle changes in trimer stability or shape, particularly as the TMEA reporter site (S366C) lies on the N-terminal side of the trimer contact region, distal to R388 and the receptor tip. Other functional defects of R388* receptors correlate well with the chemical properties of their amino acid side chains and their expected influence on trimer stability, as summarized below.
Acidic amino acid replacements at R388 (group 6) might be expected to have a drastic effect on trimer stability through charge repulsion with E385 (Fig. 6D). Indeed, R388E receptors failed to cluster, to bind CheW, or to assemble ternary signaling complexes (Fig. 5C). R388D receptors had similar but less severe defects, perhaps because the shorter aspartate side chain partially attenuates the charge interaction with E385. The clustering defects of R388D/E mutants suggest that stable trimer formation is a prerequisite for receptor clustering and possibly for subsequent CheW binding and ternary-complex assembly, as well. However, in the presence of wild-type Tar receptors, R388D/E receptors regained function (Fig. 5B), consistent with the formation of stable mixed trimers. We suggest that these rescue effects take place in mixed trimers containing one Tsr* member, whose destabilizing effects would be minimal because only one of the many trimer-promoting interactions is affected (Fig. 6E). This model of functional rescue also implies that a single R388D/E lesion on the periphery of a trimer is not deleterious to the team function.
The R388P receptor (group 7) appeared to be the most defective of all R388* mutants, perhaps due to the expected destabilizing effect of a proline replacement on the major trimer contact helix. R388P exhibited neither functional rescue nor jamming behavior and might be excluded from mixed trimers (Fig. 6E). However, R388P molecules did form TMEA cross-links and tight polar clusters detectable with a YFP-CheR reporter, and in a CheA/CheW-independent fashion. The R388P lesion probably potentiates an aberrant structural interaction, for example, strand swapping of C-terminal helices between dimers that can lead to cellular clustering.
The least defective R388* mutants had small, nonhydrophobic amino acid replacements (group 1: G, N, Q, S, T) that would not be expected to cause steric problems at the trimer interface (Fig. 6D). However, hydrophobic replacements of any size (group 3: A, C, I, L, M, V) had more drastic effects on function, indicating that the chemical nature of the 388 side chain is a critical factor. Side chain volume is evidently important as well, because lysine (group 2) was not an acceptable substitute for arginine, despite a similar basic character. R388K receptors formed ternary complexes and activated CheA with wild-type efficiency, which might reflect near-normal trimer stability. However, R388K could not mediate serine responses, presumably reflecting an altered ability to switch signaling conformations.
R388 receptors with very large amino acid replacements (groups 4 and 5: F, H/W, Y) formed clusters and bound CheW well but appeared to be defective in signal state switching. R388F/H assembled ternary complexes with reduced efficiency and did not activate CheA. However, R388Y also formed ternary complexes inefficiently but nevertheless activated CheA (Fig. 5). R388W formed ternary complexes with higher efficiency and activated CheA to very high levels (Fig. 5). We suggest that these bulky amino acids allow trimer formation but lock or distort the conformation of the resultant ternary signaling complex (Fig. 6D). R388F/H receptors are locked in the CheA-deactivating (CCW) signaling state; R388W/Y receptors are locked in the CheA-activating (CW) mode. The reduced efficiency of ternary-complex formation by R388F/H/Y may reflect a need for conformational flexibility during ternary-complex assembly. Both groups of mutants jam Tar function, suggesting that a single mutant member can prevent signaling-related conformational changes in a mixed-trimer team (Fig. 6E).
R388 is not a critical docking determinant for ternary-complex assembly.
Except for E and P replacements, R388* receptors formed ternary signaling complexes with at least 25% of wild-type efficiency (Fig. 5C). Assuming that these signaling complexes are based on trimer-of-dimer teams, we conclude that R388 is not a critical binding determinant for CheW or CheA in either of its trimer structural environments. We ascribe the inability of R388E and R388P receptors to assemble ternary signaling complexes to a failure to form stable trimers, which in turn are needed to form polar clusters and to recruit CheW and CheA.
Structural role of E391 in the Tsr dimer.
The two E391 residues in a dimer occupy similar positions in the trimer of dimers (Fig. 6A) and play no direct role in stabilizing that arrangement (22). Rather, E391 comprises the turn in the hairpin loop connecting the N- and C-terminal helices (Fig. 6B). In both subunits, the E391 side chain is oriented away from the dimer axis and is extensively solvent exposed (Fig. 7). The E391 carboxyl group does not participate in structural interactions with neighboring residues but is nevertheless a nearly invariant feature of this position in MCPs (1). However, despite its highly conserved acidic character, E391 tolerated a variety of basic, polar, and hydrophobic amino acid replacements with only modest effects on performance (Fig. 5). Below, we summarize evidence to suggest that the primary role of E391 is to modulate the rate or range of subunit motions within the dimer through its influence on the structural stability of the tip region.
FIG. 7.
Structural detail of residues at the hairpin tip of a Tsr dimer. The white backbone trace depicts the subunit that would be on the outside in the trimer arrangement; the gray trace depicts the subunit that would be at the trimer interface. Black, R388; white, G390 or G393; light gray, A389; gray, E391; dark gray, Q392. (A) Front view. (B) Side view. (C) End-on view.
Altered dimer dynamics can account for E391* signaling behaviors.
Replacement of E391 with nonpolar amino acids (groups 2, 3, and 4) produced a fast-switching output behavior. The small hydrophobic replacements exhibited normal efficiencies of CheW binding and, presumably, ternary-complex formation (Fig. 5C) and activated CheA at near-normal levels. The elevated switching rates had only modest effects on Tsr function (less than 50% reduction in the soft-agar expansion rate). Larger hydrophobic (L) and aromatic (F,W) replacements caused similar fast-switching behavior but had more severe effects on Tsr performance (over 50% reduction in the expansion rate with altered chemotactic ring morphology). Fast-switching behavior could reflect instability of both the kinase-on and kinase-off signaling states. Hydrophobic residues at the solvent-exposed E391 position would probably destabilize the tip, which in turn could accommodate increased dynamic motions in other parts of the receptor molecule. For example, the trimer contact region is flanked on the tip-distal side by a “flexible bundle” (1) containing a glycine hinge that plays a key role in signal state switching (13). This region of the receptor may stiffen or bend in response to repellent and attractant stimuli, which have been shown to trigger relative movements of the dimers within a trimer (45, 46). Other large, but less hydrophobic, replacements at E391 (group 5: M, Y, H) also impaired Tsr function but did not cause rapid switching behavior (Fig. 5D), highlighting the role of hydrophobic residues in the fast-switching phenotype. Group 5 mutant receptors had CCW-biased outputs and might be shifted toward the attractant-occupied signaling conformation, which has been suggested to be a more dynamic structural state (23).
Mutant E391 receptors with large-volume side chains (F, H, M, W, Y), whether fast switching or not, exhibited both rescue and jamming effects in Tar coexpression tests (Fig. 5B). Rescue occurred at low Tsr*/Tar stoichiometry, suggesting that in mixed trimers two Tar molecules can moderate the destabilizing influence of a single E391* member. Jamming occurred at higher Tsr*/Tar ratios, suggesting that a single Tar member cannot overcome the destabilizing influence of two E391* molecules in a mixed trimer. The Tsr defects of large-residue E391* mutants were recessive in heterodimer tests, indicating that one wild-type subunit can compensate for the destabilizing influence of the other. Because E391 residues do not directly interact in the dimer (Fig. 7), recessiveness implies that structural interactions between the two subunits can overcome the E391* defect. These functional behaviors favor models in which E391 influences both intrasubunit and intersubunit interactions in the four-helix bundle of the signaling tip. These motions could, in turn, modulate the stability of trimer signaling teams, or, alternatively, the trimer interface might serve as a fulcrum, directing stimulus-induced conformational changes to the nonaxial subunit.
E391 is not a critical docking determinant for ternary-complex assembly.
All E391* mutants formed ternary signaling complexes with apparently normal efficiency, indicating that E391 is not critical for cluster formation or for CheW or CheA binding. Other residues in the hairpin loop have not been as extensively tested but are also unlikely to be important docking determinants. A389, G390, and G393 are conserved tip residues, but probably for their backbone flexibility and small side chains rather than as specific binding determinants. Q392, a less conserved loop residue, faces the subunit interface in the dimer and is not very exposed (Fig. 7).
Signaling roles of residues at the receptor tip.
The principal role of R388 appears to be stabilization of the trimer of dimers. E391 appears to control dynamic motions of the receptor tip. Neither of these conserved residues, or their neighboring residues in the hairpin tip, seems likely to function as a docking determinant for CheW/CheA. Some amino acid replacements at R388 do disrupt ternary-complex assembly, but most likely through their effects on trimer stability and receptor clustering, which appear to be prerequisites for signal complex formation. These findings favor models of the ternary signaling complex in which CheW/CheA interact with the flanks of the receptor, possibly in the clefts between dimers in the trimer arrangement, but not directly with the hairpin tips.
Acknowledgments
We thank Khoosheh Gosink for doing some of the mutant characterization experiments, Victor Sourjik (University of Heidelberg) for several plasmids used in this work, and Claudia Studdert (National University, Mar del Plata, Argentina) and Peter Ames (University of Utah) for helpful discussions and comments on the manuscript.
This work was supported by research grant GM19559 from the National Institute of General Medical Sciences (J.S.P.) and by a Ruth L. Kirschstein National Institutes of Health fellowship, 5F32GM072406 (P.M.). J.B.O. received support from the Bioscience Undergraduate Research Program, Department of Biology, University of Utah. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 1042885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., and J. S. Parkinson. 2006. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. USA 1039292-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 1766340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 997060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ames, P., Y. A. Yu, and J. S. Parkinson. 1996. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol. Microbiol. 19737-746. [DOI] [PubMed] [Google Scholar]
- 6.Ames, P., Q. Zhou, and J. S. Parkinson. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the E. coli serine chemoreceptor. J. Bacteriol. 1906676-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 1794075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., A. C. Miller, K. K. Gosink, and J. S. Parkinson. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 1863730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. Gene 295-113. [PubMed] [Google Scholar]
- 10.Boukhvalova, M., R. VanBruggen, and R. C. Stewart. 2002. CheA kinase and chemoreceptor interaction surfaces on CheW. J. Biol. Chem. 27723596-23603. [DOI] [PubMed] [Google Scholar]
- 11.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 1852354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman, M. D., R. B. Bass, R. S. Mehan, and J. J. Falke. 2005. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry 447687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, N. R., P. M. Wolanin, J. B. Stock, D. J. Derosier, and D. R. Thomas. 2004. Three-dimensional structure and organization of a receptor/signaling complex. Proc. Natl. Acad. Sci. USA 10117480-17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosink, K. K., M. Buron-Barral, and J. S. Parkinson. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 1883487-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harayama, S., E. T. Palva, and G. L. Hazelbauer. 1979. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol. Gen. Genet. 171193-203. [DOI] [PubMed] [Google Scholar]
- 17.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 339-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwama, T., I. Kawagishi, S. Gomi, M. Homma, and Y. Imae. 1995. In vivo sulfhydryl modification of the ligand-binding site of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 1772218-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasuja, R., Y. Lin, D. R. Trentham, and S. Khan. 1999. Response tuning in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 9611346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kentner, D., and V. Sourjik. 2006. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 9619-624. [DOI] [PubMed] [Google Scholar]
- 21.Kentner, D., S. Thiem, M. Hildenbeutel, and V. Sourjik. 2006. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 61407-417. [DOI] [PubMed] [Google Scholar]
- 22.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400787-792. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: Structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 9911611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261568-585. [DOI] [PubMed] [Google Scholar]
- 25.Levit, M. N., T. W. Grebe, and J. B. Stock. 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 27736748-36754. [DOI] [PubMed] [Google Scholar]
- 26.Lin, L. N., J. Li, J. F. Brandts, and R. M. Weis. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 336564-6570. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J. D., and J. S. Parkinson. 1989. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 868703-8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, X., and R. E. Parales. 2008. Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer Tap. J. Bacteriol. 190972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 2591717-1723. [DOI] [PubMed] [Google Scholar]
- 30.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321253-256. [DOI] [PubMed] [Google Scholar]
- 31.Miller, A. S., S. C. Kohout, K. A. Gilman, and J. J. Falke. 2006. CheA kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry 458699-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, S. Y., P. P. Borbat, G. Gonzalez-Bonet, J. Bhatnagar, A. M. Pollard, J. H. Freed, A. M. Bilwes, and B. R. Crane. 2006. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 13400-407. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8116-121. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 9410541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segall, J. E., S. M. Block, and H. C. Berg. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 838987-8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, T. S., N. Le Novere, M. Daniel Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2792-796. [DOI] [PubMed] [Google Scholar]
- 39.Sourjik, V., and H. C. Berg. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428437-441. [DOI] [PubMed] [Google Scholar]
- 40.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37740-751. [DOI] [PubMed] [Google Scholar]
- 41.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer, M. S., M. F. Goy, and J. Adler. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. USA 743312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA 1012117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studdert, C. A., and J. S. Parkinson. 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. USA 10215623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaknin, A., and H. C. Berg. 2006. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc. Natl. Acad. Sci. USA 103592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaknin, A., and H. C. Berg. 2007. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 3661416-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 354984-4993. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Y., H. Park, and M. Inouye. 1993. Ligand binding induces an asymmetrical transmembrane signal through a receptor dimer. J. Mol. Biol. 232493-498. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, P., C. M. Khursigara, L. M. Hartnell, and S. Subramaniam. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 1043777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhulin, I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45157-198. [DOI] [PubMed] [Google Scholar]