Abstract
Little is known regarding the biological roles of archaeal proteases. The haloarchaeon Haloferax volcanii is an ideal model for understanding these enzymes, as it is one of few archaea with an established genetic system. In this report, a series of H. volcanii mutant strains with markerless and/or conditional knockouts in each known proteasome gene was systematically generated and characterized. This included single and double knockouts of genes encoding the 20S core α1 (psmA), β (psmB), and α2 (psmC) subunits as well as genes (panA and panB) encoding proteasome-activating nucleotidase (PAN) proteins closely related to the regulatory particle triple-A ATPases (Rpt) of eukaryotic 26S proteasomes. Our results demonstrate that 20S proteasomes are required for growth. Although synthesis of 20S proteasomes containing either α1 or α2 could be separately abolished via gene knockout with little to no impact on growth, conditional depletion of either β alone or α1 and α2 together rendered the cells inviable. In contrast, the PAN proteins were not essential based on the robust growth of the panA panB double knockout strain. Deletion of genes encoding either α1 or PanA did, however, render cells more sensitive to growth on organic versus inorganic nitrogen sources and hypo-osmotic stress and limited growth in the presence of l-canavanine. Abolishment of α1 synthesis also had a severe impact on the ability of cells to withstand thermal stress. This contrasted with what was seen for panA knockouts, which displayed enhanced thermotolerance. Together, these results provide new and important insight into the biological role of proteasomes in archaea.
Detailed three-dimensional structures of archaeal proteasomes have provided a firm foundation for understanding how these elaborate nanocompartmentalized complexes mediate protein degradation (3, 11). While their thermostable properties make archaeal proteasomes ideal for structural studies, the optimum growth requirements of archaea, which often include extreme pH, salt, and/or temperature, have limited our fundamental knowledge of how these proteolytic enzymes work in the ubiquitin-free archaeal cell.
Halophilic archaea or haloarchaea have developed into model organisms that are used to study many biological processes. Recent advances in haloarchaeal genome sequencing (4, 6, 14, 15) and the development of new genetic tools, such as the targeted and markerless deletion of chromosomal genes (1), have made this group of microbes ideal for providing insight into cell physiology.
Haloferax volcanii is a haloarchaeon which encodes at least five protein components associated with the proteasome system. These include the α1, β, and α2 proteins, encoded by psmA, psmB, and psmC, respectively, which form at least two 20S proteasome subtypes of differing subunit compositions (α1β and α1α2β) (8, 20). H. volcanii also encodes two proteasome-activating nucleotidase (PAN) proteins, PanA (panA) and PanB (panB), closely related to the regulatory particle triple A-ATPase (Rpt) proteins of eukaryal 26S proteasomes (17). These five proteasomal genes are separately dispersed throughout the major chromosome (17, 20) and, based on Northern blotting, are all translated from single-gene transcripts (7). The α1 and α2 proteins are also translated from the most distal and proximal ends of 2.1- and 1.6-kb polycistronic transcripts, respectively (7).
The only proteasome mutant strains generated in an archaeon to date are those of H. volcanii. These mutant strains include one with a novobiocin resistance marker inserted into the coding region of the psmC gene, encoding the α2 protein (8), and a second strain with a deletion of ∼100 bp and an insertion of a mevinolin resistance marker into the panA gene, encoding the Rpt-like PanA protein (9). The former strain (WFD11 [psmC1::gyrB*-gyrA′]) was used to purify 20S proteasomes of α1β composition, and the (phospho)proteome of the latter strain (GG102 [ΔpanA::hmgA*]) was investigated by tandem mass spectrometry based the robust number of phosphoproteins detected by two-dimensional electrophoresis in its proteome compared to what is the case for the parent strain, DS70 (9). Together, these mutant strains have provided insight into proteasome structure and function, but they are limited in number and scope.
In this report, we present the first systematic generation and phenotypic characterization of mutants with markerless knockouts and/or conditional promoter fusions of each gene associated with the proteasome system in an archaeal genome (H. volcanii psmA, psmB, psmC, panA, and panB genes). The same parent strain H26 (ΔpyrE) was used for the construction of all mutant strains to minimize nonrelated variables and enable comparison. Double mutants were also generated and characterized, including those deficient in both α-type subunits (α1 and α2), in both Pan proteins (PanA and PanB), and in the various combinations of PAN proteins with the α-type subunits. The phenotypes of these strains were investigated under defined growth conditions in the presence and absence of stress agents including the addition of the amino acid analogue l-canavanine, low salt, and high temperature. The results provide new and important insight into proteasome function in archaeal cells.
MATERIALS AND METHODS
Materials.
Biochemicals were purchased from Sigma-Aldrich (St. Louis, MO). Other organic and inorganic analytical-grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA). Desalted oligonucleotides were from Integrated DNA Technologies (Coralville, IN). 2′-Deoxyuridine-5′-triphosphate coupled by an 11-atom spacer to digoxigenin (DIG-11-dUTP), alkaline phosphatase-conjugated antibody raised against DIG, disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate (CSPD), and other DIG-related biochemicals were from Roche Molecular Biochemicals (Indianapolis, IN). Positively charged membranes for Southern hybridization were from Ambion (Austin, TX). Phusion and Vent DNA polymerases, restriction enzymes, T4 polynucleotide kinase, and T4 DNA ligase were from New England Biolabs (Ipswich, MA). AccuPrime GC-rich DNA polymerase was from Invitrogen (Carlsbad, CA). Standard agarose used for the separation of DNA for Southern blotting and routine analysis was from Bio-Rad Laboratories (Hercules, CA). SeaKem GTG agarose used for the separation and isolation of DNA fragments for ligation was from FMC Bioproducts (Rockland, ME).
Strains, media, and plasmids.
Strains, oligonucleotide primers used for cloning, and plasmids are summarized in Table 1 and also in Table S1 in the supplemental material. Escherichia coli DH5α was used for routine recombinant DNA experiments. H. volcanii strains were transformed (5) using plasmid DNA isolated from E. coli GM2163. Liquid cultures were aerated at 200 rpm, and solid cultures were grown on 15% (wt/vol) agar plates. E. coli strains were grown at 37°C in Luria-Bertani medium. H. volcanii strains were grown at 42°C in various media including yeast extract-peptone-Casamino Acids (YPC), Casamino Acids, glycerol-minimal medium (GMM), and GMM with alanine (GMM-Ala). Medium formulae were according to The Halohandbook (5) with the following modifications: 20 mM glycerol served as the sole carbon source for GMM and 25 mM alanine replaced the 5 mM NH4Cl in GMM to generate GMM-Ala. Media were supplemented as needed with novobiocin (0.1 μg per ml), 5-fluoroorotic acid (5-FOA) (50 μg per ml), and uracil (10 and 50 μg per ml for growth in the presence and absence of 5-FOA, respectively). Uracil was solubilized in 1 M NaOH or 100% dimethyl sulfoxide at 50 mg per ml prior to addition to the growth medium.
For growth assays, cells were grown in YPC, GMM-Ala, and GMM as indicated. Cells were freshly inoculated from −80°C glycerol stocks onto appropriate agar-based media on plates. Cells were grown twice to log phase in 2-ml portions of media and used as an inoculum for final analysis of growth under various conditions as described below. Each subculture was inoculated to a final optical density at 600 nm (OD600) of 0.01 to 0.02. For analysis of growth rate and cell yield, cells were grown in 20 ml of media in 250-ml baffled Erlenmeyer flasks. For stress studies, cells were grown in 2 ml of media in capped 13- by 100-mm2 culture tubes. For hypo-osmotic stress studies, cells were grown in GMM-Ala with the standard 2.46 M NaCl as well as concentrations of NaCl reduced as low as 1.0 M. GMM-Ala with no NaCl was added to standard medium for these assays. For canavanine sensitivity assays, cells were grown in GMM-Ala supplemented with sterile-filtered l-canavanine sulfate solution (10 mg/ml in water) at final concentrations of 0, 11.4, 22.7, 34.1, 45.4, 56.8, and 85.15 μM. For heat stress, cells were diluted to OD600 of 0.04 units and transferred in aliquots (200 μl per 1.7-ml Eppendorf tube) to a 65°C water bath for 0 to 7 h. Aliquots were removed, diluted, and grown on GMM-Ala agar at 42°C. For liquid cultures, growth was monitored by an increase in OD600 (where 1 OD600 unit equals approximately 1 × 109 CFU per ml for all strains used in this study). For solid cultures, growth was determined by CFU per ml by spotting 20-μl aliquots of dilution series. All experiments were performed at least three times with triplicate cultures for each.
DNA isolation and analysis.
DNA was separated by electrophoresis using 0.8% (wt/vol) agarose gels in 1× TAE electrophoresis buffer (40 mM Tris acetate, 2 mM EDTA, pH 8.5). Plasmid DNA was isolated from E. coli strains by use of the QIAprep spin miniprep kit (Qiagen, Valencia, CA). DNA fragments were isolated from agarose gels by use of the QIAquick gel extraction kit (Qiagen) as needed. PCRs were purified by MinElute (Qiagen) prior to modification by restriction enzymes (BamHI, HindIII, SpeI, XbaI, or NdeI) or T4 DNA polynucleotide kinase. For rapid PCR screening, template DNA was released from H. volcanii mutant strains and recombinant E. coli DH5α strains as follows. Isolated colonies were transferred to 30 μl of sterile deionized H2O by use of toothpicks, boiled (5 min), chilled on ice (10 min), and centrifuged (14,000 × g; 10 min at room temperature). Supernatant (5 to 10 μl) was used as the template for PCR. For Southern blotting, H. volcanii genomic DNA was isolated by DNA spooling (5).
PCRs.
High-fidelity double-stranded DNA, used for the construction of plasmids listed in Table 1, was amplified by PCR using Phusion, Vent, or a mixture of AccuPrime GC-rich and Vent DNA polymerase at a 9:1 ratio. Taq DNA polymerase was used for screening mutant strains and for the generation of the DIG-labeled double-stranded DNA probes that were used for Southern blotting. All PCRs were performed according to the instructions of the suppliers with the following modifications: 3% (vol/vol) dimethyl sulfoxide was included as needed and 0.1 mM deoxyribonucleoside triphosphate mix was added to the standard DIG-labeling reaction mixture, which included 1× DIG deoxyribonucleoside triphosphate (catalog no. 1277065; Roche). Primer pairs and template DNA used for the PCRs are outlined in Table S1 in the supplemental material. PCR was performed using an iCycler or GeneCycler (Bio-Rad Laboratories), and products were analyzed on 0.8% (wt/vol) agarose gels in TAE buffer. Gels were photographed after being stained with ethidium bromide at 0.5 μg · ml−1 with a Mini visionary imaging system (Fotodyne, Hartland, WI). Sizes of the fragments were estimated using the Hi-Lo DNA molecular weight marker (Minnesota Molecular, Minneapolis, MN).
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Descriptionb | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5α | F−recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | Life Technologies |
| GM2163 | F−ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL136 dam13::Tn9 xylA5 mtl-1 thi-1 mcrB1 hsdR2 | New England Biolabs |
| H. volcanii strainsa | ||
| DS70 | Wild-type isolate DS2 cured of plasmid pHV2 | 19 |
| H26 | DS70 pyrE2 | 2 |
| GZ108 | H26 panB (devoid of PanB) | This study |
| GZ109 | H26 panA (devoid of PanA) | This study |
| GZ114 | H26 psmC (devoid of α2) | This study |
| GZ120 | GZ114 panB (devoid of α2 and PanB) | This study |
| GZ130 | H26 psmA (devoid of α1) | This study |
| GZ131 | GZ114 panA (devoid of α2 and PanA) | This study |
| GZ132 | GZ108 panA (devoid of PanA and PanB) | This study |
| GZ133 | GZ108 psmA (devoid of α1 and PanB) | This study |
| GZ134 | GZ130 panA (devoid of α1 and PanA) | This study |
| GZ136 | H26 PtnaA-psmA (devoid of α1 in the absence of Trp) | This study |
| GZ137 | GZ114 (psmC) PtnaA-psmA (devoid of α1 in the absence of Trp and α2) | This study |
| GZ138 | H26 PtnaA-psmB (devoid of β in the absence of Trp) | This study |
| GZ112 | H26 psmB-pJAM202 | This study |
| Plasmids | ||
| pTA131 | Apr; pBluescript II containing Pfdx-pyrE2 | 2 |
| pET24b | Kmr; E. coli expression vector | Novagen |
| pJAM202c | Apr Nvr; control plasmid derived from pBAP5010 | This study |
| pJAM202 | Apr Nvr; pBAP5010 containing P2rrn-psmB-His6 | 8 |
| pJAM204 | Apr Nvr; pBAP5010 containing P2rrn-psmA-His6 | 8 |
| pJAM205 | Apr Nvr; pBAP5010 containing P2rrn-psmC-His6 | 8 |
| pJAM650 | Apr Nvr; pBAP5010 containing P2rrn-panA-His6 | Maupin-Furlow, unpublished |
| pJAM648 | Apr Nvr; pBAP5010 containing P2rrn-panA | Maupin-Furlow, unpublished |
| pJAM503 | Apr Nvr; pBAP5010 containing P2rrn-His6-panA | Maupin-Furlow, unpublished |
| pJAM2027 | Apr; pTA131 containing psmA with ∼500 bp of genomic DNA flanking 5′ and 3′ of the psmA coding region | This study |
| pJAM2030 | Apr; pTA131 containing psmB with ∼500 bp of genomic DNA flanking 5′ and 3′ of the psmB coding region | This study |
| pJAM2024 | Apr; pTA131 containing psmC with ∼500 bp of genomic DNA flanking 5′ and 3′ of the psmC coding region | This study |
| pJAM2022 | Apr; pTA131 containing panB with ∼500 bp of genomic DNA flanking 5′ and 3′ of the panB coding region | This study |
| pJAM2029 | Apr; pTA131-derived psmA suicide plasmid | This study |
| pJAM2031 | Apr; pTA131-derived psmB suicide plasmid | This study |
| pJAM2025 | Apr; pTA131-derived psmC suicide plasmid | This study |
| pJAM2026 | Apr; pTA131-derived panA suicide plasmid | This study |
| pJAM2023 | Apr; pTA131-derived panB suicide plasmid | This study |
| pJAM2051 | Apr; Nvr; pJAM204 containing 323-bp PtnaA | This study |
| pJAM2053 | Apr; pTA131-derived PtnaA-psmA suicide plasmid | This study |
| pJAM2052 | Apr; Nvr; pJAM202 containing 323-bp PtnaA | This study |
| pJAM2054 | Apr; pTA131-derived PtnaA-psmB suicide plasmid | This study |
Parent strain, gene deletions, PtnaA promoter fusions, and/or replicating plasmids (pJAM202) are included for H. volcanii mutant strains.
Kmr, kanamycin resistance; Apr, ampicillin resistance; Nvr, novobiocin resistance.
DNA sequencing.
Fidelity of all PCR amplified products was confirmed by sequencing the DNA of the plasmid inserts listed in Table 1. Sequence fidelity was also confirmed for the DNA fragments amplified by PCR from the genomic DNA of H. volcanii proteasomal mutant strains using ‘BamHI Forward’ and ‘HindIII Reverse’ primers. Both strands of DNA were sequenced by Sanger automated DNA sequencing using an Applied Biosystems model 3130 genetic analyzer (ICBR Genomics Division, University of Florida).
Southern blotting.
Genomic DNA isolated from H. volcanii parent and mutant strains (∼10 μg) was digested for 6 to 8 h with restriction enzymes, which generated DNA fragments of 1.1 to 2.7 kb, based on the most recent version of the genome sequence (http://archaea.ucsc.edu/, April 2007 version). The cleaved DNA was separated by agarose gel electrophoresis (20 V, 15 to 16 h) and capillary transferred and UV cross-linked to nylon membranes. Double-stranded DNA probes specific for either of the ∼500-bp regions flanking 5′ or 3′ of the coding region of the target gene were labeled with DIG by PCR (see above and Table S1 in the supplemental material for details). DIG-labeled probes were hybridized to the Southern blots and detected by CSPD-mediated chemiluminescence as recommended by the supplier (Roche) with the following modification. An increase in stringency from 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate to 0.1× SSC, 0.1% sodium dodecyl sulfate was included in the washing of the membranes at 65°C after hybridization as needed. Sizes of the fragments were estimated by methylene blue staining of the Hi-Lo DNA molecular weight markers (Minnesota Molecular) on the membrane.
Chromosomal knockout of proteasomal genes.
Proteasomal genes were targeted for markerless deletion from the chromosome of H. volcanii H26, GZ114, GZ108, or GZ130 by the pyrE2-based “pop-in/pop-out” method described in reference 2. In brief, primer pairs (see Table S1 in the supplemental material for details) were designed to PCR amplify ∼500 bp of genomic DNA flanking the 5′ and 3′ ends of the coding region for each target gene. BamHI and HindIII sites were included in the primers (′BamHI Forward' and ‘HindIII Reverse') to facilitate cloning of the PCR-generated DNA fragments into plasmid vector pTA131. For all genes with the exception of panA, an intermediate plasmid which carried the target gene in addition to the ∼500 bp 5′ and 3′ of this gene was generated and used as a template in inverse PCRs with ‘Inverse Forward’ and ‘Inverse Reverse’ primer pairs. Products from the inverse PCRs were phosphorylated using T4 polynucleotide kinase, ligated, and transformed into E. coli DH5α for the generation of the suicide plasmid. For the generation of the panA suicide plasmid, the ‘BamHI Forward’ and ‘Inverse Reverse’ primer pairs and ‘Inverse Forward’ and HindIII Reverse’ primer pairs were used in two separate PCRs, respectively. These ∼500-bp PCR products were purified, phosphorylated, and cloned into the BamHI and HindIII sites of pTA131 by three-way ligation to generate the suicide plasmid. Suicide plasmids which carried the target gene deletion were identified by PCR screening of isolated colonies, sequenced to confirm fidelity, and used for homologous recombination with the H. volcanii genome by the pop-in/pop-out method with 5-FOA (2). Colonies were screened for the absence of a readily generated ∼500-bp PCR product by use of primers specific for the coding region of the gene (′Negative-Forward' and ‘Negative-Reverse’ primer pairs). Strains which did not generate this PCR product were confirmed to be mutant strains by Southern blotting (as described above) and PCR with ‘Confirm-Forward’ and ‘Confirm-Reverse’ primer pairs. These latter PCR products were separated by agarose gel electrophoresis and sequenced to confirm DNA fidelity (as described above).
PtnaA promoter fusions of proteasomal genes.
The tryptophan-dependent tryptophanase (PtnaA) promoter region was inserted upstream of psmA and psmB on the genome of H26 and upstream of psmA on the genome of GZ114 ΔpsmC by use of the following approach. A 337-bp XbaI-to-NdeI DNA fragment carrying the PtnaA promoter region was isolated from the genome of H. volcanii DS70 by PCR using the XbaI_PtnaA up and NdeI_PtnaA down primer pairs as listed in Table S1 in the supplemental material. This PCR product was inserted in the NdeI and XbaI sites of plasmids pJAM202 and pJAM204 to replace the P2rrn with the PtnaA promoter upstream of psmB and psmA and generate plasmids pJAM2052 and pJAM2051, respectively (Table 1). The 1.0- to 1.7-kb XbaI-to-StuI fragments of pJAM2051 and pJAM2052 were inserted into the XbaI to EcoRV sites of pTA131 to generate “half-suicide” plasmids, which were completed by respectively cloning (into the XbaI and AleI sites) the ∼500- to 800-bp region immediately flanking the 5′ end of psmA and psmB on the genome. These 5′ regions were generated by PCR using the following primer pairs: SpeI_PtnaA_psmB and 00375-Confirm-Forward (for psmB) and XbaI_PtnaA_psmA and 00857-Confirm-Forward (for psmA) (see Table S1 in the supplemental material for details). The PCR products were treated with T4 polynucleotide kinase and SpeI (for psmB) or XbaI (for psmA) prior to cloning into the half-suicide plasmids. The final suicide plasmids for insertion of the PtnaA promoter upstream of psmA and psmB on the H. volcanii genome were pJAM2053 and 2054, respectively. Colonies were screened by patching onto GMM plates plus and minus tryptophan. Those strains exhibiting growth only in the presence of tryptophan were confirmed by Southern blotting and PCR using primer pairs listed in Table S1 in the supplemental material.
RESULTS
Generation of H. volcanii strains deficient in the synthesis of proteasomal proteins with a genetic approach.
The proteasome genes of H. volcanii were targeted for markerless deletion to examine the phenotypic consequences of proteasome dysfunction with genetic methods. The genes targeted for deletion included those encoding the α1 (psmA), β (psmB), and α2 (psmC) subunits of 20S core proteasomes and the PanA (panA) and PanB (panB) components of PAN complexes. The pyrE2 mutant strain H26 served as the parent for this initial series of knockouts to facilitate the use of the “pop-in/pop-out” method described by Allers et al. (2). Up to 205 colonies per target gene were isolated and screened for generation of the desired chromosomal deletion. Initial screening was performed using PCRs with primers specific for the coding region of the target gene (′Negative-Forward' and ‘Negative-Reverse’ primer pairs; see Table S1 in the supplemental material). This enabled robust and reproducible high-throughput screening to detect colonies no longer able to generate the ∼500-bp PCR product in contrast to the parent strain. Putative mutant strains were confirmed by Southern blotting using a probe specific for the regions flanking the gene deletion and PCR (see Fig. S1 and S2 in the supplemental material) using primer pairs annealing ∼200 bp outside (5′ and 3′) of the genomic DNA region carried on the suicide plasmid (′Confirm-Forward' and ‘Confirm-Reverse’; see Tables S1 and S2 in the supplemental material). With exception of psmB, all four proteasomal genes were deleted singly from the chromosome, with knockout frequencies ranging from 1.6 to 14% with an average of ∼6% (see Table S3 in the supplemental material). The psmB gene, encoding the sole β subunit which harbors the central proteolytic active site of the 20S core of the proteasome system, was deleted only in part when a wild-type copy of the gene was included in trans on the self-replicating plasmid pJAM202 (see Table S3 and Fig. S1 in the supplemental material).
By use of a similar “pop-in/pop-out” strategy, markerless double mutations of the proteasome system were also generated and confirmed by Southern blotting and PCR (see Fig. S1 and S2 in the supplemental material). Using the ΔpsmC strain GZ114, double mutant strains no longer encoding α2 and either PanA or PanB were generated (GZ131 or GZ120, respectively). Likewise, using the ΔpanB strain GZ108 as the parent, double mutants no longer encoding PanB and either α1 or PanA were generated (GZ133 or GZ132, respectively). With psmA mutant GZ130 as the parent strain, a double knockout devoid of α1 and PanA was also generated (GZ134). The knockout frequencies for these double mutations ranged from ∼2 to 16% with an average of ∼7% (see Table S3 in the supplemental material) and were similar to those frequencies obtained when generating the single-proteasomal-gene mutations, as described above. In contrast, the construction of a double mutant strain deficient in α1 and α2 production was not successful when using either GZ114 (ΔpsmC) or GZ130 (ΔpsmA) as the parent strain in combination with suicide vectors designed for the deletion of either the psmA or the psmC gene, respectively. Over 350 isolated colonies in total were screened for a psmA psmC double mutation by the high-throughput PCR method (described above and in Materials and Methods). All of these colonies generated PCR products which included 500 bp of the target gene and thus were similar to their respective parent strains.
To generate H. volcanii strains deficient in the production of all potential 20S proteasome subtypes (i.e., α1β, α1α2β, and α2β), a new genetic strategy was used in which the tryptophanase promoter PtnaA, which is tightly controlled by tryptophan, was inserted upstream of the psmA and psmB genes (Table 1). This type of PtnaA promoter fusion strategy was recently used to demonstrate the consequences of depletion of the essential chaperonin protein CCT1 from H. volcanii (10). In the presence of tryptophan, the PtnaA promoter inserted upstream of the CCT1-encoding gene was transcribed and cells were viable. In contrast, when cct1 gene transcription was off, cells were no longer able to grow in the absence of tryptophan (10). For our work, the PtnaA promoter was fused upstream of the psmA (α1) genes of both H26 parent and GZ114 (ΔpsmC) strains with efficiencies ranging from ∼3 to 4% (see Table S3 in the supplemental material). Likewise, the PtnaA promoter was fused upstream of the H26 psmB gene, encoding the sole β subunit of H. volcanii, at a frequency of ∼7%. All strains were confirmed by Southern blotting and PCR (see Fig. S1 and S2 in the supplemental material).
Growth of proteasome mutant strains on defined media.
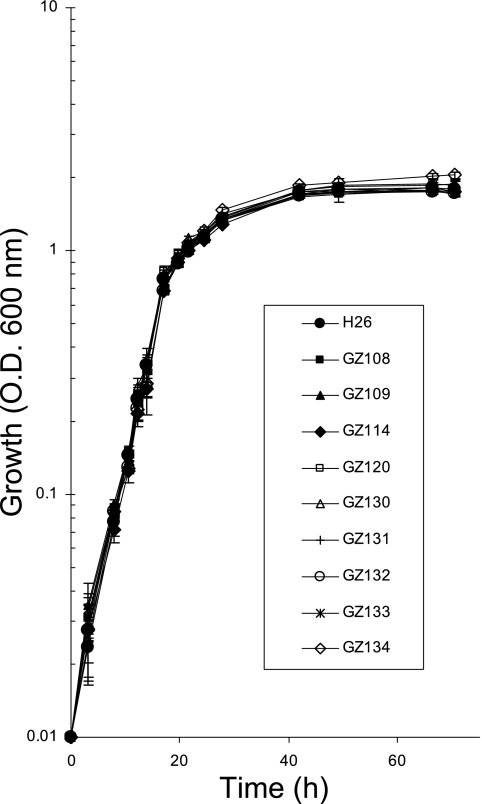
To initiate phenotypic analysis, growth of the proteasome gene knockout strains was analyzed and compared to that of the parent strain in various media under standard laboratory conditions (42°C and 200 rpm). All of the single and double deletion strains displayed relatively normal growth rates and overall cell yield compared to the H26 parent strain on YPC medium (Fig. 1) and/or GMM (Table 2). Thus, singular deficiencies in the synthesis of α1, α2, PanA, or PanB did not significantly limit the growth of H. volcanii under these conditions. Likewise, psmA panA (GZ134), psmA panB (GZ133), psmC panA (GZ131), psmC panB (GZ120), and panA panB (GZ132) double knockouts did not show impaired growth under these conditions. When the source of nitrogen in GMM (ammonium chloride) was replaced with alanine (GMM-Ala), strains with deficiencies in α1 and/or PanA displayed a two- to threefold reduction in their growth rates, which revealed that these strains are sensitive to an organic versus an inorganic source of nitrogen. However, the α1 and/or PanA deficiencies did not significantly impact the overall cell yield of these strains (Table 2). In a previous study, we reported a ΔpanA strain (GG102) that displayed a reduced growth rate and cell yield compared to its parent, DS70, in ATCC 974 medium (9). ATCC 974 is a complex medium that contains a reduced level of sodium chloride, i.e., 2.1 M rather than to the 2.5 M present in YPC and GMM used in this study.
FIG. 1.
H. volcanii proteasomal mutant strains display growth rates and cell yields similar to that of parent strain H26 on complex YPC medium. Growth at 42°C (200 rpm) was monitored by an increase in OD600, where 1 unit was approximately 1 × 109 CFU per ml for all strains.
TABLE 2.
Growth of proteasomal mutant strains
| Strain no. | Proteasome gene(s) deleted | Proteasome protein deficiency | GMMa
|
GMM-Alaa
|
||
|---|---|---|---|---|---|---|
| Growth rate (k) | Cell yield | Growth rate (k) | Cell yield | |||
| H26 | None | None | 0.239 ± 0.034 | 1.61 ± 0.12 | 0.183 ± 0.070 | 1.35 ± 0.01 |
| GZ130 | psmA | α1 | 0.152 ± 0.069 | 1.58 ± 0.01 | 0.086 ± 0.016 | 0.92 ± 0.08 |
| GZ114 | psmC | α2 | 0.237 ± 0.062 | 1.65 ± 0.02 | 0.189 ± 0.044 | 1.27 ± 0.01 |
| GZ109 | panA | PanA | 0.225 ± 0.038 | 1.56 ± 0.02 | 0.052 ± 0.002 | 1.22 ± 0.09 |
| GZ108 | panB | PanB | 0.205 ± 0.047 | 1.56 ± 0.05 | 0.158 ± 0.063 | 1.36 ± 0.20 |
| GZ131 | psmC, panA | α2, PanA | 0.182 ± 0.026 | 1.51 ± 0.02 | 0.110 ± 0.020 | 1.23 ± 0.05 |
| GZ120 | psmC, panB | α2, PanB | 0.170 ± 0.004 | 1.61 ± 0.03 | 0.120 ± 0.007 | 1.29 ± 0.08 |
| GZ132 | panA, panB | PanA, PanB | 0.166 ± 0.014 | 1.58 ± 0.06 | 0.076 ± 0.012 | 1.25 ± 0.03 |
| GZ134 | psmA, panA | α1, PanA | NDb | ND | 0.087 ± 0.018 | 1.22 ± 0.22 |
| GZ133 | psmA, panB | α1, PanB | ND | ND | 0.095 ± 0.022 | 0.93 ± 0.13 |
Strains were grown on GMM or GMM-Ala at 42°C (200 rpm) as indicated. Growth rate (k) is represented as generations per h; cell yield is represented as 109 CFU per ml.
ND, not determined.
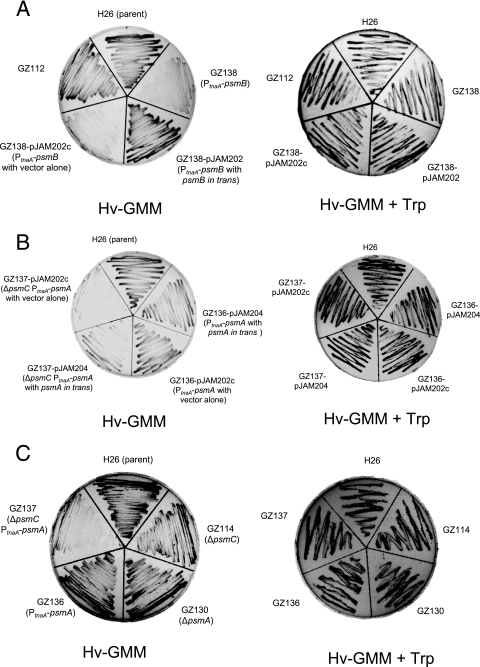
In contrast to the limited influence on cell growth of the proteasomal mutations described above (including knockout of all PAN protein synthesis, as demonstrated by ΔpanA ΔpanB of GZ132), when H. volcanii strains were depleted of all 20S proteasome subtypes, cell growth and overall cell yield were severely impaired (Fig. 2). This was demonstrated by insertion of the tryptophanase promoter, PtnaA, immediately upstream of the psmB gene in “wild-type” cells as well as upstream of psmA in a ΔpsmC strain (GZ138 and GZ137, respectively; Table 1). Since transcription from the PtnaA promoter requires tryptophan, cells can be depleted of the downstream gene product by growth on GMM, a medium devoid of amino acids. The GZ137 and GZ138 strains were constructed to deplete all α-type and β-type subunits of the 20S core particles, respectively, when grown on GMM alone. To analyze the phenotypic consequences of these gene fusions, cells were grown on GMM plus tryptophan and transferred to GMM plus or minus Trp. In the presence of tryptophan (GMM plus Trp), which turns on the transcription of the PtnaA-controlled genes, all strains tested were fully viable and displayed robust growth (Fig. 2). When tryptophan was omitted from the medium (GMM alone), the growth of strains depleted of all 20S proteasome core particle subtypes was severely impaired (GZ137 and GZ138). Control strains including H26 (parent) and GZ114 (ΔpsmC) as well as GZ130 (ΔpsmA) and GZ136 (PtnaA-psmA) were all viable and displayed robust growth in GMM whether or not tryptophan was present. Likewise, the growth of the 20S proteasome core particle-depleted cells was complemented by copies of the proteasome genes provided in trans. That is, GZ137 (ΔpsmC PtnaA-psmA) was complemented for growth on tryptophan minus medium by the self-replicating pHV2-based plasmid pJAM204, which carries the psmA gene in trans. In addition, GZ138 (PtnaA-psmB) cells were fully viable on tryptophan-minus medium when provided a copy of psmB (pJAM202). Both GZ137 and GZ138 strains were not complemented by the plasmid vector alone (pJAM202c).
FIG. 2.
20S proteasomes are required for growth of H. volcanii. H. volcanii strains were grown on solid GMM supplemented with 2.5 mM tryptophan. Cells were transferred by loop to solid GMM with or without 2.5 mM tryptophan (Hv-GMM + Trp or Hv-GMM, respectively) and grown at 42°C as indicated. See Table 1 for strain genotype details.
Based on these results, production of at least a subgroup of the 20S core proteasome population is essential for H. volcanii growth. Although only α1β and α1α2β 20S proteasome subtypes have been purified and characterized to date (8, 20), the viability of the ΔpsmA strain compared to that of the PtnaA-psmB fusion strain suggests that an α2β complex is also present. The essential role of 20S proteasome particles in cell growth and/or division provides substantial support toward our working model in which proteasomes are proposed to be central to the quality control and regulated turnover of proteins in archaea and thus are major players in posttranscriptional control within this group of organisms (12). This premise was originally based on the nanocompartmentalized structure and energy-dependent proteolytic mechanism conserved between archaeal proteasomes and known regulatory proteases (e.g., bacterial Clp and Lon proteases, eukaryal 26S proteasome) as well as the universal distribution of the 20S core particle and absence of other predictable, energy-dependent proteases in the cytosol of archaeal cells.
The essential function of the 20S core particles of proteasomes contrasts with what is seen for PAN complexes, which do not appear essential based on the robust growth of GZ132 (ΔpanA ΔpanB). These results, however, are consistent with the sporadic distribution of PAN (Rpt-like) coding sequences among archaea, including their absence from the genomes of Thermoplasma and Pyrobaculum spp. (12). This also lends support to our working model in which other triple-A ATPase proteins (in addition to PAN) facilitate the proteasome-mediated degradation of proteins in archaea. Likely candidates include Cdc48-like proteins, which appear universal among archaea and are linked to proteasome function in eukaryotes (12).
Proteasome components are important for survival during and after stress.
To further investigate the phenotypes of the H. volcanii proteasome mutant strains, cells were grown under a variety of environmental stresses. These included growth under a low-salt condition (Fig. 3), growth in the presence of the amino acid analogue l-canavanine (Fig. 4), and growth after exposure to high temperature (Fig. 5).
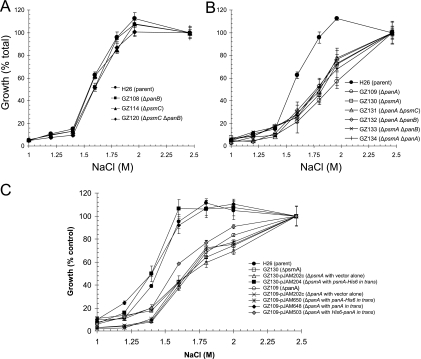
FIG. 3.
Strains deficient in 20S proteasomal α1 or PanA are sensitive to hypo-osmotic stress. Growth of H. volcanii parent (H26) and proteasomal mutant strains in GMM-Ala with NaCl concentrations as indicated and described in Materials and Methods was determined. Growth is represented as a percentage relative to growth for that strain at 2.46 M NaCl (100% growth was 0.92 to 1.36 × 109 CFU·ml−1). Experiments and plating were performed in triplicate and the mean ± standard deviation (SD) was calculated.
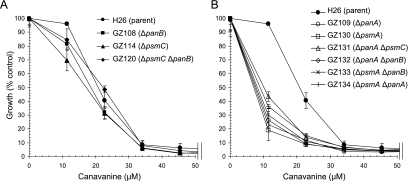
FIG. 4.
Strains deficient in 20S proteasomal α1 or PanA are sensitive to the amino acid analogue l-canavanine. Growth of H. volcanii parent (H26) and proteasomal mutant strains in GMM-Ala supplemented with l-canavanine as indicated and described in Materials and Methods was determined. Growth is represented as a percentage relative to growth at 0 μM canavanine (100% growth similar to that for Fig. 3). Experiments were performed in triplicate and the mean ± SD was calculated.
FIG. 5.
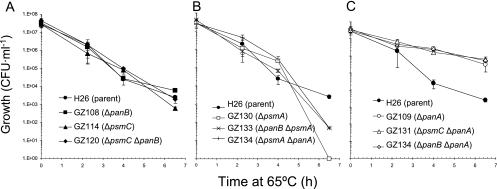
Strains deficient in 20S proteasomal α1 are sensitive to heat stress, whereas strains deficient in PanA are “superthermotolerant.” Parent (H26) and proteasomal mutant strains were grown to log phase at 42°C in GMM-Ala. Cells were diluted to an OD600 of 0.04 units and the effects of heat on cell viability were measured by a shift to 65°C (see Materials and Methods for details). Aliquots of cells were removed at 0-, 2-, 4-, 6-, and 8-h intervals. Cells were diluted, plated on GMM-Ala, and incubated at 42°C for 5 days. Experiments were performed in triplicate and the mean ± SD was calculated.
H. volcanii is a halophilic archaeon that is sensitive to hypo-osmotic stress, with requirements for sodium chloride in the range of 1.7 to 2.5 M for optimal growth (13). The GMM-Ala used in this study includes a variety of salts, most notably sodium chloride at 2.46 M and magnesium salts totaling 0.23 M (88 mM MgCl2 and 142 mM MgSO4). GMM-Ala also includes 20 mM glycerol, which is a common osmolyte of bacteria and eukaryotes, as a carbon source. To assess the ability of H. volcanii parent and mutant strains to grow under a low-salt condition, the sodium chloride concentration of GMM-Ala was varied. Consistent with the salt requirements of this halophilic archaeon, little to no growth was observed for all H. volcanii strains when the NaCl was reduced to 1.25 M and below, even in the presence of the 20 mM glycerol used in this medium (Fig. 3). When the sodium chloride concentrations of GMM-Ala were reduced to 1.6 to 1.8 M, a reduced yet comparable level of growth was observed for parent H26 and psmC and/or panB deletion strains (GZ108, GZ114, and GZ120). Likewise, all four of these strains grew to comparable levels in medium with 2 to 2.46 M concentrations of sodium chloride. Thus, deficiencies in either α2 or PanB appeared to have little to no influence on the ability of H. volcanii to grow under a low-salt condition (Fig. 3A). In contrast, strains with deficiencies in α1 and/or PanA were more sensitive to hypo-osmotic stress than their parent, H26 (Fig. 3B). All strains carrying deletions in psmA and/or panA displayed an approximately twofold reduction in overall cell yield when grown in low-salt medium (1.6 to 2 M NaCl) compared to what was seen for parent strain H26 (Fig. 3B). Growth of the single ΔpsmA and ΔpanA deletion strains was similar to that of the double ΔpsmA ΔpanA knockout strain under all sodium chloride concentrations examined. Thus, mutations in psmA and panA were not additive, suggesting PanA and α1 are associated in the same hypo-osmotic stress response pathway.
Both psmA and panA deletion strains were complemented to wild-type levels of growth under a low-salt condition by gene copies provided in trans (Fig. 3C). The psmA deletion strain GZ130 was readily complemented to wild-type levels of growth under a low-salt condition by transformation of plasmid pJAM204, encoding the α1 protein with a C-terminal polyhistidine tag (His6) and was not complemented by plasmid vector pJAM202c alone. Thus, the hypo-osmotic stress phenotype observed for the psmA deletion strain is due to an α1 deficiency. Furthermore, the His6 tag, which is fused to the C terminus of α1 and predicted to be localized to the external wall of the 20S core cylinder (8), does not impair the ability to complement the psmA knockout strain. Similar to the psmA strain, the panA deletion strain GZ109 was complemented in trans by plasmid pJAM648, which encodes the PanA protein, and GZ109 was not complemented by plasmid vector pJAM202c alone (Fig. 3C). In contrast to what was seen for the psmA strain, however, plasmids pJAM650 and pJAM503, encoding PanA with C- and N-terminal His6 tags, respectively, were unable to fully restore the growth of the panA mutant to wild-type levels under low-salt conditions (Fig. 3C). Both His6-tagged PanA proteins synthesized from these plasmids are purified as soluble complexes from H. volcanii cells (unpublished results). The N terminus of PanA is predicted to form a coiled-coil structure important in substrate recognition (17), and the C terminus of PanA includes the hydrophobic tyrosine X (HbYX) motif, which triggers 20S proteasome gate opening after ATP binding (16). These functions may be disrupted by the addition of the His6 tags. Thus, although speculative, the ability to complement the panA mutant with only the unmodified PanA protein suggests that the recognition of substrates and 20S gate opening are important functions of PanA during hypo-osmotic stress.
Growth of H. volcanii strains was also examined in the presence of l-canavanine (Fig. 4), an amino acid analogue related to arginine that is incorporated into nascent polypeptides during translation and promotes protein unfolding. In general, all H. volcanii strains examined, including parent H26, were sensitive to this amino acid analogue with little to no growth at concentrations of ≥34 μM l-canavanine (Fig. 4). Deletions in psmC and/or panB had little to no influence on this sensitivity. In contrast, strains with deletions in either psmA or panA were hypersensitive to l-canavanine and displayed impaired growth at concentrations as low as ∼10 μM, in contrast to what was seen for parent strain H26, the growth of which was not impaired at this concentration. The ΔpsmA ΔpanA double mutant grew to a cell density similar to that of the single ΔpsmA or ΔpanA mutant strains at all concentrations of l-canavanine examined. Thus, similar to hypo-osmotic stress, mutations in psmA and panA were not additive in the l-canavanine-hypersensitive response.
The viability of H. volcanii proteasomal mutant strains was also compared to that of parent H26 after exposure to heat stress at 65°C (Fig. 5). Strains with deletions in panB and/or psmC genes displayed reductions in viability similar to that seen for parent H26 throughout the 6.5-h time course, with an average reduction of 4 log units from ∼4 × 107 to 3 × 103 CFU per ml (Fig. 5A). In contrast, all strains which harbored a deletion in the psmA gene were severely impaired in their thermotolerance (Fig. 5B). Few if any of the ΔpsmA strains were viable after the 6.5-h exposure to 65°C. Surprisingly, strains deficient in panA, whether in the presence or in the absence of a functional psmC or panB gene, displayed greater thermotolerance than parent strain H26 (Fig. 5C). Although a significant decrease in CFU per ml was observed for these ΔpanA strains after the 6.5-h heat stress, this reduction in cell viability was less than 2 log units from an average of ∼4 × 107 to 4.1 × 105 CFU per ml. Introduction of the psmA deletion into the ΔpanA background reversed this “superthermotolerance” and resulted in cells severely impaired in their ability to overcome heat stress, similar to what was seen for the other psmA deletion strains (Fig. 5B).
DISCUSSION
The ability to generate markerless deletions or conditional promoter (PtnaA) fusions in genes encoding all of the known components of the proteasome system of H. volcanii has provided tremendous insight into the central role that this proteolytic system plays in the biology of archaea. Our results demonstrate that H. volcanii requires 20S core particles for growth. Although the synthesis of 20S proteasome subtypes containing either α1 or α2 can be separately abolished with little to no impact on cell viability under standard “nonstressful” laboratory conditions, depletion of either the β subunit alone or α1 and α2 subunits together had a profound impact on cell growth. Synthesis of an α-type or β-type 20S proteasome subunit was essential for growth. Thus, of all the individual components of the proteasomal system, the β subunit, which is needed for the generation of the active site of 20S core particles, is the most crucial for cell viability. The α1 and α2 proteins, which form the outer rings of the 20S core particles, provide some redundancy of function. The depletion of either one of these α-type subunits did not impair growth under standard laboratory conditions; however, the double deletion was lethal.
Interestingly, the psmA and panA genes, each of which were required to overcome the hypersensitive responses to l-canavanine and low salt, encode the respective α1 and PanA proteasomal proteins, shown to be predominant throughout the growth of H. volcanii (17). The depletion of α1 via the psmA mutation also had a severe impact on the ability of H. volcanii to withstand thermal stress. In contrast, the psmC and panB genes, which had little if any impact on growth under the various conditions examined, encode the α2 and PanB proteasomal proteins, respectively, which are at relatively low levels during exponential or log-phase growth and increase severalfold during the stationary phase of H. volcanii (17). This is consistent with our model, which suggests that α2 and PanB have ancillary roles in proteasome function, whereas α1, β, and PanA are the central proteasome components of the growing cell.
In contrast to the approach and findings of our study, Ruepp et al. (18) used the tripeptide carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone inhibitor (Z-L3VS) to study the in vivo function of 20S proteasomes in Thermoplasma acidophilum. This archaeon differs from H. volcanii in that it is thermoacidophilic and does not encode PAN proteins. The addition of Z-L3VS to cell culture irreversibly modified 75 to 80% of the β subunits of 20S proteasomes in T. acidophilum. Although the effects of this proteasome inhibition impaired thermotolerance, inhibition had only a marginal influence on cell growth under normal conditions. Whether these differences are biological remains to be determined. The use of Z-L3VS limited the ability of Ruepp et al. (18) to completely abolish proteasome function in T. acidophilum.
It was somewhat surprising that H. volcanii cells harboring deletions in the panA gene (with exception of ΔpanA ΔpsmA cells) displayed enhanced thermotolerance compared to that of parent strain H26 and other proteasome mutant strains. The reason for this finding remains to be determined. Our recent characterization of the (phospho)proteome of the related panA mutant strain GG102, which harbors a mevinolin resistance marker insertion and deletion within the panA gene, reveals not only a global increase in the number of different phosphoproteins but also an increase in the abundance of proteins related to stress responses as well as Cdc48-related triple-A ATPase homologs (9). Whether any of these proteins compensate in part for the loss of the PanA function and render the cell more resistant to thermal stress than wild-type cells remains speculative.
Supplementary Material
Acknowledgments
Thanks to P. Lund, J. Soppa, and T. Allers for providing strains, plasmids, and helpful discussion. Thanks also to Jonathan Eisen and The Institute of Genomic Research for completion and annotation of the H. volcanii genome sequence.
This research was funded in part by the following grants to J.A.M.-F.: from NIH, R01 GM057498; and from DOE, DE-FG02-05ER15650.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allers, T., and M. Mevarech. 2005. Archaeal genetics—the third way. Nat. Rev. Genet. 658-73. [DOI] [PubMed] [Google Scholar]
- 2.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumeister, W., J. Walz, F. Zühl, and E. Seemüller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92367-380. [DOI] [PubMed] [Google Scholar]
- 4.Bolhuis, H. H., P. P. Palm, A. A. Wende, M. M. Falb, M. M. Rampp, F. F. Rodriguez-Valera, F. F. Pfeiffer, and D. D. Oesterhelt. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyall-Smith, M. 2008. The Halohandbook: protocols for halobacterial genetics, p. 67. http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf.
- 6.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 151336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil, M. A., K. E. Sherwood, and J. A. Maupin-Furlow. 2007. Transcriptional linkage of Haloferax volcanii proteasomal genes with non-proteasomal gene neighbours including RNaseP, MOSC domain and SAM-methyltransferase homologues. Microbiology 1533009-3022. [DOI] [PubMed] [Google Scholar]
- 8.Kaczowka, S. J., and J. A. Maupin-Furlow. 2003. Subunit topology of two 20S proteasomes from Haloferax volcanii. J. Bacteriol. 185165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkland, P. A., M. A. Gil, I. M. Karadzic, and J. A. Maupin-Furlow. 2008. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J. Bacteriol. 190193-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Large, A., C. Stamme, C. Lange, Z. Duan, T. Allers, J. Soppa, and P. A. Lund. 2007. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol. Microbiol. 661092-1106. [DOI] [PubMed] [Google Scholar]
- 11.Löwe, J., D. Stock, B. Jap, P. Zwickl, W. Baumeister, and R. Huber. 1995. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268533-539. [DOI] [PubMed] [Google Scholar]
- 12.Maupin-Furlow, J. A., M. A. Humbard, P. A. Kirkland, W. Li, C. J. Reuter, A. J. Wright, and G. Zhou. 2006. Proteasomes from structure to function: perspectives from archaea. Curr. Top. Dev. Biol. 75125-169. [DOI] [PubMed] [Google Scholar]
- 13.Mullakhanbhai, M. S., and H. Larsen. 1975. Halobacterium volcanii spec. nov., a Dead Sea Halobacterium with a moderate salt requirement. Arch. Microbiol. 104207-214. [DOI] [PubMed] [Google Scholar]
- 14.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, and H. Dale. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 9712176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer, F., S. C. Schuster, A. Broicher, M. Falb, P. Palm, K. Rodewald, A. Ruepp, J. Soppa, J. Tittor, and D. Oesterhelt. 2008. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 91335-346. [DOI] [PubMed] [Google Scholar]
- 16.Rabl, J., D. M. Smith, Y. Yu, S. C. Chang, A. L. Goldberg, and Y. Cheng. 2008. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reuter, C. J., S. J. Kaczowka, and J. A. Maupin-Furlow. 2004. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J. Bacteriol. 1867763-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruepp, A., C. Eckerskorn, M. Bogyo, and W. Baumeister. 1998. Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett. 42587-90. [DOI] [PubMed] [Google Scholar]
- 19.Wendoloski, D., C. Ferrer, and M. L. Dyall-Smith. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147959-964. [DOI] [PubMed] [Google Scholar]
- 20.Wilson, H. L., H. C. Aldrich, and J. A. Maupin-Furlow. 1999. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J. Bacteriol. 1815814-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.