Abstract
Denitrification is a well-studied respiratory system that is also important in the biogeochemical nitrogen cycle. Environmental signals such as oxygen and N-oxides have been demonstrated to regulate denitrification, though how denitrification is regulated in a bacterial community remains obscure. Pseudomonas aeruginosa is a ubiquitous bacterium that controls numerous genes through cell-to-cell signals. The bacterium possesses at least two N-acyl-l-homoserine lactone (AHL) signals. In our previous study, these quorum-sensing signals controlled denitrification in P. aeruginosa. In addition to the AHL signals, a third cell-to-cell communication signal, 2-heptyl-3-hydroxy-4-quinolone, referred to as the Pseudomonas quinolone signal (PQS), has been characterized. In this study, we examined the effect of PQS on denitrification to obtain more insight into the respiratory regulation in a bacterial community. Denitrification in P. aeruginosa was repressed by PQS, which was partially mediated by PqsR and PqsE. Measuring the denitrifying enzyme activities indicated that nitrite reductase activity was increased by PQS, whereas PQS inhibited nitric oxide reductase and the nitrate-respiratory chain activities. This is the first report to demonstrate that PQS influences enzyme activities, suggesting this effect is not specific to P. aeruginosa. Furthermore, when iron was supplied to the PQS-added medium, denitrifying activity was almost restored, indicating that the iron chelating property of PQS affected denitrification. Thus, our data indicate that PQS regulates denitrification primarily through iron chelation. The PQS effect on denitrification was relevant in a condition where oxygen was limited and denitrification was induced, suggesting its role in controlling denitrification where oxygen is present.
Bacteria regulate their metabolism by sensing environmental signals in order to adapt to various environmental conditions. In the environment, a number of bacteria are capable of using N-oxides as alternative electron acceptors of oxygen. Denitrification is a mode of anaerobic respiration in which nitrate (NO3−) or nitrite (NO2−) is reduced to gaseous N-oxides, such as nitric oxide (NO), nitrous oxide (N2O), and nitrogen (N2), concomitant with energy generation (61). The switch from aerobic respiration to denitrification is usually known to be regulated by a response to N-oxides and oxygen levels (2, 28, 31). These N-oxides and oxygen levels are sensed through the CRP/FNR (cAMP receptor protein/fumarate and nitrate reductase regulator) family, the members of which are global regulators that activate transcription of denitrifying genes (49). In Pseudomonas aeruginosa, a ubiquitous gram-negative environmental bacterium, denitrifying genes are also regulated by N-oxides and oxygen levels through a regulatory network requiring ANR, DNR regulatory proteins, and a nitrate-responding two-component regulator, NarXL (45).
Although how respiration is regulated by physicochemical factors such as oxygen and N-oxide concentrations is well studied, how respiration is regulated in a bacterial community remains obscure. The availability of electron acceptors will change due to the bacterial population or community. For instance, when a bacterium forms a biofilm, the cells outside of the biofilm can utilize oxygen, whereas inside the biofilm, the oxygen is depleted and bacterial metabolism changes (5). Where bacteria exist in high density, they can interact with each other through cell-to-cell communication signaling molecules, enabling the bacteria to coordinate diverse biological functions. These signal molecules in gram-negative bacteria are typically N-acyl-l-homoserine lactones (AHLs). P. aeruginosa possesses at least two of these AHL-dependent quorum-sensing systems, the LasR-LasI (las) and RhlR-RhlI (rhl) systems (42). In order to investigate how bacterial respiration is regulated as a community develops, we have previously examined denitrification regulation by the AHL quorum-sensing systems (52). The results demonstrated that the las and rhl quorum-sensing systems repress denitrification, and the regulation by the las quorum-sensing system is dependent on the rhl quorum-sensing system.
In addition to the AHL quorum-sensing signals, a non-AHL quorum-sensing signal, 2-heptyl-3-hydroxy-4-quinolone, referred to as the Pseudomonas quinolone signal (PQS), has been characterized in P. aeruginosa (41). PQS regulates the production of itself and virulence factors such as elastase, rhamnolipids, and pyocyanin mediated by a LysR-type regulator PqsR (MvfR) and PqsE (19, 22, 54). The function of PqsE is not known. The direct precursor of PQS is 2-heptyl-4-quinolone (HHQ), which is converted to PQS by a putative monooxygenase, PqsH, that is regulated by the LasR quorum-sensing system (16). HHQ has also been reported to act as a ligand of PqsR, indicating its role in cell-cell communication as well as PQS (58). In addition, HHQ is produced in other pathogenic bacteria besides P. aeruginosa, suggesting a role in interspecies communication (17).
To obtain more insight into how energy production is regulated in a bacterial community, we examined the effect of PQS on denitrification. Based on our results, PQS affects denitrifying enzyme activities primarily due to the chelating activity of PQS. These results suggest that PQS not only affects respiration in P. aeruginosa but may also affect respiration in other species of bacteria. The low production of PQS in anaerobic conditions points out that this effect is relevant in interfaces of oxygen respiration and denitrification.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacterial strains were routinely grown at 37°C in Luria-Bertani (LB) medium or on LB agar plates. When necessary, gentamicin was added at the concentration of 10 μg/ml for Escherichia coli and 80 μg/ml for P. aeruginosa. PQS and N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) were synthesized and purchased from NARD institute, Ltd. (Hyogo, Japan). A total of 20 mM PQS and 400 μM 3-oxo-C12-HSL stock solution in dimethyl sulfoxide was prepared and added to the culture as a final concentration of 50 μM or 1 μM, respectively. An equivalent amount of dimethyl sulfoxide was added to control cultures. Before beginning experimental cultures, P. aeruginosa was grown aerobically in 24-ml test tubes containing 4 ml of LB medium and was used to inoculate cultures at a starting optical density at 600 nm (OD600) of 0.01. The culture that examined the effect of 3-oxo-C12-HSL on denitrification was inoculated at a starting OD600 of 0.1. For anaerobic cultures, the air of butyl-rubber-sealed Hungate tubes or Erlenmeyer flasks was replaced with argon by flushing gas through a needle. Anaerobic growth, denitrification activity, and transcriptional activity were measured using 17-ml Hungate tubes containing 5 ml LBN medium (LB medium supplemented with 100 mM KNO3) incubated at 37°C with shaking at 200 rpm. Oxygen-limiting experiments were carried out by using butyl-rubber-sealed 17-ml Hungate tubes containing 2 ml LBN medium incubated at 37°C with shaking at 170 rpm. Cells used to determine denitrifying enzyme activities were anaerobically cultured in a 500-ml Erlenmeyer flask containing 80 ml LBN at 37°C and 200 rpm. The pqsA transcriptional fusion plasmid was constructed by cloning the promoter region of pqsA (59), using pmpqsAF3 and pmpqsAR PCR primers (Table 1), into the multicloning site of pMEX9 reporter plasmid. Plasmids were transformed into P. aeruginosa strain PAO1 by electroporation (20). Complementation plasmids carrying pqsR or pqsE were constructed by cloning each gene into the multicloning site of pUCP24. cpqsRF/cpqsRR or cpqsEF/cpqsER primer pairs (Table 1) were used to amplify pqsR and pqsE, respectively.
TABLE 1.
Bacterial strains, plasmids, and primers
| Strains, plasmids, and primers | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | E. coli strain for transformation | TaKaRa |
| DH5α | E. coli strain for transformation | TaKaRa |
| S17-1 | Mobilizer strain | 48 |
| P. aeruginosa | ||
| PAO1 | WT | 27 |
| ΔpqsA | PAO1 mutant with a deletion in the pqsA gene | This study |
| ΔpqsH | PAO1 mutant with a deletion in the pqsH gene | This study |
| ΔpqsR | PAO1 mutant with a deletion in the pqsR gene | This study |
| ΔlasI | PAO1 mutant with a deletion in the lasI gene | 52 |
| ΔrhlI | PAO1 mutant with a deletion in the rhlI gene | 52 |
| ΔpqsA ΔpqsR | PAO1 mutant with deletions in the pqsA and pqsR genes | This study |
| ΔpqsA ΔpqsE | PAO1 mutant with deletions in the pqsA and pqsE genes | This study |
| ΔpqsA ΔlasI | PAO1 mutant with deletions in the pqsA and lasI genes | This study |
| ΔpqsA ΔrhlI | PAO1 mutant with deletions in the pqsA and rhlI genes | This study |
| Plasmids | ||
| pHSG398 | Cloning vector; Cpr | TaKaRa |
| pG19II | pK19mobsac derived suicide vector; sacB Gmr | 36 |
| pG19pqsA | pqsA deletion cassette in pG19II | This study |
| pG19pqsR | pqsR deletion cassette in pG19II | This study |
| pG19pqsE | pqsE deletion cassette in pG19II | This study |
| pG19pqsH | pqsH deletion cassette in pG19II | This study |
| pUCP24 | Broad-host-range vector; Gmr | 55 |
| pUCPpqsR | pqsR in pUCP24 | This study |
| pUCPpqsE | pqsE in pUCP24 | This study |
| pMEX9 | pME4510 derived promoter-probe vector; xylE Gmr | 52 |
| pMEXpqsA | pqsA promoter region in pMEX9 | This study |
| pMEXnarK | nark1 promoter region in pMEX9 | 52 |
| pMEXnirS | nirS promoter region in pMEX9 | 52 |
| pMEXnorC | norC promoter region in pMEX9 | 52 |
| pMEXnosR | nosR promoter region in pMEX9 | 52 |
| Primers | ||
| pmpqsAF3 | 5′-GGAATTCGGGGCATGTAGGTGTCCTCTTCGG-3′ | |
| pmpqsAR | 5′-CCCAAGCTTGCGGGACTTGGGATTGATCACG-3′ | |
| ΔpqsAF1 | 5′-GGTCTAGAGGCAAGGTGCAACAATGGACAGTGG-3′ | |
| ΔpqsAR1 | 5′-GACTAGTGACAGAACGTTCCCTCTTCAGCG-3′ | |
| ΔpqsAF2 | 5′-CACTAGTGAGGAACGGGCATGTTGATTCAGGC-3′ | |
| ΔpqsAR2 | 5′-GCGAAGCTTGGAAGTTCACAGGTGATCGCTGCC-3′ | |
| ΔpqsHF1 | 5′-CCCAAGCTTCTTGTCCTGCAGGTCGATATCC-3′ | |
| ΔpqsHR1 | 5′-GAAGATCTCGGTCATCCGTTGCTCCTTAGC-3′ | |
| ΔpqsHF2 | 5′-GAAGATCTTTCACCGGTGTCGGTGAGATGG-3′ | |
| ΔpqsHR2 | 5′-GCTCTAGATCGAGAGCTTCTCGAAGATGCG-3′ | |
| ΔpqsRF1 | 5′-CCCAAGCTTGCCCACCAGTACAGCATTTTGCG-3′ | |
| ΔpqsRR1 | 5′-GAAGATCTTCCTGCGCTTTCTCGAAAGCGC-3′ | |
| ΔpqsRF2 | 5′-GAAGATCTCCCTTATTCCTTTTATTGGGTGGC-3′ | |
| ΔpqsRR2 | 5′-GCTCTAGAGGCAGTACGAAGATAGCTATCGC-3′ | |
| ΔpqsEF1 | 5′-CCCAAGCTTGTAGGGCGCATCAATACGTCGG-3′ | |
| ΔpqsER1 | 5′-GAAGATCTGGTCATCATCCAGTTGACCGG-3′ | |
| ΔpqsEF2 | 5′-GAAGATCTGCCGGATGCTGGAGATTCTCTCCC-3′ | |
| ΔpqsER2 | 5′-TGCTCTAGACAGGTCGACCTGGTAGTTGCC-3′ | |
| cpqsRF | 5′-GCTCTAGAACCCAATAAAAGGAATAAGGGATGC-3′ | |
| cpqsRR | 5′-CCCAAGCTTGAACGCTCTACTCTGGTGCGG-3′ | |
| cpqsEF | 5′-GCTCTAGACGCAAACCTGAGGAGGTGAACC-3′ | |
| cpqsER | 5′-CCCAAGCTTGTCCCGTCTCAGTCCAGAGGC-3′ |
Construction of P. aeruginosa mutants.
The primers used in this study are listed in Table 1. pG19pqsA, pG19pqsH, pG19pqsR, and pG19pqsE plasmids carrying deletion cassettes of pqsA, pqsH, pqsR, and pqsE were constructed with the same procedure described previously (36). The PAO1 chromosome was amplified with pqsAF1/pqsAR2, pqsHF1/pqsHR2, pqsRF1/pqsRR2, or pqsEF1/pqsER2 primer pairs and then ligated into the XbaI/HindIII-treated multicloning site of a cloning vector, pHSG398. The DNA flanking fragments of pqsA, pqsH, pqsE, and pqsR on pHSG398 were amplified using inverse PCR with pqsAF2/pqsAR1, pqsHF2/pqsHR1, pqsRF2/pqsRR1, or pqsEF2/pqsER1 PCR primers. The amplified pqsA flanking fragment was digested with SpeI, and the amplified pqsH, pqsR, and pqsE flanking fragment was digested with BglII, as restriction sites had been attached to the primers. The pqsA-, pqsH-, pqsR-, and pqsE-deleted DNA fragments were subcloned into the XbaI-HindIII site in the multicloning site of pG19II, generating pG19pqsA, pG19pqsH, pG19pqsR, and pG19pqsE. The pG19II-derived plasmids were transferred into PAO1, ΔpqsA, ΔlasI, and ΔrhlI strains by conjugating with E. coli S17-1 (48), followed by homologous recombination previously described elsewhere (36). The mutants were analyzed by PCR.
Preparation of cell fractions.
Cells grown in LBN medium were collected in the mid-logarithmic phase (6-h incubation time), centrifuged at 7,000 × g at 4°C, and washed twice with 100 mM potassium phosphate buffer, pH 7.5, containing 10% glycerol. The cells were suspended in the same buffer and sonicated. The sonicated cells were centrifuged for 10 min at 2,000 × g at 4°C to remove unbroken cells and then centrifuged at 104,000 × g at 4°C for 60 min. The supernatants were collected as soluble fractions of the cells, and the pellets were resuspended in the same buffer and collected as membrane fractions.
Enzyme activity assays.
NAR activity was assayed by determining its NO2− product colorimetrically as described by Nicholas and Nason (39).
Benzylviologen-dithionite-related NAR activity was assayed according to a method previously described elsewhere (30) by using benzylviologen instead of methylviologen. One hundred microliters of Na2S2O4 was added at a final concentration of 10 mM into a 400 μl 100 mM potassium phosphate buffer (pH 7.5) containing 10 mM NaNO3, 0.2 mM benzylviologen, and an appropriate concentration of the cell membrane fraction of anaerobically cultured P. aeruginosa. The air in the reaction mixtures was replaced with N2 before adding Na2S2O4. The reaction mixture was incubated at 30°C for 5 min and was vortexed until the color of the mixture became clear, indicating methylviologen oxidation.
NAR activity associated with the respiratory chain was measured using NADH as an electron donor (43). Five-hundred-microliter reaction mixtures for NADH-derived NAR activity assays contained 10 mM NaNO3 and the membrane fraction of anaerobically cultured P. aeruginosa. The air in the reaction mixture was replaced with N2. The reaction was started by the addition of NADH at a final concentration of 1 mM at 30°C and was stopped by being boiled at 100°C for 15 min.
NIR activity was assayed by measuring NO2− consumption (32). The reaction was started by the addition of 100 μl NADH at a final concentration of 1 mM into a 400-μl mixture containing 200 μM NaNO2, 200 μM phenazine methosulfate, and the soluble fraction of anaerobically cultured P. aeruginosa. The air in the reaction mixtures was replaced with N2 before adding NADH, and the reactions were carried out at 30°C for 10 min.
NOR activity was assayed by a modified method measuring its product, N2O, with a gas chromatograph (50). One hundred microliters of 20 mM NO donor sodium nitroprusside was injected into 900-μl reaction mixtures in which the air had been replaced with N2 and which contained 4 mM NADH, 0.2 mM phenazine methosulfate, and the membrane fraction of anaerobically cultured P. aeruginosa. The reactions were carried out at 30°C for 6 min.
Catechol 2,3-dioxygenase (C23O) specific activity (the xylE gene product) was determined by the method described previously (36, 44) by monitoring A375.
Analytical methods.
Bacterial growth in liquid cultures was monitored based on the OD600. The protein amount was quantified using the method of Bradford (6). NO3− concentrations were determined using the Brucine method described in reference 39. N2O, N2, and O2 concentrations were measured with a gas chromatograph (GC-8AIT; Shimadzu) equipped with a gas thermal conductivity detector. Helium was used as the carrier gas. A Shincarbon ST column was used for N2O detection, and a Molecular Sieve 5A column was used for the detection of N2 and O2.
PQS production assays.
PQS was collected from the supernatant and detected by thin-layer chromatography (TLC) analysis, following the method previously described (37). Stationary-phase samples cultured for 16 h were centrifuged for 1 min at 13,000 × g to collect the supernatants. PQS was extracted from 1 ml of supernatants twice with 600 μl acidified ethyl acetate. The ethyl acetate portion was collected into a new tube and dried. Extracts were resuspended in 20 μl of 1:1 acidified ethyl acetate/acetonitrile. Aliquots of extracts were loaded on TLC plates (silica gel 60 F254; Merck) which had been soaked in 5% KH2PO4 for 30 min and activated at 100°C for 1 h. The extracts were separated using 17:2:1 methylene chloride/acetonitrile/1,4-dioxane as the solvent. Synthetic PQS was used as a control. Photographs were taken under long-wave UV light by using a digital camera. The spot intensity was evaluated by using ImageMaster 1D Elite version 4.2 software (Amersham Pharmacia Biotech), following the method previously published (37).
RESULTS
Anaerobic growth is repressed by PQS.
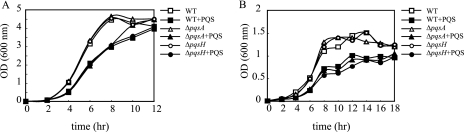
First, we examined the effect of PQS on the growth of P. aeruginosa. When grown aerobically in LB medium or anaerobically in LBN medium, no change in growth was observed between the wild-type (WT) strain and the ΔpqsA or ΔpqsH mutants (Fig. 1). However, when PQS was added to the medium, a longer lag phase was observed in the aerobic culture, as had been observed previously (19) (Fig. 1A). This may be due to the prooxidant effect of PQS, as it was demonstrated in a recent paper (24). When PQS was added to anaerobic cultures (Fig. 1B), it resulted in a lower stationary-phase OD than that of the growth in LBN medium alone. To investigate the mechanism of PQS suppression of anaerobic growth, we further examined the effect of PQS on denitrification.
FIG. 1.
PQS effect on aerobic growth and anaerobic growth. Aerobic growth (A) and anaerobic growth (B) of P. aeruginosa with or without exogenous PQS. PQS was added at a final concentration of 50 μM in all experiments. Three independent experiments were carried out, and representative data are shown.
PQS affects denitrifying enzyme activities.
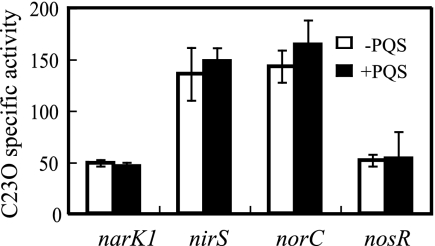
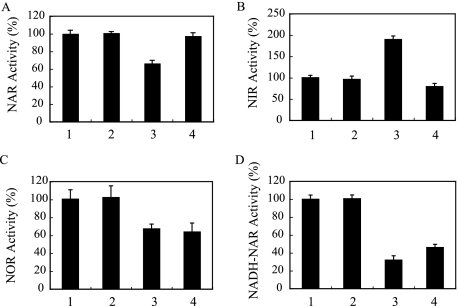
Denitrification is a process known to reduce NO3− to N2O or N2 gases and is physiologically important for acquiring energy under anaerobic conditions (61). During the denitrification process, NO3− reductase (NAR), NO2− reductase (NIR), NO reductase (NOR), and N2O reductase (NOS) are involved (61). Each denitrifying enzyme is encoded by genes that are organized into distinct operons (2, 3, 29, 47). In order to examine the effect of PQS on transcription of each denitrifying enzyme, promoter fusion plasmids (52) with the xylE gene fused with promoter regions of narK1, nirS, norC, and nosR denitrifying genes were each transferred to a ΔpqsA mutant. As a control, the basal C23O activity was measured in a ΔpqsA mutant transformed with a pMEX9 plasmid with or without added PQS. C23O activity was measured during the mid-logarithmic phase (a 6-h incubation). The promoter-dependent C23O activity was determined by dividing its C23O activity by the basal C23O activity. When PQS was added to the culture of each strain at a final concentration of 50 μM, no significant effect on the promoter activity could be observed (Fig. 2). This result indicates that the denitrification regulation by PQS is not due to transcriptional regulation but is caused by other posttranscriptional mechanisms. To examine whether denitrification is regulated in posttranscriptional levels, denitrifying enzyme activities were measured during the mid-logarithmic phase. A ΔpqsA mutant was cultured with or without added PQS, and then NAR, NIR, and NOR denitrifying enzyme activities were measured. As a result, NAR activity was suppressed (66% activity compared to the NAR activity of a ΔpqsA mutant cultured without added PQS) in the culture incubated with PQS (Fig. 3A). Interestingly, NIR activity increased 1.8-fold in the culture incubated with PQS (Fig. 3B). NOR activity was suppressed to 64% when PQS was added to the culture (Fig. 3C). The changes in these denitrifying enzyme activities may be due to posttranscriptional regulation, or PQS may have affected enzyme activities. To further examine whether PQS affects denitrifying enzyme activities, an assay was carried out by adding PQS to each enzyme assay reaction mixture with ΔpqsA mutant cell fractions collected from cells cultured without PQS. As shown in Fig. 3C, NOR activity was inhibited by the PQS addition in vitro. On the other hand, NAR and NIR activity was not affected by the addition of PQS in vitro (Fig. 3A and B). Taken with the result that transcription of denitrifying genes was not regulated by PQS, our results indicate that NAR and NIR activities are regulated posttranscriptionally, requiring growth with PQS, and NOR activity is inhibited by PQS.
FIG. 2.
PQS effect on denitrifying gene expression. C23O activity was measured in cells cultured for 6 h (mid-logarithmic phase). C23O activity (nanomoles of product/minute/milligram of protein) of denitrifying gene promoters was measured and was normalized by the C23O activity of promoterless control plasmids. +PQS, cultured with exogenous PQS; −PQS, cultured without exogenous PQS. The data displayed are the means ± standard deviations of more than three independent experiments.
FIG. 3.
PQS effect on denitrifying enzyme activities and NO3− respiratory chain. NAR (A), NIR (B), NOR (C), and NADH-dependent NAR (D) activities were measured in cells cultured for 6 h (mid-logarithmic phase). PQS was added at a final concentration of 50 μM for the culture medium or the reaction mixture. Bar 1, PAO1; bar 2, ΔpqsA mutant cultured without added PQS; bar 3, ΔpqsA mutant cultured with PQS; bar 4, PQS added to the reaction mixture with cell fractions collected from a ΔpqsA mutant cultured without added PQS. More than three independent experiments were carried out, and the data represented are means ± standard deviations of triplicate assays.
PQS inhibits the NO3− respiratory chain.
Denitrifying enzymes are associated with the respiratory chain in which energy is generated. During NO3− respiration, the electron donated from NADH is transferred through the respiratory chain and finally accepted by NO3−. The final step is mediated by NAR (61). In the NAR activity assay (Fig. 3A), an electron donor (benzylviologen) that donates electrons directly to NAR was used, and the activity of the NO3− respiratory chain was not taken into account. In order to examine whether the NO3− respiratory chain is affected by PQS, a NADH-derived NAR activity assay was carried out. When NADH was used as an electron donor, NADH-NAR activity was repressed in the culture with added PQS and was also inhibited in vitro when PQS was added to the reaction mixture (Fig. 3D). These results indicate that NO3− respiration is inhibited by PQS. The fact that PQS inhibited NADH-derived NAR activity (Fig. 3D), but not benzylviologen-derived NAR activity (Fig. 3A), indicates that PQS's inhibition occurs somewhere in the electron transfer from NADH to NO3−.
Iron-chelating property of PQS affects denitrification.
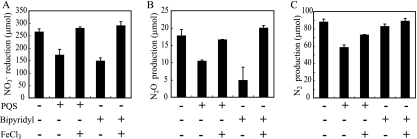
In this study, our results addressed that PQS influences denitrification by affecting the denitrifying enzyme activities without regulating their transcription. PQS has been reported not only to regulate transcription in P. aeruginosa but also to chelate iron (7, 18). We considered that this chelating effect on iron may affect the denitrification activity. To test this hypothesis, FeCl3 was added to the medium in addition to PQS. The iron addition fully restored suppression of NO3− reduction and N2O production in the culture with exogenous PQS (Fig. 4A and B) and indicated that the PQS regulation of denitrification is due to iron chelation. To confirm that iron chelation affects denitrification, an iron chelator, 2,2′-bipyridyl, was added to the culture, and denitrification activity was measured. NO3− reduction and N2O production were suppressed (as is the case of PQS); NO3− reduction and N2O production were then restored by supplementing FeCl3 (Fig. 4A and B). Interestingly, N2 production was restored only partially by FeCl3 in the PQS-added culture, and 2,2′-bipyridyl did not repress N2 production (Fig. 4C). These results suggest that N2O reduction to N2, which is catalyzed by NOS, is regulated by mechanisms other than iron chelation by PQS.
FIG. 4.
Effect of iron chelation on denitrification. NO3− reduction (A), N2O production (B), and N2 production (C) of ΔpqsA mutant. PQS, 2,2′-bipyridyl, and FeCl3 were added at final concentrations of 50 μM, 120 μM, and 100 μM, respectively. Data displayed are means ± standard deviations of three independent experiments.
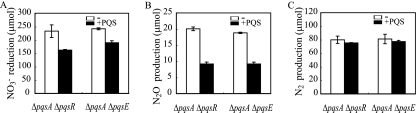
PQS regulates N2 production through PqsR and PqsE.
A number of PQS-regulated genes require the cognate response regulator PqsR (11, 15, 54) or a functionally unknown PqsE, which is known to facilitate the response to PQS (19, 22). To further examine if the N2O reduction to N2 is regulated by this pathway, ΔpqsA ΔpqsR and ΔpqsA ΔpqsE double mutants were constructed. PQS was added to the mutants, and the amounts of NO3− that was reduced and N2O and N2 that were produced during a 12-h incubation were measured. The denitrifying activities of ΔpqsA ΔpqsR and ΔpqsA ΔpqsE mutants (Fig. 5) were identical to that of ΔpqsA (Fig. 4). As shown in Fig. 5C, the repression of N2 production by PQS was not observed in either the ΔpqsA ΔpqsR or the ΔpqsA ΔpqsE mutants, while PQS repressed N2 production in ΔpqsA (Fig. 4C), indicating that N2 production is regulated by PQS through PqsE and the PqsR transcriptional regulator. Together with the result that N2 production was not affected by an iron chelator (Fig. 4), our data indicate that PQS regulates N2 production by a pathway that mediates PqsR and PqsE and does not depend on the iron chelation property of PQS. This regulation is likely to be involved in the transcriptional regulation of factors involved in N2 production other than the nos operon, since PQS did not regulate nosR transcription (Fig. 2). The NO3− reduction and N2O production were repressed by PQS even in the absence of PqsE or PqsR (Fig. 5A and B), as had been observed for ΔpqsA (Fig. 4A and B), supporting our suggestion that NO3− reduction to N2O is regulated by PQS iron chelation. To confirm that the results observed for ΔpqsA ΔpqsR and ΔpqsA ΔpqsE strains are not due to polar effects, an experiment complementing each mutant with its appropriate WT copy (ΔpqsA ΔpqsR/pUCP-pqsR, ΔpqsA ΔpqsE/pUCP-pqsE) was carried out. Surprisingly, NO3− reduction and N2O and N2 production were all strongly repressed when pqsR and pqsE were expressed on a plasmid, even in the absence of PQS (data not shown).
FIG. 5.
PqsR and PqsE involvement in N2 production regulation by PQS. NO3− reduction (A), N2O production (B), and N2 production (C) of ΔpqsA ΔpqsR and ΔpqsA ΔpqsE mutants with or without exogenous PQS, represented as black bars or white bars, respectively. pUCP24 plasmid was transferred into each strain as a control for the complementation experiments. The data displayed are the means ± standard deviations from three independent experiments. PQS was added at a final concentration of 50 μM in all experiments.
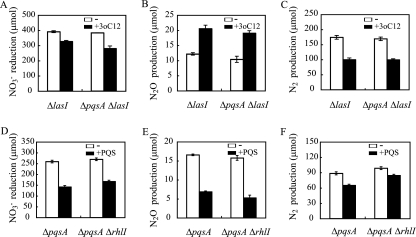
Relation of AHLs and PQS in denitrification regulation.
In our previous study, AHLs regulated transcriptions of denitrifying genes (52). PQS is known to be incorporated in the AHL signaling pathway where the las quorum-sensing system regulates the PQS system and the PQS system regulates the rhl quorum-sensing system (38). To further investigate the relationship between AHL and PQS signaling systems in denitrification regulation, we first examined whether the las quorum-sensing requires PQS for denitrification regulation. When 3-oxo-C12-HSL was added to the culture, 3-oxo-C12-HSL regulated denitrifying activity at the same extent in a ΔlasI mutant and a ΔpqsA ΔlasI double mutant (Fig. 6A to C), indicating that PQS is not required for the 3-oxo-C12-HSL denitrification regulations. This result correlates with the fact that PQS is lacking under anaerobic conditions (later discussed in Fig. 7), suggesting that PQS has less effect on las quorum-sensing regulated phenotypes under anaerobic conditions. The N2O accumulation induced by 3-oxo-C12-HSL may be due to the imbalance of NOR and NOS activity, in which NOS activity may have been repressed by 3-oxo-C12-HSL to a greater extent than NOR activity, as is the case of NO accumulation caused by rhlR deletion (23). Furthermore, it was examined whether the rhl quorum-sensing system is incorporated in the effect of PQS on denitrification. When PQS was added to a ΔpqsA ΔrhlI double mutant, NO3− reduction to N2O was repressed, as had been observed in a ΔpqsA mutant (Fig. 6D and E). However, N2 production was repressed only partially by the PQS addition in the ΔpqsA ΔrhlI double mutant compared to the ΔpqsA mutant (Fig. 6F), indicating that the rhl quorum-sensing system is partially required for the repression of N2 production by PQS. Since N2 production was not fully repressed by the PQS addition to a ΔpqsA ΔpqsR mutant, these data suggest that a transcriptional regulation mediated by PqsR and the rhl quorum-sensing system is involved in N2 production. According to our previous study (52), the rhl quorum-sensing system repressed nosR transcription; therefore, it may simply be considered that PQS induced the rhl quorum-sensing system through PqsR, leading to the repression of nosR transcription. However, repression of nosR transcription by PQS was not observed in this study (Fig. 2). The effect of PQS on nosR transcription may have been too little to be detected by the transcriptional assay; otherwise, there may be a complicated regulatory system in N2 production. The result that the N2 production in the ΔpqsA ΔpqsR strain was not repressed by PQS, while N2 production in the ΔpqsA ΔrhlI strain was partially repressed by PQS, indicates that there are other factors regulated by PqsR that are involved in the N2 production in addition to the rhl quorum-sensing system.
FIG. 6.
Relation between AHLs and PQS in denitrification regulation. NO3− reduction (A), N2O production (B), and N2 production (C) of ΔlasI and ΔpqsA ΔlasI mutants with (+3oC12) or without (−) exogenous 3-oxo-C12HSL. 3-Oxo-C12HSL was added at a final concentration of 1 μM in all experiments. NO3− reduction (D), N2O production (E), and N2 production (F) of ΔpqsA and ΔpqsA ΔrhlI mutants with (+PQS) or without (−) exogenous PQS are shown. PQS was added at a final concentration of 50 μM in all experiments. The data displayed are the means ± standard deviations from three independent experiments.
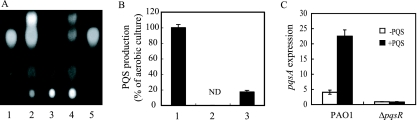
FIG. 7.
PQS signaling under anaerobic conditions. (A) PQS production in the supernatant of aerobic, anaerobic, or oxygen-limited cultures. PQS was detected by TLC analysis. Lane 1, synthetic PQS; lane 2, aerobic culture; lane 3, anaerobic culture; lane 4, oxygen-limited culture with LBN medium; lane 5, synthetic PQS. (B) Comparison of PQS production between aerobic, anaerobic, and oxygen-limited cultures. Bar 1, aerobic culture; bar 2, anaerobic culture; bar 3, oxygen-limited culture with LBN medium. PQS production was compared by measuring the band intensity of the TLC plates with an image analyzer. ND, not detected. (C) pqsA expression in PAO1 and a ΔpqsR mutant under anaerobic conditions. pMEXpqsA or pMEX9 was introduced in each cells. C23O activity (nanomoles of product/minute/milligram of protein) was measured in cells cultured for 12 h (stationary phase). C23O activity of pMEXpqsA-introduced cells was measured and was normalized by the C23O activity of pMEX9-introduced cells. Data displayed are means ± standard deviations of three independent experiments.
PQS signaling under anaerobic conditions.
In order to reveal the PQS effect on denitrification, our experiments were carried out under anaerobic conditions. Interestingly, although PQS suppressed denitrification, no changes in growth between the WT and the ΔpqsA ΔpqsH mutants were observed (Fig. 1), and no differences in denitrification activity were observed between WT and non-PQS-producing strains (data not shown). To examine whether PQS is produced under anaerobic conditions, PQS was extracted from culture supernatants and was detected by TLC. When grown aerobically, PQS was detected but it could not be detected under anaerobic conditions (Fig. 7A and B). Although PQS was not produced under anaerobic conditions, the result that the addition of PQS to a ΔpqsR mutant did not repress N2 production (Fig. 5C) suggests that PQS can regulate gene transcriptions in a PqsR-dependent manner under anaerobic conditions. To examine whether the PQS signaling system mediated by PqsR could be activated under anaerobic conditions, pqsA expression, which is known to be regulated by PQS through PqsR under aerobic conditions (37), was measured in the WT P. aeruginosa stain PAO1 and a ΔpqsR mutant. When PQS was added to PAO1, pqsA was induced, while it was not induced in the absence of PqsR (Fig. 7C). These results, along with results demonstrating the lack of PQS under anaerobic conditions (Fig. 7), indicate that cells under anaerobic conditions can respond to PQS through PqsR but cannot produce PQS. The reason why PQS is lacking under anaerobic conditions could be due to the final step of PQS synthesis, which is predicted to require diatomic oxygen (22).
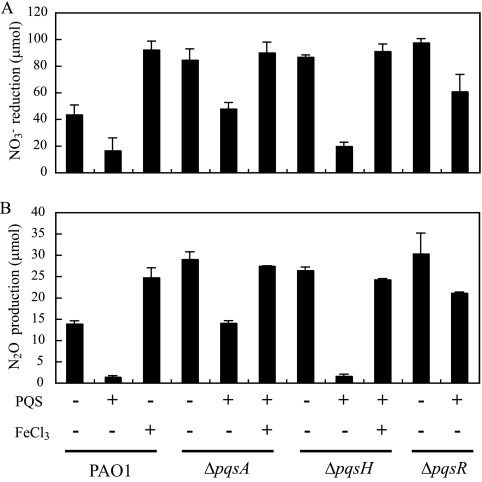
Effect of PQS on denitrification is relevant under oxygen-limiting conditions.
When considering the environment, oxygen concentration is not always constant and changes according to several factors, including cell population. The PQS effect on denitrification may be relevant under such conditions. To examine whether PQS affects denitrification under conditions where oxygen is gradually depleted and denitrification is induced, we cultured P. aeruginosa in a Hungate tube with a rubber cap. The oxygen concentration was 7.5 ppm at the start of the culture and became less than 1.7 ppm at the end of the culture (16 h). Under these conditions, PQS was detected in the supernatant (Fig. 7A and B), and the ΔpqsA, ΔpqsH, and ΔpqsR mutants reduced more NO3− and produced more N2O than PAO1 (Fig. 8). These results indicate that denitrification is repressed by PQS when PQS is produced. Interestingly, the PQS production was lower compared to the aerobic conditions (Fig. 7A and B), suggesting that oxygen or N-oxides related to denitrification regulate PQS production. When PQS was added to the medium, PQS repressed NO3− production and N2O production in PAO1 and the ΔpqsH mutant to a greater extent than in the ΔpqsA and ΔpqsR mutants (Fig. 8). The transcriptional regulator PqsR is known to regulate the pqsABCDE operon (54). Therefore, under oxygen-limiting conditions, other 2-alkyl-4-quinolones (AHQs), such as 2-heptyl-4-quinolone N-oxide (HQNO) or HHQ, which are produced as a result of the transcription of the pqsABCDE operon (16), may be involved in denitrification regulation. The denitrification repression by PQS in the ΔpqsR mutant indicates that PQS represses denitrification without mediating PqsR, as had been observed under anaerobic conditions. To confirm if this denitrification suppression by PQS was a result of the iron-chelating activity, FeCl3 was added to the medium. FeCl3 addition increased denitrifying activity in PAO1 to the level of the non-PQS-producing mutants and restored denitrifying activity in the PQS-added ΔpqsA and ΔpqsH mutants (Fig. 8). Taken together, PQS affects denitrification by chelating iron under oxygen-limiting environments where denitrification is induced and a sufficient amount of PQS is produced to influence denitrification.
FIG. 8.
PQS effect on denitrification under oxygen-limited conditions. NO3− reduction (A) and N2O production (B) of PAO1, ΔpqsA, ΔpqsH, and ΔpqsR mutants in LBN medium in the presence of oxygen. PQS and FeCl3 were added at final concentrations of 50 μM and 100 μM, respectively. Data displayed are means ± standard deviations of three independent experiments.
DISCUSSION
Bacterial energy production is well known to be regulated by the environment. On the other hand, the environment surrounding the bacteria changes due to the bacterial population or community. However, the impact of cell-to-cell communication on regulating energy production has not been well documented. This research was carried out to gain more insight into respiration regulation in a bacterial population. Some studies in Rhizobium species have reported that cell-to-cell communication affects the growth rate, although the mechanisms remain obscure (14, 25, 46, 56). Another study demonstrated that butanediol fermentation in Serratia species is affected by quorum-sensing as a result of changes in acidic end products (53). We have demonstrated previously that AHL signal molecules (C4-HSL and 3-oxo-C12HSL) repress denitrification in P. aeruginosa, which was mediated by regulatory proteins RhlR or LasR exerting denitrifying gene expression (52). In contrast to the AHL regulation of denitrification in P. aeruginosa, the PQS effect on denitrification does not necessarily require the regulatory protein PqsR but depends primarily on its iron-chelating property and has a potential to inhibit denitrification in other species. PQS may have depleted iron from the medium, and besides, results from our in vitro assay adding PQS to the cell lysates (Fig. 3) suggest that PQS can affect enzyme activities, presumably by chelating iron. According to our knowledge, this is the first report to demonstrate that PQS affects enzyme activities. Thus, PQS is the third cell-cell communication molecule to be reported to control denitrification in P. aeruginosa, and this receptor-independent effect on denitrification by PQS may have an impact on P. aeruginosa interactions with other bacteria species.
Another fact suggesting the role of PQS in interspecies interactions is the presence of HHQ producers, which could induce PQS production. HHQ has been reported to be produced in the supernatant of some human pathogens (17). Once HHQ is produced in the supernatant, it can be converted to PQS by P. aeruginosa (16). The fact that PQS can affect denitrification by chelating iron, and that PQS production could be induced by other HHQ-producing bacteria, leads us to postulate that PQS may have an impact on a bacterial community of HHQ producers, and further studies in this area are expected.
The biosynthetic pathway that produces PQS also generates diverse AHQs besides HHQ (16). HQNO is one of the AHQs, known as a classical respiratory inhibitor that suppresses growth in gram-positive bacteria but not in gram-negative bacteria (34). The spectrum of HQNO is broader in vitro, where it was demonstrated to bind to a nitrate reductase subunit (NarI) of Escherichia coli (35). Although PQS and HQNO inhibit respiration, their mechanisms seem to differ. HQNO inhibits respiration by binding to quinone-reacting cytochromes and inhibits respiratory electron transfer from quinone to cytochromes (35), while PQS depends on iron chelating. According to the mechanism, PQS may inhibit respiration in other species, and it will be interesting to examine the difference in spectrum of HQNO in future research.
In this study, PQS did not decrease aerobic growth, but it did decrease anaerobic growth (Fig. 1). The amount of iron required for denitrification may be greater than that for aerobic respiration. Supporting this hypothesis, Diggle et al. (18) have demonstrated that PQS would decrease aerobic growth under iron-deficient conditions. In P. aeruginosa, the aerobic respiratory chain is predicted to be well branched, possessing five putative terminal oxidases (13), which may be another reason why PQS had less effect on aerobic growth than on anaerobic growth. Still, the regulation and the role of these terminal oxidases are poorly understood, and it will be interesting to examine whether these terminal oxidases are affected by PQS and to investigate the role of PQS in the aerobic respiratory chain.
The suppression of NO3− reduction and N2O production by PQS iron chelation (Fig. 4), along with PQS's inhibition of NO3− respiration and NOR activities (Fig. 3), suggests that PQS inhibits NO3− respiration and NOR activity by chelating iron. Iron is related to the NO3− respiratory chain during the transfer of electrons from NADH to NO3− (21, 61). The NOR enzyme in P. aeruginosa is a cytochrome bc-type enzyme consisting of cytochrome c and cytochrome b (1, 61). Therefore, it will be reasonable to consider that PQS inhibits their activities by chelating iron. Conversely, it was surprising to us that NIR activity was not inhibited by PQS (Fig. 3B), because the P. aeruginosa NIR enzyme is a cytochrome cd1-type enzyme (60) that requires iron for its activity. There is a considerable difference in the hydrophobicity between the enzymes inhibited by PQS and the noninhibited enzymes. The PQS-affected NO3− respiration and NOR enzyme are associated with the cell membrane, while the NIR enzyme is soluble and located in the periplasm (61). Together with the fact that PQS is a highly hydrophobic molecule (10), it can be assumed that the protein hydrophobicity is one of the factors that determine whether PQS affects the activity or not. The result that PQS is associated with the cell membrane (33) could also support our study that PQS inhibits NO3− respiration and NOR enzyme activities. It will be interesting to further investigate whether the PQS effect on NO3− respiration and the NOR enzyme activities is due to direct interactions of PQS with the enzymes.
Denitrification regulation by PQS was due not only to the iron-chelating property of PQS but was also partially regulated through PqsE and PqsR regulatory proteins. The result that an iron chelator, 2,2′-bipyridyl, did not suppress N2 production, while NO3− reduction and N2O were suppressed (Fig. 4), suggests that N2 production is regulated by PQS but by mechanisms other than iron chelation. This suggestion was confirmed by the result that PqsE and PqsR were required for PQS regulation of N2 suppression (Fig. 5C). Results from the experiment in which PQS was added to a ΔpqsA ΔrhlI mutant indicate that several factors, including the rhl quorum-sensing system, are incorporated in this regulation (Fig. 6D to F). Collectively, our study demonstrates that denitrification is regulated by PQS by at least two pathways; one pathway is dependent on the iron-chelating property of PQS, and the other is dependent on transcriptional regulation by PQS mediated through PqsE and PqsR (Fig. 9). The fact that iron chelators inhibit N2O production without inhibiting N2 production may serve as a basis for development of a more efficient water treatment system, since N2O emission during wastewater treatment is known to contribute to global warming and has become a problem. Previously, microarray data by Déziel et al. (15) comparing transcriptional profiles of WT with a ΔmvfR (pqsR) mutant suggested that denitrifying genes are controlled by PqsR. When pqsR was expressed on a plasmid in our experiment, denitrification activity was repressed in a ΔpqsA ΔpqsR mutant background that does not produce AHQs, including PQS (data not shown). Along with the result that PQS did not affect transcription of denitrifying genes (Fig. 2), these results suggest that PqsR regulates denitrifying genes alone without PQS when it is expressed in a sufficient amount. Still, the experiments of Déziel et al. (15) were carried out under aerobic conditions without NO3− added, and it will not be that simple to compare their results with ours using anaerobic conditions.
FIG. 9.
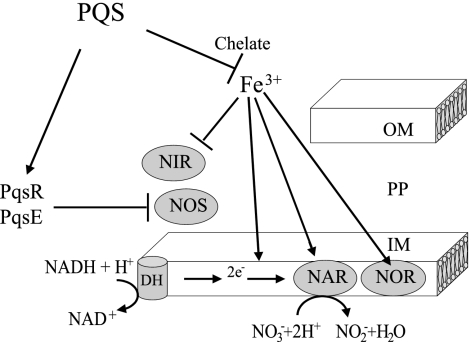
Proposed model for PQS regulation of NO3− respiration and denitrifying enzymes. The NO3− respiratory chain and NOR enzyme activities are inhibited by PQS through iron chelation. NAR activity is repressed, and NIR activity is increased by PQS indirectly, presumably through iron chelation. PqsE and PqsR are involved in the NOS activity repression. DH, NADH dehydrogenase; OM, outer membrane; PP, periplasmic space; IN, inner membrane; e−, electron flow.
Our data indicate that the effect of PQS on denitrification is relevant where oxygen is present or was present and denitrification is induced. The regulation may be important in the transition from aerobic respiration to denitrification. Also, the regulation may be important in an environment where oxygen is localized, such as in biofilms, where oxygen is consumed at the surface and conditions become anaerobic inside (5). Our in vitro results (Fig. 3) suggest that PQS promotes NO accumulation, since NIR activity (which reduces NO2− to NO) was elevated, while NOR activity (which further reduces NO to N2O) was suppressed after the addition of PQS into the medium. These results may explain the mechanism of NO accumulation in biofilms, which in turn regulates biofilm formation by upregulating bacterial motility (4). PQS has also been detected in the lungs of P. aeruginosa-infected individuals with cystic fibrosis, conditions which are considered to be O2 limited (57) and where NO3− levels are sufficient for anaerobic growth (40). In addition, P. aeruginosa is known to denitrify even in the presence of oxygen (12, 51). Although the ecological role of aerobic denitrification is still uncertain, PQS denitrification regulation may be involved.
Regarding the significance of respiratory regulation by cell-to-cell communication molecules, a number of studies have demonstrated that respiratory chains are not only coupled to energy generation but also serve other functions. Considering the advantage of repressing growth, there are studies reporting that inhibiting respiration results in increased aminoglycoside resistance (9, 26). A study in P. aeruginosa has suggested that NAR-related functions are involved in this resistance, since resistance to aminoglycosides was increased in a NAR activity-deficient mutant and the susceptibility was increased in a strain with increased NAR activity (8). The denitrification regulation by PQS may not be restricted to the regulation of energy generation but may have significance for bacterial physiology. The biological roles of denitrification regulation by cell-to-cell communication signaling molecules, such as AHL and PQS, remain elusive, and it is reasonable to assume that there are biological functions yet to be uncovered.
Acknowledgments
We thank Michael A. Kertesz for providing us with the pME4510 plasmid and Herbert P. Schweizer for providing us with the pUCP24 plasmid.
This study was partially supported by a grant to N.N. from the Industrial Technology Research Grant Program 2003-2005 of the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Arai, H., Y. Igarashi, and T. Kodama. 1995. The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 1261279-284. [DOI] [PubMed] [Google Scholar]
- 2.Arai, H., T. Kodama, and Y. Igarashi. 1999. Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 17019-24. [DOI] [PubMed] [Google Scholar]
- 3.Arai, H., M. Mizutani, and Y. Igarashi. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 14929-36. [DOI] [PubMed] [Google Scholar]
- 4.Barraud, N., D. J. Hassett, S. H. Hwang, S. A. Rice, S. Kjelleberg, and J. S. Webb. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 1887344-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 482659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bredenbruch, F., R. Geffers, M. Nimtz, J. Buer, and S. Häussler. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 81318-1329. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, L. E., T. Nicas, B. W. Holloway, and C. Crowther. 1980. Aminoglycoside-resistant mutation of Pseudomonas aeruginosa defective in cytochrome c552 and nitrate reductase. Antimicrob. Agents Chemother. 1771-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan, L. E., and H. M. Van Den Elzen. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob. Agents Chemother. 12163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calfee, M. W., J. G. Shelton, J. A. McCubrey, and E. C. Pesci. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 9814613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, F., Q. Xia, and L. K. Ju. 2003. Aerobic denitrification of Pseudomonas aeruginosa monitored by online NAD(P)H fluorescence. Appl. Environ. Microbiol. 696715-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comolli, J. C., and T. J. Donohue. 2002. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol. Microbiol. 45755-768. [DOI] [PubMed] [Google Scholar]
- 14.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277462-468. [DOI] [PubMed] [Google Scholar]
- 15.Déziel, E., S. Gopalan, A. P. Tampakaki, F. Lépine, K. E. Padfield, M. Saucier, G. Xiao, and L. G. Rahme. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol. Microbiol. 55998-1014. [DOI] [PubMed] [Google Scholar]
- 16.Déziel, E., F. Lépine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 1011339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diggle, S. P., P. Lumjiaktase, F. Dipilato, K. Winzer, M. Kunakorn, D. A. Barrett, S. R. Chhabra, M. Cámara, and P. Williams. 2006. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13701-710. [DOI] [PubMed] [Google Scholar]
- 18.Diggle, S. P., S. Matthijs, V. J. Wright, M. P. Fletcher, S. R. Chhabra, I. L. Lamont, X. Kong, R. C. Hider, P. Cornelis, M. Cámara, and P. Williams. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 1487-96. [DOI] [PubMed] [Google Scholar]
- 19.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Cámara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 5029-43. [DOI] [PubMed] [Google Scholar]
- 20.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 70221-226. [DOI] [PubMed] [Google Scholar]
- 21.Fewson, C. A., and J. D. Nicholas. 1961. Nitrate reductase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 49335-349. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 1846472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. S. Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S. H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 541425-1443. [DOI] [PubMed] [Google Scholar]
- 24.Häussler, S., and T. Becker. 2008. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman, L. R., E. Déziel, D. A. D'Argenio, F. Lépine, J. Emerson, S. McNamara, R. L. Gibson, B. W. Ramsey, and S. I. Miller. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10319890-19895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 4373-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ka, J. O., J. Urbance, R. W. Ye, T. Y. Ahn, and J. M. Tiedje. 1997. Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol. Lett. 15655-60. [DOI] [PubMed] [Google Scholar]
- 29.Kawasaki, S., H. Arai, T. Kodama, and Y. Igarashi. 1997. Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: sequencing and identification of a locus for heme d1 biosynthesis. J. Bacteriol. 179235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, M., Y. Matsuo, A. Takimoto, S. Suzuki, F. Maruo, and H. Shoun. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 27116263-16267. [DOI] [PubMed] [Google Scholar]
- 31.Körner, H., and W. G. Zumft. 1989. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 551670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam, Y., and D. J. Nicholas. 1969. A nitrite reductase with cytochrome oxidase activity from Micrococcus denitrificans. Biochim. Biophys. Acta 180459-472. [DOI] [PubMed] [Google Scholar]
- 33.Lépine, F., E. Déziel, S. Milot, and L. G. Rahme. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 162236-41. [DOI] [PubMed] [Google Scholar]
- 34.Machan, Z. A., G. W. Taylor, T. L. Pitt, P. J. Cole, and R. Wilson. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30615-623. [DOI] [PubMed] [Google Scholar]
- 35.Magalon, A., R. A. Rothery, D. Lemesle-Meunier, C. Frixon, J. H. Weiner, and F. Blasco. 1998. Inhibitor binding within the NarI subunit (cytochrome bnr) of Escherichia coli nitrate reductase A. J. Biol. Chem. 27310851-10856. [DOI] [PubMed] [Google Scholar]
- 36.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 481320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 23027-34. [DOI] [PubMed] [Google Scholar]
- 38.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1822702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas, D. J. D., and A. Nason. 1957. Determination of nitrate and nitrite. Methods Enzymol. 3981-984. [Google Scholar]
- 40.Palmer, K. L., S. A. Brown, and M. Whiteley. 2007. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 1894449-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 9611229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radcliffe, B. C., and D. J. Nicholas. 1970. Some properties of a nitrate reductase from Pseudomonas denitrificans. Biochim. Biophys. Acta 205273-287. [DOI] [PubMed] [Google Scholar]
- 44.Sawada, I., H. Maseda, T. Nakae, H. Uchiyama, and N. Nomura. 2004. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa. Microbiol. Immunol. 48435-439. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber, K., R. Krieger, B. Benkert, M. Eschbach, H. Arai, M. Schobert, and D. Jahn. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 1894310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schripsema, J., K. E. de Rudder, T. B. van Vliet, P. P. Lankhorst, E. de Vroom, J. W. Kijne, and A. A. van Brussel. 1996. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J. Bacteriol. 178366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma, V., C. E. Noriega, and J. J. Rowe. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vector for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118640-659. [DOI] [PubMed] [Google Scholar]
- 49.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie van Leeuwenhoek 6623-36. [DOI] [PubMed] [Google Scholar]
- 50.Takaya, N., M. A. Catalan-Sakairi, Y. Sakaguchi, I. Kato, Z. Zhou, and H. Shoun. 2003. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl. Environ. Microbiol. 693152-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, K. L., D. Lloyd, and L. Boddy. 1994. Effects of oxygen, pH and nitrate concentration on denitrification by Pseudomonas species. FEMS Microbiol. Lett. 118181-186. [DOI] [PubMed] [Google Scholar]
- 52.Toyofuku, M., N. Nomura, T. Fujii, N. Takaya, H. Maseda, I. Sawada, T. Nakajima, and H. Uchiyama. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1894969-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Houdt, R., P. Moons, M. Hueso Buj, and C. W. Michiels. 2006. N-Acyl-l-homoserine lactone quorum sensing controls butanediol fermentation in Serratia plymuthica RVH1 and Serratia marcescens MG1. J. Bacteriol. 1884570-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wade, D. S., M. W. Calfee, E. R. Rocha, E. A. Ling, E. Engstrom, J. P. Coleman, and E. C. Pesci. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1874372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson, A., V. Danino, F. Wisniewski-Dye, J. K. Lithgow, and J. A. Downie. 2002. N-Acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 1844510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao, G., E. Déziel, J. He, F. Lépine, B. Lesic, M. H. Castonguay, S. Milot, A. P. Tampakaki, S. E. Stachel, and L. G. Rahme. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 621689-1699. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, G., J. He, and L. G. Rahme. 2006. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 1521679-1686. [DOI] [PubMed] [Google Scholar]
- 60.Yamanaka, T., S. Kijimoto, and K. Okunuki. 1963. Biological significance of Pseudomonas cytochrome oxidase in Pseudomonas aeruginosa. J. Biochem. 53416-421. [DOI] [PubMed] [Google Scholar]
- 61.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]