Abstract
Growth experiments showed that adenine and hypoxanthine can be used as nitrogen sources by several strains of K. pneumoniae under aerobic conditions. The assimilation of all nitrogens from these purines indicates that the catabolic pathway is complete and proceeds past allantoin. Here we identify the genetic system responsible for the oxidation of hypoxanthine to allantoin in K. pneumoniae. The hpx cluster consists of seven genes, for which an organization in four transcriptional units, hpxDE, hpxR, hpxO, and hpxPQT, is proposed. The proteins involved in the oxidation of hypoxanthine (HpxDE) or uric acid (HpxO) did not display any similarity to other reported enzymes known to catalyze these reactions but instead are similar to oxygenases acting on aromatic compounds. Expression of the hpx system is activated by nitrogen limitation and by the presence of specific substrates, with hpxDE and hpxPQT controlled by both signals. Nitrogen control of hpxPQT transcription, which depends on σ54, is mediated by the Ntr system. In contrast, neither NtrC nor the nitrogen assimilation control protein is involved in the nitrogen control of hpxDE, which is dependent on σ70 for transcription. Activation of these operons by the specific substrates is also mediated by different effectors and regulatory proteins. Induction of hpxPQT requires uric acid formation, whereas expression of hpxDE is induced by the presence of hypoxanthine through the regulatory protein HpxR. This LysR-type regulator binds to a TCTGC-N4-GCAAA site in the intergenic hpxD-hpxR region. When bound to this site for hpxDE activation, HpxR negatively controls its own transcription.
Purines play a key role in nucleic acid and nucleotide metabolism in all cells. In addition, they can be used as nitrogen sources by many microorganisms when ammonia, the preferred nitrogen source, is limiting (50). Transport of these nucleobases across the cell membrane is mediated by transmembrane proteins, which are distributed in three basic families, designated nucleobase-ascorbate transporters, purine-related transporters, and plant urine related transporters (23).
The first step in the assimilation of adenine or guanine as a nitrogen source is usually a deamination reaction catalyzed by specific enzymes, yielding one molecule of ammonia and hypoxanthine or xanthine, respectively. The catabolic pathway for hypoxanthine assimilation can be divided into two parts. In the first part, this compound is transformed to allantoin, and in the second part, allantoin degradation is completed to CO2 and ammonia. Although the first part of this pathway is common to all species, the degradation of allantoin can follow different routes according to the microbial species (11, 43, 50, 54).
Hypoxanthine is sequentially oxidized by xanthine dehydrogenase to xanthine and uric acid, which is further converted to allantoin (Fig. 1A). Most of the reported xanthine dehydrogenases or oxidoreductases are molybdenum-containing hydroxylases. This group of enzymes catalyzes the hydroxylation of carbon atoms using oxygen derived ultimately from water, rather than O2, and does not require an external source of reducing equivalents (19). Recently, the uric acid oxidation reaction has been studied in great detail, leading to the identification of other enzymatic activities associated with uric acid degradation (24, 37). Conversion of uric acid to allantoin involves three enzymatic reactions. Oxidation of uric acid by uricase or urate oxidase yields 5-hydroxyisourate (HIU), which is then transformed to 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) by HIU hydrolase, and finally OHCU undergoes stereoselective decarboxylation by the action of OHCU decarboxylase to give CO2 and (S)-allantoin.
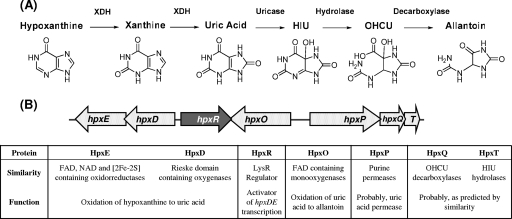
FIG. 1.
Metabolic map for hypoxanthine assimilation to allantoin (A) and gene organization of the hpx system involved in this pathway in K. pneumoniae (B). The arrows show the extents and directions of transcription of the genes. Sequence similarity and function assignment of the hpx-encoded proteins are indicated in the table. XDH, xanthine dehydrogenase.
Purine catabolism in enterobacteria has been poorly characterized at the genetic level, except in Escherichia coli. In E. coli, aerobic adenine catabolism to allantoin has been indirectly demonstrated by the production of 14CO2 from [14C]adenine, which implies the existence of functional xanthine dehydrogenase (encoded by genes xdhABC and xdhD at min 65 of the genome) and uricase activities (53). Nevertheless, nitrogen assimilation from adenine is not complete, since in the presence of oxygen allantoin is not further degraded. In E. coli, expression of genes encoding allantoate amidohydrolase (allB) and ureidoglycolate dehydrogenase (allD), both of which are required for allantoin assimilation, takes place only under anaerobic conditions (11, 39). Thus, aerobic growth of E. coli in adenine as the sole nitrogen source is supported only by the ammonia liberated by adenosine deaminase during the conversion to hypoxanthine. Regarding Klebsiella pneumoniae, there are a few reports on the ability of certain strains to grow on adenine or hypoxanthine as nitrogen sources (16, 51, 53). However, at present there are no reported descriptions of the genetic systems involved in this catabolic pathway.
In the presence of ammonia, there is a strong repression of many systems that allow enterobacteria such as E. coli or K. pneumoniae to use alternative nitrogen sources such as amino acids or purines. When ammonia is limiting, the ability of these cells to obtain nitrogen from these compounds requires in almost every case a two-component system in which a phosphorylated activator, NtrC-P, binds to an enhancer and interacts with RNA polymerase bearing the sigma factor σ54. The Ntr (nitrogen regulation) system activates a set of genes involved in the catabolism of nitrogenous compounds whose degradation products include ammonia or glutamate (31, 38). The Ntr system also activates the transcription of another transcriptional regulator, the nitrogen assimilation control protein (NAC) (3, 15, 28). NAC regulates a subset of genes that are dependent on RNA polymerase bearing σ70 for their transcription (20, 44). Thus, NAC serves to couple the σ54-dependent transcription of the Ntr system with that of the σ70-dependent catabolic genes.
In this study we identified and characterized the gene cluster (named hpx) responsible for the oxidation of hypoxanthine to allantoin in K. pneumoniae. This genetic system, which is essential for the aerobic assimilation of hypoxanthine as a nitrogen source, is regulated by both nitrogen limitation and the presence of specific substrates. However, the nitrogen control of the operon encoding the enzyme catalyzing the oxidation of hypoxanthine to uric acid is not mediated by the Ntr system.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The genotypes and sources of the bacterial strains, plasmids, and promoter fusions are given in Table 1. Genetic crosses were performed by P1-mediated transduction (17).
TABLE 1.
Strains, fusions, and plasmids used in this study
| Strain, fusion, or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| K. pneumoniae strains | ||
| KC2653 | hutC515 Δ[bla]-2 dadA1 str-6 | 26 |
| 52145 | Serotype O1:K2; carries the virulence plasmid pKP100 | 33 |
| ATCC 13882 | K. pneumoniae subsp pneumoniae, deposited as K. aerogenes | American Type Culture Collection |
| ATCC 13883 | K. pneumoniae, subsp pneumoniae (Schroeter) Trevisan | American Type Culture Collection |
| JA-K10 | KC2653 hpxD::mini-Tn5 Km | This study |
| JA-K11 | KC2653 glnD::mini-Tn5 Km | This study |
| JA-K12 | KC2653 hpxE::mini-Tn5 Km | This study |
| JA-K13 | KC2653 gltD::mini-Tn5 Km | This study |
| JA-K14 | KC2653 hpxO::kan | This study |
| JA-K15 | KC2653 hpxP::kan | This study |
| JA-K16 | KC2653 hpxR::kan | This study |
| KC2562 | hutC515 rpoN5018 | 26 |
| KC5249 | hutC515 Δ[bla]-2 nac-2 | R. A. Bender |
| KC2738 | hutC515 ntrC2::Tn5-131 | 4 |
| KC6483 | hutC515 Δ[bla]-2 str-6 [Δrbs(A′BC′K)::pCB1583] | R. A. Bender |
| E. coli strains | ||
| DH5αF′ | φ80d lacZΔM15 recA1 endA1 λ−gyrA96 thi-1 hsdR17 (rK− mK+) phoA supE44 relA1 deoR Δ(lacZYA-argF)U169 | Gibco BRL |
| ECL1 | HfrC phoA8 relA1 tonA22 T2r (λ) | 25 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tcr)] | Stratagene |
| S17(λ pir) | Tpr SmrrecA thi pro hsdR−hsdM+ RP4::2-Tc::Mu::Km Tn7 λ | Biomedal |
| EB6193 | RP4-2 tet Mu-1 Kan::Tn7 integrant; leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5 uidA(ΔMuI)::pir+thi Spr/Smr | R. A. Bender |
| Fusionsa | ||
| Φ(hpxD-lacZ) | hpxD (−346 to +97) fused to lacZ | This study |
| Φ(hpxR-lacZ) | hpxR (−212 to +93) fused to lacZ | This study |
| Φ(hpxO-lacZ) | hpxO (−267 to +288) fused to lacZ | This study |
| Φ(hpxP-lacZ) | hpxP (−258 to +297) fused to lacZ | This study |
| Plasmids | ||
| pUC18Not | Apr; identical to pUC18 but with NotI in the multiple-cloning site | Biomedal |
| pGEMT | Apr; cloning vector for PCR products | Promega |
| pUT mini-Tn5 Km | Apr Kmr; tnp* gene of Tn5-IS50R inserted in SalI site of pGP704; mini-Tn5 Km transposable element | Biomedal |
| pKAS32 | Apr; pGP704, rpsL | 46 |
| pRS415 | Apr; promoterless lacZYA reporter for operon fusions with replication origin of pBR322 | 45 |
| pCB1583 | Apr Kmr; promoterless lacZ reporter for integration of operon fusions on host genome with oriR6K replication origin; rpsL | R. A. Bender |
| pMAL-c2x | Apr; vector for cytoplasmic expression of maltose binding protein fusion proteins | New England Biolabs |
| pKD4 | Apr Kmr | 12 |
Nucleotide sequences are given in the 5′-to-3′ direction for the coding strand of each gene and are numbered relative to the gene transcription initiation nucleotide at position +1.
Growth conditions.
Cultures were grown at 30°C with aeration in lysogeny broth (LB) (5) or in W4 minimal medium (47) supplemented with glucose at 0.4% as the sole carbon source. NaCl was removed from LB plates for selection of directed mutants. For limiting nitrogen conditions, freshly made glutamine (Gln) was used at 0.04%. Ammonium sulfate and Gln (NGln), both at 0.2%, were used for nitrogen excess (2, 3). Purines were routinely used as nitrogen sources at 0.1%. When indicated, they were used at 0.6 mM. Hypoxanthine and uric acid were prepared as described by Rouf and Lomprey (40). The selection medium in the conjugation experiments consisted of W4 minimal medium supplemented with sodium citrate at 0.4% as the carbon source and ammonium sulfate at 0.2% as the nitrogen source. In experiments in which urease was measured, the growth medium was supplemented with 1 μM NiSO4. When required, the following antibiotics were used at the indicated concentrations: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 50 μg/ml; streptomycin (Sm), 50 μg/ml; and tetracycline, 30 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) and isopropyl-β-d-thiogalactoside (IPTG) were used at 30 and 10 μg/ml, respectively.
Preparation of cell extracts and enzyme activities.
Cultures were grown at 30°C to an optical density at 600 nm (OD600) of 0.5. Cells were collected by centrifugation, washed in 1% KCl, and suspended at a concentration that contained 1 to 1.5 mg of protein per ml (1). Urease was assayed as described by Liu and Bender (26). β-Galactosidase activity was assayed in detergent-treated whole cells using ο-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate and was expressed as U/mg of cell protein (32). One unit of β-galactosidase activity corresponds to the amount of enzyme that hydrolyzes 1 nmol of ONPG per min. The data reported are the averages from at least four separate experiments performed in triplicate with a standard error of <15%. Protein concentration was determined by the method of Lowry et al. (27) with bovine serum albumin as a standard.
DNA manipulation and sequencing.
Bacterial genomic DNA was obtained using the Wizard genomic DNA purification kit (Promega), and plasmid DNA was prepared using the Wizard Plus SV Midipreps DNA purification system (Promega). DNA manipulations were performed as described by Sambrook and Rusell (41). DNA fragments were amplified by PCR using chromosomal DNA as a template. When necessary, specific restriction sites were incorporated at the 5′ ends of the primers to facilitate the cloning of the fragments in the appropriate vector. PCRs were performed with Pfu DNA polymerase under standard conditions. DNA was sequenced using an automated ABI 377 DNA sequencer and fluorescent dye termination methods. The primers used in this study are given in Table S1 in the supplemental material.
Mapping of the 5′ ends of the hpx transcripts.
The 5′ regions of the hpxDE, hpxR, hpxO, and hpxPQR transcripts were determined by the rapid amplification of cDNA 5′ ends (5′-RACE) (41) using a commercial 5′-RACE kit (Roche Diagnostics, GmbH). Total RNA was isolated from KC2653 cells grown aerobically to an OD600 between 0.5 and 1 in glucose minimal medium with hypoxanthine as the nitrogen source using the Qiagen RNeasy total RNA kit and then treated with RNase-free DNase (Ambion). The cDNAs were transcribed from the RNA with specific hpxD, hpxR, hpxO, or hxpP antisense oligonucleotides. A homopolymeric dA tail was added (via terminal transferase) to the 3′ termini of the corresponding cDNAs. Amplification of reverse transcription products was performed with nested gene-specific primers and an oligo(dT) anchor primer. The double-stranded cDNAs obtained were cloned into a pGEMT vector for sequencing.
Mini-Tn5 Km-1 random mutagenesis.
Random mini-Tn5 insertion mutants of K. pneumoniae that did not use hypoxanthine as a nitrogen source were obtained by conjugation using the pUTmini-Tn5 Km delivery vector (Biomedal). K. pneumoniae strain KC2653 was used as the recipient. E. coli strain S17.1(λ pir) harboring pUTmini-Tn5 Km was used as the donor, and conjugation was carried out following the supplier's instructions. Different amounts of the conjugation mix were plated on citrate minimal medium plates containing Km. Since E. coli cells do not use citrate as a carbon source, this medium counterselects the donor strain and selects recipient cells carrying the Km transposon marker. Hypoxanthine mutants were subsequently selected by replica plating on glucose minimal medium with hypoxanthine as the nitrogen source.
Mini-Tn5 insertions were mapped by inverse PCR. This protocol involved the isolation of genomic DNA; digestion with the restriction enzyme HhaI or TaiI, which cut within the transposon and numerous times within the genome; ligation to circularize all linear genomic fragments; and then PCR using two outward-facing transposon-specific primers. Amplified products were sequenced, and the similarity to genes and open reading frames deposited in databases was determined using the BLAST search algorithm at the National Center for Biotechnology Information. Following this computational analysis, the sites of transposon insertion in the isolated mutants were identified.
Directed mutagenesis of K. pneumoniae hpxR, hpxO, and hpxP genes.
Knockout mutants of K. pneumoniae KC2653 were generated by antibiotic marker exchange using the suicide plasmid pKAS32 (46). This vector contains the R6K origin of replication, which functions only in bacteria that produce the replication protein π. In addition, this vector expresses the E. coli rpsL gene, encoding ribosomal protein S12, which provides a positive selection for bacteria that have exchanged cloned plasmid sequences with the corresponding chromosomal sequences.
To clone the hpxR, hpxO, or hpxP gene, primers were designed to amplify the corresponding open reading frames plus their flanking regions by PCR. A Km cassette was obtained for each gene by inserting the Km resistance gene kan, which was obtained by PCR amplification from plasmid pKD4 (12), into the corresponding coding region. In all the Km cassettes, the kan gene was flanked by K. pneumoniae-specific genomic sequences of at least 500 bp. Mutagenesis of chromosomal genes was carried out by homologous recombination between the Km cassette and the wild-type gene after mating E. coli S17.1(λ pir) harboring the recombinant pKAS32 derivative containing the Km cassette with K. pneumoniae strain KC2653 as the recipient.
To construct the hpxO knockout mutant, the Km-O cassette was obtained as follows. A 2,297-bp fragment encompassing hpxO and its flanking regions was amplified by PCR from genomic DNA of strain KC2653. Restriction sites for EcoRI or XbaI were incorporated at the 5′ ends of the primers to facilitate directed cloning of the amplified fragment into pKAS32. Disruption of the cloned hpxO gene was performed by insertion of the kan gene into the MluI site located 365 bp downstream of the ATG codon. To construct the hpxR knockout mutant, a 1,127-bp fragment encompassing hpxR and its flanking regions was amplified by PCR and cloned into the EcoRI and XbaI restriction sites of pUC18Not. Disruption of the hpxR cloned gene was performed by insertion of the kan gene into the AscI site located 591 bp downstream of the ATG codon. The insert containing the disrupted hpxR gene was further subcloned into the NotI restriction site of the suicide plasmid pKAS32. A similar approach was followed to construct the Km-P cassette used to obtain the hpxP knockout mutant. The genomic region encompassing this gene was first cloned into pUC18Not. Disruption of the cloned hpxP gene was performed by insertion of the kan gene into the SphI site (made blunt) located 255 bp downstream of the ATG codon. This recombinant plasmid was digested with NotI and the corresponding insert subsequently subcloned into pKAS32.
The obtained pKAS32 recombinant plasmids were propagated into strain EB6193 and introduced into E. coli S17.1(λ pir) by electroporation for mating with the recipient Smr strain KC2653 as described previously. Transconjugants were selected for resistance to Km on citrate plates and further purified in this medium. Isolated colonies were then grown on LB plates without NaCl containing Km and Sm (1 mg/ml) to facilitate homologous recombination. After several rounds of growth at 30°C in this medium, colonies were screened for sensitivity to Ap in order to identify which transconjugants had undergone allelic exchange and therefore did not carry the plasmid still integrated (merodiploids of the target gene). Gene disruption of the correct target gene was verified by PCR and Southern blot analysis.
Construction of lacZ transcriptional fusions.
Transcriptional fusions were constructed by inserting the promoter fragments into plasmid pRS415 (45). This plasmid carries a cryptic lacZYA operon and confers resistance to Ap. To construct the hpxD-lacZ fusion, a 443-bp PhpxD promoter fragment was obtained by PCR and was cloned into plasmid pRS415 using the EcoRI and BamHI sites. To construct the hpxR-lacZ fusion, a 305-bp PhpxR promoter fragment, obtained by PCR, was cloned into plasmid pRS415 using the same restriction sites.
To construct the hpxO-lacZ fusion, a 555-bp fragment encompassing the hpxO-hpxP intergenic region was amplified by PCR and cloned into the pRS415 vector using the EcoRI and SmaI sites. The same promoter fragment was cloned in the opposite direction to construct the hpxP-lacZ fusion. For all constructs, plasmid DNA was sequenced to ensure that the fragment was inserted in the correct orientation and that no mutations had been introduced during the amplification reaction.
To transfer the lacΖ fusions into the K. pneumoniae chromosome as a single copy, the recombinant plasmid was first digested with EcoRI and SacI, and the fragment containing the promoter fusion was subcloned into plasmid pCB1583 (26). This is a pir-dependent plasmid whose lacZ gene is flanked by genes of the K. pneumoniae d-ribose operon, thus allowing the integration of the cloned fusion by homologous recombination into the d-ribose operon of the K. pneumoniae recipient strain. The recombinant plasmids containing the different lacZ fusions were selected after transformation of strain EB6193 as blue colonies on LB-X-Gal-Km plates and then introduced into strain KC2653 by electroporation. After several rounds of selection in different growth media, stable recombinants in the rbs locus were isolated as those displaying the Aps Kms Smr d-ribose negative phenotype (26).
Expression and purification of HpxR.
HpxR was purified using the malE gene fusion system. For this purpose, hpxR was amplified by PCR and cloned into the BamHI and EcoRI restriction sites of plasmid pC2x, yielding plasmid pC2x-hpxR. The forward primer was designed to fuse the ATG start codon of hpxR in frame with the gene encoding MalE. Overproduction of the MalE-HpxR fusion protein was achieved in strain XL1Blue carrying the recombinant plasmid pC2x-hpxR after induction by IPTG (0.3 mM) in LB-glucose-Ap medium for 3 h at 37°C. The fusion protein was then purified by affinity chromatography with amylose resin (New England BioLabs) according to the manufacturer's instructions. Elution was performed with column buffer containing 10 mM maltose, and 100 μg of the fusion protein was digested with 1 U of factor Xa by incubation at room temperature for 12 h. The cleaved HpxR protein was used in gel shift experiments.
DNA binding studies.
For electrophoretic mobility shift assays, fragments corresponding to the different promoters of the hpx genetic system were generated by PCR from KC2653 chromosomal DNA. After purification from acrylamide gels, fragments were labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham). Labeled DNA fragments were incubated with either purified HpxR or crude extracts obtained as described by Nunoshiba et al. (35) in 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol in a total volume of 20 μl. Poly(dI-dC) was used as a nonspecific competitor. The binding mixtures were incubated for 15 min at 37°C and electrophoresed in a prerun gel of 5% native polyacrylamide containing 10% glycerol in 1× Tris-borate-EDTA buffer at 4°C. The gels were then dried under vacuum, and the bands were visualized by autoradiography.
Nucleotide sequence accession number.
The sequence of the 6,744-bp gene cluster from K. pneumoniae strain KC2653 that was sequenced in this study has been deposited in GenBank under accession number EU653284.
RESULTS
Utilization of adenine and hypoxanthine as nitrogen sources by K. pneumoniae.
We analyzed the ability of K. pneumoniae strain KC2653 to grow on adenine and the intermediates of its catabolic pathway, hypoxanthine and allantoin. This strain can utilize all these compounds as a nitrogen source under aerobic conditions, which indicates that an entire adenine catabolic pathway exists in K. pneumoniae strain KC2653. The final cell density of cultures grown with equimolecular amounts (0.6 mM) of NH4Cl, adenine, hypoxanthine, or allantoin as the sole nitrogen sources indicated that all nitrogens in these purines and derivatives were assimilated (Table 2). E. coli strain ECL1 was analyzed in parallel as a negative control (53). Other strains of K. pneumoniae, such as ATCC 13882, ATCC 13883 and 52145, displayed the same purine and allantoin phenotypes as strain KC2653. These compounds cannot support growth of these strains when used as a carbon source.
TABLE 2.
Final cell densities of cultures grown in glucose minimal medium with different nitrogen sources at 0.6 mM
| Strain | Relevant genotype | OD600 of cultures grown with:
|
||||
|---|---|---|---|---|---|---|
| NH4Cl | Adenine | Hypoxanthine | Uric acid | Allantoin | ||
| ECL1 | E. coli wild type | 0.22 | 0.26 | NGa | NG | NG |
| KC2653 | Wild type | 0.23 | 0.86 | 0.81 | 0.86 | 0.98 |
| JA-K10 | hpxD | 0.21 | 0.24 | NG | 0.88 | 0.95 |
| JA-K12 | hpxE | 0.23 | 0.22 | NG | 0.89 | 0.89 |
| JA-K14 | hpxO | 0.22 | 0.22 | NG | NG | 0.85 |
| JA-K15 | hpxP | 0.25 | 0.83 | 0.90 | 0.83 | 0.81 |
| JA-K16 | hpxR | 0.23 | 0.20 | NG | 0.86 | 0.80 |
| KC2738 | ntrC | NG | NG | NG | NDb | ND |
| KC5249 | nac | 0.20 | 0.90 | 0.86 | ND | ND |
NG, no growth.
ND, not determined.
Identification of the genetic system for hypoxanthine assimilation.
In the assimilation pathway of adenine as a nitrogen source, this nucleobase is converted to hypoxanthine. Thus, we focused our study on hypoxanthine metabolism. To search for genes of K. pneumoniae involved in hypoxanthine assimilation as a nitrogen source under aerobic conditions, we generated a set of mutants selected by their inability to utilize this purine as a nitrogen source. Random mini-Tn5 insertion mutants of strain KC2653 were obtained as described in Materials and Methods. Four mutants were selected by their inability to assimilate hypoxanthine as the sole nitrogen source after replica plating on glucose-hypoxanthine; two of them (JA-K10 and JA-K12) retained the ability to use allantoin as a nitrogen source, but the other two mutants (JA-K11 and JA-K13) lost this ability.
Mapping of mini-Tn5 insertions in the selected mutants was performed by inverse PCR analysis followed by sequencing the region adjacent to the Tn5 joining site using a specific primer for mini-Tn5. Based on the genome sequence of K. pneumoniae subsp. pneumoniae MGH 78578, available in the NCBI GenBank (NC_009648), insertions in the JA-K11 and JA-K13 mutants were located within genes glnD (KPN_00180), encoding uridil transferase, and gltD (KPN_03625), encoding a glutamate synthase subunit, respectively. These genes are involved in nitrogen assimilation and its control under nitrogen-limiting conditions.
In JA-K10 and JA-K12, the mini-Tn5 insertions were closely located and disrupted genes of unknown function. In JA-K12, the disrupted gene (KPN_01661) encodes a protein displaying similarity to oxidoreductases acting on aromatic compounds. In JA-K10, the transposon insertion was located 120 bp upstream of the ATG codon of the same gene, KPN_01661, in an open reading frame displaying similarity to genes encoding oxygenases of aromatic compounds, such as vanillate O-demethylase from the genus Xanthomonas or Pseudomonas. Since mutations in these genes did not impair utilization of allantoin as a nitrogen source (Table 2), the encoded proteins may act in enzymatic reactions involved in the transformation of hypoxanthine to allantoin.
Computational analysis of the genomic region encompassing the genes disrupted in the JA-K10 and JA-K12 mutants allowed us to identify a gene cluster formed by seven genes that may be involved in the hypoxanthine assimilation pathway and for which we propose the notation hpx (Fig. 1B). An in silico analysis through the Neural Network Promoter Prediction program (http://www.fruitfly.org/seq_tools/promoter.html) identified putative promoters at the 5′ ends of genes hpxD, hpxR, hpxO, and hpxP, suggesting its organization into four transcriptional units.
This gene cluster (6,744 bp) was sequenced from K. pneumoniae strain KC2653. Comparison of this nucleotide sequence with that of strain MGH 78578 revealed some differences at the nucleotide level but the same gene cluster organization. For all genes, the encoded proteins displayed a high degree of identity (around 98% overall).
The amino acid sequences encoded by the hpx genes were used as the query sequences for BLASTP search in the GenBank database. hpxR encodes a protein of the LysR regulatory family, hpxO encodes a protein displaying high similarity to flavin adenine dinucleotide (FAD)-containing monooxygenases acting on aromatic compounds, and hpxP encodes a product that is similar to xanthine and guanine/uracil transporters. The hpxQ gene product displays similarity to OHCU decarboxylases and the hpxT product to proteins of the transthyretin family involved in the hydrolysis of HIU.
To determine the function of these proteins in the metabolic pathway of hypoxanthine dissimilation, hpxO, hpxP, or hpxR knockout mutants were constructed in the genetic background of strain KC2653, as described in Materials and Methods. The effects of these mutations on different steps in the hypoxanthine catabolic pathway were analyzed by growth of the mutant strains on glucose minimal medium containing hypoxanthine or uric acid as the sole nitrogen source. The phenotypes of the mutants with mini-Tn5 insertions in hpxD and hpxE (strains JA-K10 and JA-K12) were also analyzed. Mutations in hpxO impaired utilization of both hypoxanthine and uric acid as nitrogen sources, while mutations in hpxD or hpxE abolished only hypoxanthine assimilation (Table 2). These results suggested a putative role of hpxD- and hpxE-encoded proteins as xanthine dehydrogenase subunits and HpxO as a putative uricase. Based on sequence similarity, we propose that HpxT could hydrolyze the intermediate HIU generated in the uricase reaction, yielding OHCU, which would be transformed into allantoin by the action of HpxQ (7, 23, 36). We have been unable to directly verify these functions by assaying xanthine dehydrogenase or uricase activity in cell extracts obtained from strain KC2653 or with the hpxDE and hpxO genes expressed from a high-copy-number plasmid.
Regarding hpxP, although the encoded protein displays high similarity to purine transporters, disruption of this gene (mutant strain JA-K15) did not modify growth on hypoxanthine or uric acid (Table 2).
Mutations in hpxR (mutant strain JA-K16) did not abolish uric acid utilization but impaired assimilation of hypoxanthine as a nitrogen source (Table 2). This phenotype suggests that this LysR-type regulator could act as an activator of hpxDE expression.
Transcriptional organization of the hpx regulon.
In this study, the 5′ ends of the hpx transcripts were experimentally determined by the 5′-RACE method (Fig. 2). For the hpxDE, hpxR, and hpxO transcriptional units, inspection of the DNA sequences upstream of nucleotide +1, the mRNA start site, revealed the presence of −35 and −10 sequences, which were similar to the σ70 consensus (Fig. 2). However, for the hpxPQT operon this position was localized in a putative σ54 promoter region identified by in silico analysis using Promscan (score, 55) (http://www.promscan.uklinux.net/home.html) (48). These results suggest that in cells grown with hypoxanthine as the nitrogen source, hpxDE, hpxR, and hpxO are transcribed by the σ70-dependent RNA polymerase, while hpxPQT transcription seems to be dependent on σ54.
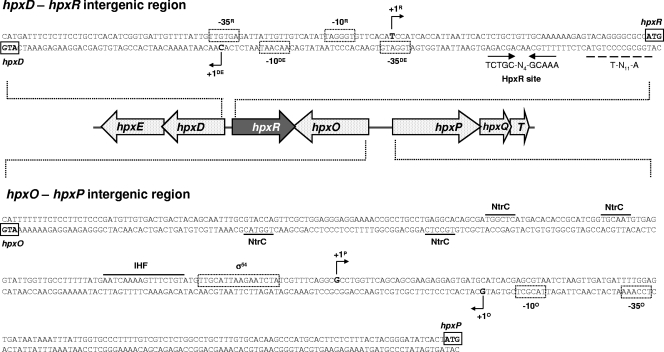
FIG. 2.
Promoter sequences of the divergently transcribed hpxD and hpxR genes and hpxO and hpxP genes. For each gene, the ATG initiation codon is boxed, the consensus for RNA polymerase (−10 and −35 sequences for σ70 recognition in hpxD, hpxR, and hpxO or the σ54 recognition sequence in hpxP) is boxed with dotted lines, and the 5′ end is shown by a black arrowhead labeled +1. Putative IHF or NtrC binding sites identified using Promscan and Virtual Footprint programs are indicated. Consensus sequences for LysR-type regulators (T-N11-A) are indicated. The sequence with partial dyad symmetry, TCTGC-N4-GCAAA, whose deletion abolished HpxR binding is indicated by arrows and labeled as the HpxR site, whereas the T-N11-A sequence, shown not to be involved in HpxR binding, is underlined with a dashed line.
Effect of the nitrogen source on expression of the hpx transcriptional units.
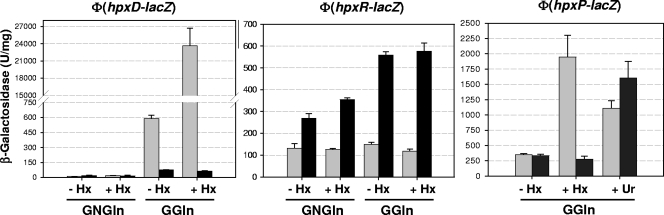
As the hpx genetic system is involved in the assimilation of an organic nitrogenous compound, expression of the hpx promoter fusions (hpxD-lacZ, hpxR-lacZ, hpxO-lacZ, and hpxP-lacZ) was analyzed in the genetic background of strain KC2653 grown with different nitrogen sources. β-Galactosidase activity was measured in glucose cultures with nitrogen excess (GNGln), with nitrogen limitation (GGln), or with hypoxanthine (GHx) or uric acid (GUr) as the sole nitrogen source.
The results presented in Table 3 show that only hpxDE and hpxPQT are regulated by nitrogen availability. These transcriptional units are repressed by nitrogen excess but induced under nitrogen-limiting conditions. Maximal induction of hpxDE was achieved by growth with hypoxanthine as the sole nitrogen source, whereas for hpxPQT, hypoxanthine and uric acid yielded similar β-galactosidase levels. These results suggest additional control of these operons, probably mediated by hypoxanthine or derived metabolites. We next examined whether this specific regulation is influenced by nitrogen availability. To this end, hypoxanthine or uric acid was added to high-ammonia (GNGlnHx and GNGlnUr) or to nitrogen-limited (GGlnHx and GGlnUr) cultures. Induction of Φ(hpxDE-lacZ) and Φ(hpxP-lacZ) by either hypoxanthine or uric acid was impaired when cells were grown in the presence of excess nitrogen (Table 3). However, addition of hypoxanthine to nitrogen-limited cultures led to a large increase in the β-galactosidase levels, although these were somewhat lower than those obtained with hypoxanthine alone (Table 3). Addition of uric acid to nitrogen-limited cultures induced expression only of Φ(hpxP-lacZ). These results strongly indicate that two levels of control drive the expression of hpxDE and hpxPQT operons: (i) nitrogen-limiting conditions derived from low ammonia levels and (ii) availability of the specific nitrogen compound.
TABLE 3.
β-Galactosidase activity from the hpx operon fusions in the genetic background of strain KC2653 grown under different conditions of nitrogen availability
| Promoter fusion | β-Galactosidase sp act (U/mg) from cells grown ina:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| GNGln | GNGlnHx | GNGlnUr | GGln | GGlnHx | GGlnUr | GHx | GUr | |
| Φ(hpxD-lacZ) | 11 | 18 | 8.0 | 589 | 23,609 | 463 | 33,184 | 1,447 |
| Φ(hpxR-lacZ) | 132 | 125 | 117 | 149 | 118 | 149 | 138 | 136 |
| Φ(hpxO-lacZ) | 624 | 579 | 489 | 820 | 729 | 706 | 999 | 916 |
| Φ(hpxP-lacZ) | 44 | 42 | 47 | 348 | 1,944 | 1,109 | 4,170 | 3,892 |
Activity values are the averages from four assays with a standard error of 15% or less. One unit of β-galactosidase activity corresponds to the amount of enzyme that hydrolyzes 1 nmol of ONPG per min. Growth media were as follows: GNGln, nitrogen-excess medium, containing 0.4% glucose and 0.2% each ammonium sulfate and l-glutamine; GNGlnHx and GNGlnUr, same as GNGln but containing 0.1% hypoxanthine or 0.1% uric acid, respectively; GGln, nitrogen-limiting medium, containing 0.4% glucose and 0.04% l-glutamine; GGlnHx and GGlnUr, same as GGln but containing 0.1% hypoxanthine or 0.1% uric acid, respectively; GHx and GUr, 0.4% glucose minimal media containing, respectively, 0.1% hypoxanthine or 0.1% uric acid as the sole nitrogen source.
Regarding the other structural gene (hpxO), as β-galactosidase levels were similar under nitrogen excess or nitrogen limitation, its regulation by nitrogen availability may be ruled out. When hypoxanthine or uric acid was the only nitrogen source, β-galactosidase levels were slightly higher, although they did not reach a twofold increase with respect to the other nitrogen conditions (Table 3).
Levels of β-galactosidase activity expressed by the hpxR-lacZ fusion were similar for all the nitrogen sources tested. These results indicate the constitutive expression of the regulatory gene hpxR, which is not influenced by nitrogen availability or the presence of hypoxanthine (Table 3).
Expression of hpxDE and hpxPQT in Ntr mutants.
We next examined whether the regulation of hpxDE and hpxPQT operons by nitrogen availability was mediated by components of the Ntr system. Since hpxDE is transcribed by the σ70-dependent RNA polymerase, nac mutants were also tested in this study. First, we analyzed the ability of the mutant strains KC2738 (ntrC) and KC5249 (nac) to grow on hypoxanthine as the sole nitrogen source. Mutations in ntrC completely abolished growth on this purine, whereas a nac mutation had no effect, with the culture yield for this strain (KC5249) being similar to that for the parental strain KC2653 (Table 2). These results pointed to the involvement of NtrC in the regulation of the hpx genetic system.
To analyze the effect of rpoN, ntrC, or nac mutations on hpxDE and hpxPQT expression, these mutations were introduced by P1 transduction into the genetic background of strain KC2653 bearing Φ(hpxD-lacZ) or Φ(hpxP-lacZ). The presence of these mutations in the selected transductants was checked by the reduction in urease activity levels (not shown). Expression of Φ(hpxD-lacZ) and Φ(hpxP-lacZ) in the derived mutant strains was determined after growth under nitrogen-limiting conditions (GGln) and compared with that of the parental strain KC2653 (Table 4). Expression of hpxDE was not modified by rpoN, ntrC, or nac mutations, thus ruling out the involvement of NtrC or NAC in the nitrogen control of the hpxDE operon. Consistent with these results, an in silico analysis of the hpxD 5′ upstream region performed with Promscan and Virtual Footprint software (http://www.prodoric.de/vfp and http://www.promscan.uklinux.net/home.htlm) failed to identify any putative binding site for NtrC or NAC. Thus, an as-yet-unidentified nitrogen-sensing system other than Ntr seems to be involved in the control of this operon.
TABLE 4.
Expression of hpxD-lacZ and hpxP-lacZ promoter fusions in mutants with different mutations of the Ntr system grown in glucose nitrogen-limiting medium
| Promoter fusion | β-Galactosidase sp act (U/mg) from cells grown in GGlna
|
|||
|---|---|---|---|---|
| KC2653 | KC2653 rpoN | KC2653 ntrC | KC2653 nac | |
| Φ(hpxD-lacZ) | 589 | 624 | 668 | 479 |
| Φ(hpxP-lacZ) | 347 | 108 | 84 | |
Activity values are the averages from four assays with a standard error of 15% or less. One unit of β-galactosidase activity corresponds to the amount of enzyme that hydrolyzes 1 nmol of ONPG per min. GGln, nitrogen-limiting medium containing 0.4% glucose and 0.04% l-glutamine.
Regarding Φ(hpxP-lacZ), β-galactosidase levels clearly showed that expression of hpxPQT was significantly reduced under nitrogen limitation in both rpoN and ntrC mutants (Table 4). In these mutants activity levels were close to those obtained under conditions of excess nitrogen. An in silico analysis of the hpxP promoter region identified four putative NtrC binding sites upstream from the σ54-recognized sequence and an integration host factor (IHF) binding site located between the NtrC and the σ54 sites (Fig. 2). These findings strongly support the hypothesis that the nitrogen regulation of the hpxPQT operon is mediated by NtrC.
Specific regulation of the hpx gene cluster mediated by hypoxanthine or uric acid.
As stated above, in addition to nitrogen control, a specific regulatory mechanism was observed for the hpxDE and hpxPQT operons in the presence of hypoxanthine, or in the presence of uric acid in the case of hpxPQT (Table 3). In an attempt to identify the signal molecule mediating this activation, expression of Φ(hpxD-lacZ) and Φ(hpxP-lacZ) was analyzed in the genetic background of an hpxD mutant (unable to transform hypoxanthine in uric acid) or an hpxO mutant (unable to degrade uric acid). Parental strain KC2653 was analyzed as the control. β-Galactosidase activity was determined in cells grown under nitrogen-limiting conditions in the presence of hypoxanthine (GGlnHx) or uric acid (GGlnUr) (Table 5). Induction of Φ(hpxP-lacZ) by hypoxanthine was abolished in the hpxD mutant but remained unchanged in the hpxO mutant. These results indicate that uric acid is required for induction of the hpxPQT operon.
TABLE 5.
Expression of hpxD-lacZ and hpxP-lacZ promoter fusions in hpxD or hpxO mutants grown in glucose nitrogen-limiting medium in the presence or absence of either hypoxanthine or uric acid
| Promoter fusion | β-Galactosidase sp act (U/mg) from cells grown ina:
|
|||
|---|---|---|---|---|
| GGlnHx
|
GGlnUr
|
|||
| KC2653 | JA-K10 (hpxD) | KC2653 | JA-K14 (hpxO) | |
| Φ(hpxD-lacZ) | 23,609 | 32,274 | 463 | 197 |
| Φ(hpxP-lacZ) | 1,944 | 266 | 1,108 | 910 |
Activity values are the averages from four assays with a standard error of 15% or less. One unit of β-galactosidase activity corresponds to the amount of enzyme that hydrolyzes 1 nmol of ONPG per min. GGlnHx and GGlnUr, nitrogen-limiting media that contain 0.4% glucose and 0.04% l-glutamine plus 0.1% hypoxanthine or 0.1% uric acid, respectively.
In contrast, an hpxD mutation did not impair induction of Φ(hpxD-lacZ) by hypoxanthine. In the presence of this compound, β-galactosidase levels were even higher than those in wild-type cells. Accordingly, in the parental strain KC2653, growth in the presence of uric acid yielded β-galactosidase levels similar to those obtained in cultures grown in low-ammonia medium in the absence of this compound (Table 3). Taken together, these results point to hypoxanthine as the signal molecule that triggers hpxDE induction. β-Galactosidase levels in the hpxO mutant grown in the presence of uric acid were somewhat lower than those displayed by the parental strain KC2653.
Role of HpxR in the regulation of hpxDE and hpxR.
The fact that mutations in hpxR did not abolish uric acid utilization but impaired assimilation of hypoxanthine as a nitrogen source (Table 2) pointed to HpxR as an activator of hpxDE expression. In order to experimentally confirm this role, expression of Φ(hpxD-lacZ) was analyzed in the hpxR mutant grown in different nitrogen conditions (Fig. 3). In this mutant, induction of Φ(hpxD-lacZ) in limited nitrogen (GGln) and in the presence of hypoxanthine (GGlnHx) was totally abolished. Under these conditions, β-galactosidase levels were lower than 100 U/mg. These results clearly implicate HpxR in the transcriptional activation of the hpxDE operon, mediated not only by hypoxanthine but also by nitrogen availability.
FIG. 3.
Effect of an hpxR mutation on the expression of Φ(hpxD-lacZ), Φ(hpxR-lacZ), and Φ(hpxP-lacZ). Cells of the parental strain KC2653 (gray bars) and the hpxR mutant strain JA-K16 (black bars) bearing the indicated promoter fusion were grown in the indicated culture media. GNGln is a nitrogen-excess medium that contains 0.4% glucose and 0.2% each ammonium sulfate and l-Gln, and GGln is a nitrogen-limiting medium that contains 0.4% glucose and 0.04% l-Gln. Where indicated, 0.1% hypoxanthine (Hx) or 0.1% uric acid (Ur) was added to these media. β-Galactosidase activity is expressed as specific activity. One unit of β-galactosidase activity corresponds to the amount of enzyme that hydrolyzes 1 nmol of ONPG per min. Error bars indicate standard deviations.
Since regulators of the LysR family usually repress their own transcription when activating the divergent structural gene (42), expression of Φ(hpxR-lacZ) was also analyzed in the hpxR mutant. In this mutant a sixfold increase in β-galactosidase activity under nitrogen-limiting conditions (GGln or GGlnHx) and a threefold increase in cultures grown in nitrogen excess (GNGln or GNGlnHx) were observed with respect to the parental strain KC2653 (Fig. 3). These results indicate that HpxR is autogenously regulated and that this mechanism may contribute to maintenance of basal levels of this regulatory protein.
As expected, expression of Φ(hpxO-lacZ) was not modified in the hpxR mutant (not shown). Regarding expression of Φ(hpxP-lacZ), the mutation in hpxR did not change the induction pattern under nitrogen-limiting conditions or in the presence of uric acid (Fig. 3). The lack of induction in cultures grown in the presence of hypoxanthine may be attributed to an indirect effect, since in this mutant formation of uric acid from hypoxanthine by the action of HpxDE is no longer possible.
Analysis of HpxR binding to promoter regions of the hpx system.
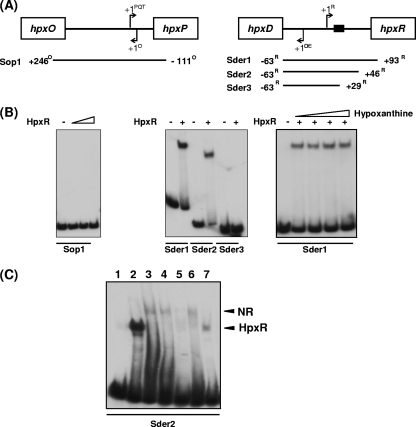
To examine the binding of HpxR to the promoter regions of the hpx gene cluster, gel shift experiments with purified HpxR were performed with fragments encompassing the intergenic region between hpxD and hpxR (Sder1) or between hpxO and hxpP (Sop1). The results presented in Fig. 4B show binding of HpxR to probe Sder1, which is consistent with the proposed role of HpxR in the regulation of hpxDE and hpxR transcription. Addition of hypoxanthine to binding reaction mixtures did not modify the number or the mobility of the complexes formed between HpxR and Sder1. In contrast, no retarded complexes were observed with probe Sop1. This is consistent with the results obtained in the expression analysis of hpxO and hpxPQT in the genetic background of an hpxR mutant and clearly eliminates a direct role of HpxR in hxpPQT control.
FIG. 4.
Binding of HpxR to promoter fragments of the hpx system. (A) Diagrams of the hpxO-hpxP and hpxD-hpxR intergenic regions showing the transcription start site for each gene. The putative HpxR binding site (TCTGC-N4-GCAAA) in the divergent hpxD and hpxR promoters is indicated by a black box. The promoter fragments used as probes (Sop1, Sder1, Sder2, and Sder3) and their end positions with respect to the +1 position of the indicated gene are shown below each intergenic region. (B) Gel shift assays performed with recombinant HpxR. The 32P-labeled Sop1 probe encompassing the hpxO-hpxP intergenic region was added to binding mixtures containing increasing amounts of HpxR (0.1, 0.4, 0.8, or 2 μg) (left panel). Sder1, Sder2, or Sder3 32P-labeled probes were added to binding mixtures containing 0.5 μg of HpxR (middle panel). Hypoxanthine from 100 to 500 μM was added to binding reaction mixtures carried out with 0.5 μg of HpxR and probe Sder1 (right panel). All mixtures contained 20 ng of the indicated probe and a 500-fold molar excess of poly(dI-dC). Reaction mixtures were incubated at 30°C for 15 min and directly subjected to polyacrylamide gel electrophoresis. (C) Gel shift assays performed with probe Sder2 and crude extracts from cells of strain KC2653 grown with different nitrogen sources. Lane 1, no protein added; lane 2, recombinant HpxR as a control; lane 3, cell extract from GNGln cultures; lane 4, cell extract from GNGlnHx cultures; lane 5, cell extract from GGln cultures; lane 6, cell extract from GGlnHx cultures; lane 7, cell extract from GHx cultures. All mixtures contained 20 ng of the indicated probe, 5 μg of the cell extract, and a 500-fold molar excess of poly(dI-dC). Reaction mixtures were incubated at 30°C for 15 min and directly subjected to polyacrylamide gel electrophoresis. Retarded complexes attributed to HpxR or to nitrogen repression conditions (NR) are indicated.
To more precisely locate the HpxR binding site in the hpxD-hpxR intergenic region, two additional probes with deletions of the intergenic region close to hpxR were obtained (Sder2 and Sder3) and used in electrophoretic mobility shift assays. A retarded complex was observed with probe Sder2 but not with probe Sder3 (Fig. 4). The sequence TCTGC-N4-GCAAA, with a partial dyad symmetry and displaying the consensus for the LysR-type regulators (T-N11-A), was identified in Sder2 (positions −54 to −66 with respect to the hpxD transcriptional start site) (Fig. 2). This location together with the fact that this consensus sequence is partially deleted in probe Sder3 gives support to its role as putative HpxR site. The sequence T-N11-A, without any dyad symmetry identified in silico and located between positions −73 to −85 with respect to hpxD (Fig. 2), does not seem to be functional, as its deletion in Sder2 did not impair HpxR binding.
Additional gel shift assays were performed with probe Sder2 and cell extracts obtained from wild-type strain KC2653 grown under different nitrogen availability conditions. In cells grown with hypoxanthine as the sole nitrogen source, only one retarded complex (HpxR), displaying the same electrophoretic mobility as the HpxR complex, was apparent (Fig. 4C, lane 7). Under conditions of nitrogen excess, a different retarded complex (NR) was observed, which may be compatible with a protein interacting with this promoter under conditions of high nitrogen availability (Fig. 4C, lanes 3 and 4). In cells grown under nitrogen-limiting conditions in the presence of hypoxanthine, faint bands corresponding to both complexes could be detected (Fig. 4C, lane 6).
DISCUSSION
At present the utilization of purine bases and their derivatives as carbon or nitrogen sources in K. pneumoniae is poorly defined. Significant discrepancies in the capacities of different isolates of the genus Klebsiella to use these compounds for growth can be found in the literature (49, 50). The great variety of diverse phenotypes observed from strain to strain has been attributed to the high frequency of horizontal transfer events within this genus (29). For instance, a 22-kb chromosomal region containing genes similar to the all regulon of E. coli (11) was present in the genomes of some K. pneumoniae strains associated with liver infection. This region, with a different G+C content, was absent from the genomes of most strains not associated with liver infection, such as MGH 78578, and confers to the pathogenic strains the ability to use allantoin as the sole source of carbon, nitrogen, and energy under both aerobic and anaerobic conditions (8).
Our results show that adenine and hypoxanthine can be used as nitrogen sources but not as carbon sources by several strains of K. pneumoniae under aerobic conditions. In contrast to what has been described for E. coli (11, 53), the assimilation of all nitrogens of these purines indicates that in K. pneumoniae the catabolic pathway is complete and proceeds past allantoin. Under anaerobic conditions these K. pneumoniae strains can grown with allantoin but fail to grow with hypoxanthine and are only able to assimilate one nitrogen from adenine. This suggests that the reactions involved in the oxidation of hypoxanthine or uric acid are impaired by the absence of oxygen.
Here we have identified and characterized the genetic system of K. pneumoniae that is responsible for the oxidation of hypoxanthine to allantoin, which is essential for nitrogen assimilation from purines under nitrogen-limiting conditions. The hpx system displays several interesting features: (i) the encoded proteins involved in the oxidation of hypoxanthine and uric acid do not display any similarity to other reported enzymes known to catalyze these reactions, and (ii) a global nitrogen control system other than Ntr is involved in the regulation of the hpxDE operon.
Oxidation of hypoxanthine to uric acid was shown to be a function of the hpxDE-encoded proteins. However, as reported by other authors (53), we were not able to detect xanthine dehydrogenase activity associated with HpxD and HpxE. In this sense, it is worth mentioning that these proteins did not display any similarity to xanthine dehydrogenases, which typically contain molybdenum cofactor (MoCo) binding domains (19). No other genes encoding MoCo-containing xanthine dehydrogenase subunits are found in the K. pneumoniae MGH78578 genome. These observations are consistent with reports that mutations in the genes encoding proteins of the MoCo synthesis pathway did not impair growth of K. pneumoniae M5a1 (now classified as Klebsiella oxytoca) with hypoxanthine as the sole nitrogen source (16). The high similarity of hpxDE-encoded proteins to oxygenases acting on aromatic compounds and the presence of Rieske domains in HpxD and of [2Fe-2S] and FAD domains in HpxE suggest that these proteins may constitute a two-component oxygenase structurally related to class IB dioxygenases, enzymes typically involved in aromatic ring hydroxylations (30). HpxD would be equivalent to the oxygenase component and HpxE to the reductase component. Oxidation of hypoxanthine to uric acid requires two sequential hydroxylation reactions, most likely catalyzed by monooxygenases. However, the distinction between dioxygenase activity and monooxygenase activity is not always absolute. For instance toluene and naphthalene dioxygenases can catalyze monooxygenase reactions, as in the oxidation of indene and indan, respectively (30). In addition, the electron transfer chain of aromatic dioxygenases is present in other oxygenases such as O-vanillate demethylase of Pseudomonas strain ATCC 19151, which catalyzes a monooxygenase reaction (30). As stated in Results, HpxD displays high similarity to one of the subunits of O-vanillate demethylase.
Likewise, the hpxO-encoded protein is essential for uric acid oxidation, although similarity was found not with known uricases but instead with FAD-dependent monooxygenases. The identification in the hpxPQT operon of genes encoding proteins similar to HIU hydrolases and OHCU decarboxylases suggests that hydroxylation of uric acid by the monooxygenase HpxO would result in a cyclic intermediate, probably HIU. Thus, the complete conversion of uric acid to allantoin would require the sequential action of HpxO, HpxQ, and HpxT. The inability to detect uricase activity in cells with hpxO expressed from a high-copy-number plasmid is probably due to the lack of HpxQ and/or HpxT function.
On the basis of amino acid sequence similarity and the induction of hpxPQT expression by uric acid, HpxP is proposed to be a permease of the nucleobase-ascorbate transporter family that may act as a xanthine and/or uric acid transporter. HpxP contains the xan_ur_permease domain present in other xanthine/uracil transporters of this family, such as UapA of Aspergillus nidulans or PbuX of Bacillus subtilis (9, 18, 23). Although disruption of hpxP did not modify growth on hypoxanthine or uric acid, a role of this protein as a purine transporter cannot be ruled out, since in a given organism, the existence of several nucleobase transporters that display wide substrate specificity is rather common (13, 14, 21). In fact, at least two genes encoding purine transporters (KPN_04013 and KPN_02555) can be identified by in silico analysis in the MGH 78578 genome.
Expression of the hpx genetic system is activated by nitrogen limitation and by the presence of specific substrates, with hpxDE and hpxPQT being the transcriptional units controlled by both signals. Repression of these operons under conditions of nitrogen excess is not relieved by the addition of hypoxanthine or uric acid. Expression of the structural gene hpxO and the regulator gene hpxR remains almost unchanged under different conditions of nitrogen availability. The low increase (less than twofold) in β-galactosidase activity in KC2653 cells bearing Φ(hpxO-lacZ) grown with hypoxanthine as a nitrogen source may be the consequence of the regulatory events operating under these conditions on the overlapping and divergent hpxP promoter.
Mutations impairing NtrC function abolish assimilation of hypoxanthine as a nitrogen source, suggesting the involvement of the Ntr system in the regulation of genes involved in this catabolic pathway. In fact, nitrogen control of hpxPQT transcription is mediated by the Ntr system, as the expression of Φ(hpxP-lacZ) is clearly reduced in cultures grown under nitrogen-limiting conditions. This is further supported by the results for the hpxP transcription initiation site, which showed that in cells grown in hypoxanthine, hpxPQT transcription seems to be dependent on σ54. Furthermore, NtrC and IHF binding sites have been identified in the promoter region in locations compatible with NtrC-mediated control, as reported for other NtrC-controlled genetic systems (6, 36, 48). The fact that hpxPQT expression was not completely turned off in the rpoN mutant suggests that another promoter may drive its transcription. In contrast, neither NtrC nor NAC is involved in the nitrogen control of the hpxDE operon, which is dependent on RNA polymerase bearing σ70 for transcription. The retarded complex observed in cell extracts obtained from cells grown under conditions of nitrogen excess suggests that an as-yet-unidentified repression mechanism may regulate hpxDE transcription under these conditions. Regulation of gene expression by means of repressors under conditions of nitrogen excess has been described for Bacillus subtilis (52) and yeast (10). Consistently, the NR retarded complex is absent in extracts of cells grown with hypoxanthine. Under these conditions, only the HpxR-DNA complex is apparent. The addition of this purine to cultures with limited nitrogen may result in a semi-excess nitrogen condition, as described elsewhere (34). In this situation, in which two nitrogen sources are present (GlnHx), the ammonia molecules liberated from hypoxanthine are added to those formed from Gln, thus increasing the intracellular ammonia levels. Under such conditions in which both NR and HpxR proteins may be present, the two retarded complexes can be observed as faint bands.
Thus, different regulators are involved in the coordinated activation of the structural hpxDE and hpxPQT operons under conditions of nitrogen limitation. Activation of these operons by the specific substrates is also mediated by different signal molecules and regulatory proteins. Our results clearly indicate that hpxPQT induction requires uric acid formation and that the regulatory protein HpxR is not involved in this mechanism. However, hpxDE expression is induced in the presence of hypoxanthine through the regulatory protein HpxR. Mobility shift experiments performed with deleted promoter fragments suggest that this LysR-type regulator binds to the TCTGC-N4-GCAAA site in the intergenic hpxD-hpxR region located upstream of the −35 promoter sequence of hpxD and downstream of the transcription initiation site of hpxR. This location is consistent with the activator role of HpxR in hpxDE transcription. When bound to this site for hpxDE activation, HpxR negatively controls its own transcription, as shown by the increased expression of Φ(hpxR-lacZ) in the hpxR mutant strain JA-K16 under all the culture conditions tested.
As in other members of the LysR family (39, 42), binding of HpxR to its recognition site does not require interaction with the effector molecule. Addition of hypoxanthine to binding reaction mixtures does not modify the pattern of the HpxR-DNA complex, suggesting that if this purine is the direct effector, it may induce a conformational change that allows interaction of HpxR with RNA polymerase or other proteins involved in the formation of the transcription complex. Alternatively, these results may indicate that induction by hypoxanthine could be attributed to an indirect effect through changes in the intracellular levels of other signal molecules. Our results rule out uric acid as the signal molecule mediating this indirect effect. Other molecules related to hypoxanthine metabolism or purine salvage could be involved, as was described for PurR from Lactococcus lactis (22). The fact that mutations in hpxR also impaired expression of Φ(hpxD-lacZ) under nitrogen-limiting conditions in the absence of hypoxanthine (GGln) strongly indicates that HpxR controls hpxDE expression under such conditions.
From these results, a model for the control of the hpxDE operon by HpxR may be derived. Under conditions of nitrogen excess, expression of hpxDE is repressed. When nitrogen is limiting, the repression is relieved, thus allowing transcription of this operon. In this situation, HpxR bound to the TCTGC-N4-GCAAA recognition site can activate hpxDE transcription to some extent. In the presence of hypoxanthine, HpxR triggers maximal transcription of the hpxDE operon, probably through a conformational change mediated by a signal molecule that facilitates interaction of HpxR with RNA polymerase or other transcriptional regulators.
Oxidation of hypoxanthine by the oxygenase HpxDE yields uric acid, which in turn induces hpxPQT expression. This regulatory mechanism operates under nitrogen limitation, conditions under which NtrC also contributes to the activation of this operon.
Supplementary Material
Acknowledgments
This research was supported by grant BFU 2007-63090/BMC from the Ministerio de Educación y Ciencia, Spain, to L.B., and by Public Health Service grant GM47156 from the National Institutes of Health to R.A.B. L.R received a predoctoral fellowship from the Generalitat de Catalunya, Spain.
We thank Miguel Regué for providing K. pneumoniae strain 52145.
Footnotes
Published ahead of print on 10 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baldauf, S. L., M. A. Cardani, and R. A. Bender. 1988. Regulation of the galactose-inducible lac operon and the histidine utilization operons in pts mutants of Klebsiella aerogenes. J. Bacteriol. 1705588-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, R. A., K. A. Janssen, A. D. Resnick, M. Bluemenberg, F. Foor, and B. Magasanik. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J. Bacteriol. 1201001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, R. A., P. M. Snyder, R. Bueno, M. Quinto, and B. Magasanik. 1983. Nitrogen regulation system of Klebsiella aerogenes: the nac gene. J. Bacteriol. 156444-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, R. A., and B. Friederich. 1990. Regulation of assimilatory nitrate reductase formation in Klebsiella aerogenes W70. J. Bacteriol. 1727256-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmona, M., F. Calverie-Martín, and B. Magasanik. 1997. DNA bending and the initiation of transcription at sigma54-dependent bacterial promoters. Proc. Natl. Acad. Sci. USA 949568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cendron, L., R. Berni, C. Folli, I. Ramazzina, R. Percudani, and G. Zanotti. 2007. The structure of 2-oxo-4-hydroxi-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation. J. Biol. Chem. 28218182-18189. [DOI] [PubMed] [Google Scholar]
- 8.Chou, H. C., C. Z. Lee, L. C. Ma, C. T. Fang, S. C. Chang, and J. T. Wang. 2004. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 723783-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen, L. C., S. Schou, P. Nygaard, and H. H. Saxild. 1997. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 1792540-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman, J. A., R. Rai, D. M. Loprete, T. Cunningham, V. Svetlov, and T. G. Cooper. 1997. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J. Bacteriol. 1793416-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 1817479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diallinas, G., L. Gorfinkiel, H. N. Arst, G. Cecchetto, and C. Scazzocchio. 1995. Genetic and molecular characterization of a gene encoding a wide specifity purine permease of Aspergillus nidulans reveals a novel family of transporters conserved in prokaryotes and eukaryotes. J. Biol. Chem. 2708610-8622. [DOI] [PubMed] [Google Scholar]
- 14.Diallinas, G., and C. A. Scazzocchio. 1989. A gene coding for the uric acid-xanthine permease of Aspergillus nidulans: inactivational cloning, characterization and sequence of a cis-acting mutation. Genetics 122341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, J., T. J. Goss, R. A. Bender, and A. J. Ninfa. 1995. Repression of the Klebsiella aerogenes nac promoter. J. Bacteriol. 1775535-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzón, A., J. Li, A. Flores, J. Casadesús, and V. Stewart. 1992. Molybdenum cofactor (chlorat resistant) mutants of Klebsiella pneumoniae M5a1 can use hypoxanthine as the sole nitrogen source. J. Bacteriol. 1746298-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg, R. B., R. A. Bender, and S. L. Streicher. 1974. Direct selection for P1-sensitive mutants of enteric bacteria. J. Bacteriol. 118810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorfinkiel, L., G. Diallinas, and C. Scazzochio. 1993. Sequence and regulation of the uapA gene encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans. J. Biol. Chem. 26823376-23381. [PubMed] [Google Scholar]
- 19.Hille, R. 2005. Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433107-116. [DOI] [PubMed] [Google Scholar]
- 20.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karatza, P., and S. Frillingos. 2005. Cloning and functional characterization of two bacterial members of the NAT/NCS2 family of Escherichia coli. Mol. Membr. Biol. 22251-261. [DOI] [PubMed] [Google Scholar]
- 22.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 1803907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koning, H., and G. Diallinas. 2000. Nucleobase transporters. Mol. Membr. Biol. 7575-94. [DOI] [PubMed] [Google Scholar]
- 24.Lee, Y., D. H. Lee, C. W. Kho, A. Y. Lee, M. Jang, S. Cho, C. H. Lee, J. S. Lee, P. K. Myung, B. C. Park, and S. G. Park. 2005. Transthyretin-related proteins functions to facilitate the hydrolysis of 5-hydroxyisourate, the end product of the uricase reaction. FEBS Lett. 5794769-4774. [DOI] [PubMed] [Google Scholar]
- 25.Lin, E. C. C. 1976. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30535-579. [DOI] [PubMed] [Google Scholar]
- 26.Liu, Q., and R. A. Bender. 2007. Complex regulation of urease formation from the two promoters of the ure operon of Klebsiella pneumoniae. J. Bacteriol. 1897593-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-273. [PubMed] [Google Scholar]
- 28.Macaluso, A., E. A. Best, and R. A. Bender. 1990. Role of nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J. Bacteriol. 1727249-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez, J., L. Martínez, M. Rosenblueth, J. Silva, and E. Martínez- Romero. 2004. How are sequence analyses modifying bacterial taxonomy? The case of Klebsiella. Int. Microbiol. 7261-268. [PubMed] [Google Scholar]
- 30.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46277-305. [DOI] [PubMed] [Google Scholar]
- 31.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Nassif, X., J. M. Fournier, J. Arondel, and P. J. Sansonetti. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nygaard, P., S. M. Bested, K. A. K. Andersen, and H. H. Saxils. 2000. Bacillus subtilis guanine deaminase is encoded by the yknA gene and is induced during growth with purines as the nitrogen source. Microbiology 1463061-3069. [DOI] [PubMed] [Google Scholar]
- 35.Nunoshiba, T., E. Hidalgo, C. F. Amábile-Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1746054-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter, S. C., A. K. North, A. B. Wedel, and S. Kustu. 1993. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 72258-2273. [DOI] [PubMed] [Google Scholar]
- 37.Ramazzina, I., C. Folli, A. Scchi, R. Berni, and R. Percudani. 2006. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat. Chem. Biol. 2144-148. [DOI] [PubMed] [Google Scholar]
- 38.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57155-176. [DOI] [PubMed] [Google Scholar]
- 39.Rintoul, M. R., E. Cusa, L. Baldoma, J. Badia, L. Reitzer, and J. Aguilar. 2002. Regulation of the Escherichia coli allantoin regulon: coordinated function of the repressor AllR and activator AllS. J. Mol. Biol. 324599-610. [DOI] [PubMed] [Google Scholar]
- 40.Rouf, M. A., and R. F. Lomprey. 1968. Degradation of uric acid by certain aerobic bacteria. J. Bacteriol. 96617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Rusell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 42.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 43.Schultz, A. C., P. Nygaard, and H. H. Saxild. 2001. Functional analysis of 14 genes that consitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J. Bacteriol. 1833293-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 1752116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 46.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 16947-52. [DOI] [PubMed] [Google Scholar]
- 47.Smith, G. R., S. H. Yeheskel, and B. Magasanik. 1971. Genetic and metabolic control of enzymes responsible for histidine degradation in Salmonella typhimurium. J. Biol. Chem. 2463320-3329. [PubMed] [Google Scholar]
- 48.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 1851757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyler, B. 1978. Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 471127-1162. [DOI] [PubMed] [Google Scholar]
- 50.Vogels, G. D., and C. Van der Drift. 1976. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40403-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, S. H. 1988. The use of purines compounds as sole sources of carbon and nitrogen by Klebsiella species. Microbios 5657-62. [PubMed] [Google Scholar]
- 52.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors, including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 1795494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xi, H., B. L. Schneider, and L. Reitzer. 2000. Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J. Bacteriol. 1825332-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo, H. S., F. S. Genbauffe, and T. G. Cooper. 1985. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol. Cell. Biol. 52279-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.