Abstract
In previous work (E. E. Smith, D. G. Buckley, Z. Wu, C. Saenphimmachack, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson, Proc. Natl. Acad. Sci. USA 103:8487-8492, 2006) it was shown that Pseudomonas aeruginosa undergoes intense genetic adaptation during chronic respiratory infection (CRI) in cystic fibrosis (CF) patients. We used the same collection of isolates to explore the role of hypermutation in this process, since one of the hallmarks of CRI is the high prevalence of DNA mismatch repair (MMR) system-deficient mutator strains. The presence of mutations in 34 genes (many of them positively linked to adaptation in CF patients) in the study collection of 90 P. aeruginosa isolates obtained longitudinally from 29 CF patients was not homogeneous; on the contrary, mutations were significantly concentrated in the mutator lineages, which represented 17% of the isolates (87% MMR deficient). While sequential nonmutator lineages acquired a median of only 0.25 mutation per year of infection, mutator lineages accumulated more than 3 mutations per year. On the whole-genome scale, data for the first fully sequenced late CF isolate, which was also shown to be an MMR-deficient mutator, also support these findings. Moreover, for the first time the predicted amplification of mutator populations due to hitchhiking with adaptive mutations in the course of natural human infections is clearly documented. Interestingly, increased accumulation of mutations in mutator lineages was not a consequence of overrepresentation of mutations in genes involved in antimicrobial resistance, the only adaptive trait linked so far to hypermutation in CF patients, demonstrating that hypermutation also plays a major role in P. aeruginosa genome evolution and adaptation during CRI.
Chronic respiratory infection (CRI) with Pseudomonas aeruginosa is the main driver of morbidity and mortality in cystic fibrosis (CF) patients (4, 15). The establishment of P. aeruginosa CRI is mediated by a complex adaptive process that includes physiological changes produced by the activation of specific regulatory pathways, including the induction of the biofilm mode of growth or the differential expression of virulence genes (39), and genetic changes leading to selection of an important number of adaptive mutations required for long-term persistence (19, 27, 34).
Although CF patients with CRI are generally infected by a single P. aeruginosa strain that in most cases persists throughout the patient's life (32), one of the hallmarks of such infections is the emergence and fixation over time of multiple phenotypic variants of the underlying clonal populations (24), a process known as adaptive radiation (30). Many of the selected phenotypes have been clearly linked to adaptation to the lung environment that favors lifelong persistence of CRI (27). Indeed, once the adaptation stage is reached and the CRI is fully established, eradication is generally no longer possible. At this point, with resignation, the therapeutic goals change from attempting to cure the infection to slowing the decline of lung function and improving the patient's quality of life.
The intense genetic adaptation process that takes place during the establishment of CRI has recently been quantified (34). Whole-genome sequencing of early (6 months of age) and late (96 months of age) P. aeruginosa isolates from a CF patient revealed that up to 68 mutations were acquired during establishment of the CRI. A clear signal for positive selection was demonstrated, since an extraordinarily high ratio of nonsynonymous mutations per site to synonymous mutations per site was documented. Many of the mutations detected led to loss of function of the affected genes, which were frequently involved in bacterial virulence. The work was completed by analysis of a collection of sequential P. aeruginosa isolates from 29 additional CF patients; 24 of the genes that had been mutated early in patient 1 and 10 additional genes that had been shown to be candidates for mutation in many CF infections were sequenced. Again, the signal for positive selection of mutations in these genes was overwhelming, since only 5 synonymous mutations and 103 nonsynonymous mutations were found.
One of the hallmarks of CRI with P. aeruginosa is the high prevalence of DNA mismatch repair (MMR) system-deficient hypermutable (or mutator) strains, which are detected in 30 to 60% of the patients, in contrast to acute infections, for which a prevalence of less than 1% has been reported (1, 6, 16, 21, 24, 25). While the presence of mutator strains has been found to be linked to the high rates of antibiotic resistance of P. aeruginosa clinical isolates recovered from patients with CRI (1, 7, 16, 24), the role of these strains in genetic adaptation to the CRI setting remains uncertain. Furthermore, despite the fact that the effect of hypermutation on genetic adaptation and bacterial evolution has been explored for a number of pathogens using in vivo models of colonization or infection (5, 12, 20, 23), there is still little information for natural human infectious diseases. Therefore, the goal of this work was to explore the contribution of hypermutation to the genetic adaptation of P. aeruginosa to the airways of CF patients, using the collection of clinical strains described above that were analyzed previously by Smith et al. (34).
MATERIALS AND METHODS
Bacterial strains.
Laboratory strains and plasmids used or constructed in this work are listed in Table 1. The collection of P. aeruginosa CF isolates used was the collection previously analyzed by Smith et al. (34). This collection includes 35 longitudinal isolates from one CF patient (patient 1). The complete genomes (6.5 Mb) of the earliest available isolate (isolate 1, collected at 6 months of age) and the latest available isolate (isolate 35, collected at 96 months of age) have been sequenced, and the mutations have been detected and traced by PCR-based genotyping in the remaining intermediate isolates (34). The collection also included 90 P. aeruginosa clinical isolates obtained longitudinally from respiratory samples from 29 additional CF patients (patients 2 to 30). There were at least two clonal isolates from each patient; the earliest isolate was collected from each patient before he or she was 11 years old, and the latest isolate was collected >5 years after the recovery of the earliest isolate. The isolates were designated by using a number for identification of a patient (2 to 30), followed by the age of the patient (in years) when the isolate was collected. For this set of 90 CF isolates a total of 34 genes and their regulatory regions have been fully sequenced (34). The 34 genes sequenced include 24 of the genes that were found to be mutated early in the isolates from patient 1 and 10 additional genes that have previously been shown to be candidates for mutation in many CF infections. The list of sequenced genes includes mexZ (PA2020), lasR (PA1430), PA0313, mexA (PA0425), accC (PA4848), vfr (PA0652), mexS (PA2491), exsA (PA1713), PA0506, wspF (PA3703), rpoN (PA4462), fleQ (PA1097), mexT (PA2492), nalD (PA3574), ampD (PA4522), PA0366, cyaB (PA3217), P1-001 (in a 45-kb genomic island), PA3817, pilB (PA4526), PA1333, PA3565, PA2312, anr (PA1544), rhlR (PA3476), phoP (PA1179), rhlI (PA3476), PA4796, pqsB (PA0997), toxR (PA0707), PA2435, PA4420, ORF C/SG114, and PA2121. For each isolate, we recorded the total number of mutations for the 34 genes. We also recorded the number of new mutations that accumulated per year of infection in the sequential isolates, which was determined by dividing the number of mutations detected in an isolate that were not present in the preceding isogenic isolate by the time period (in years) between the collection times for the two isolates.
TABLE 1.
Strains, plasmids, and primers used in this work
| Strain, plasmid, or primer | Genotype, relevant characteristics, or sequence (5′-3′) | Reference, source, or use |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Laboratory strain, completely sequenced | 36 |
| PAOMS | PAO1 ΔmucS::lox | Submitted for publication |
| PAOMA | PAO1 ΔmutA::lox | Submitted for publication |
| PAOMSA | PAO1 ΔmutS::lox ΔmucA::lox | Submitted for publication |
| E. coli XL-1 Blue | F′::Tn10 proAB lacIq Δ(lacZ)M15 recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK−) mcrB1 | Laboratory collection |
| Plasmids | ||
| pUCP24 | Gmr; pUC18-based Escherichia-Pseudomonas shuttle vector | 41 |
| pJMML | mutL from PAO1 cloned in pJMS6αlac | 25 |
| pUCPMS | mutS from PAO1 cloned in pUCP24 | 26 |
| pUCPML | mutL from PAO1 cloned in pUCP24 | This study |
| Primers | ||
| mutSF1 | TTAACATTACCCTCTTTTGCAC (−50 to −29)a | Amplification and sequencing of mutS |
| mutSR1 | TCATTTTCTAGTTCTCTCCTCA (2637 to 2616) | Amplification and sequencing of mutS |
| mutSF4 | CGCTCCGCTCCAGGACAGCGC (1611 to 1633) | Sequencing of mutS |
| mutSF5 | CGGCTGCCTGCTCGCCTAC (701 to 719) | Sequencing of mutS |
| mutSF6 | CATTCGGCGGAGGGCTACCTG (210 to 230) | Sequencing of mutS |
| mutSR6 | TGGCGGTTTCGCTCATCTCCAC (2031 to 2009) | Sequencing of mutS |
| mutSF11 | TTCCTGATGGACCTGGAAGCG (1331 to 1350) | Sequencing of mutS |
| mutLF | CGATGATCGCCCAGCGCT (−124 to −107) | Amplification and sequencing of mutL |
| mutLR | TCCGCCGGGTCGCGGATA (2175 to 2158) | Amplification and sequencing of mutL |
| mutLF2 | TAGCGCGCCTGACCATGA (330 to 347) | Sequencing of mutL |
| mutLF3 | GCGCATGGTGCGCGACAA (797 to 814) | Sequencing of mutL |
| mutLF4 | GCCTCCGGCGGCTCCTCCG (1178 to 1197) | Sequencing of mutL |
| mutLR2 | GCAGGTCGGCGATGACAT (1680 to 1663) | Sequencing of mutL |
The numbers in parentheses are positions based on the mutS or mutL start codons.
Determination of mutation frequencies.
The frequencies of mutation to rifampin (300 μg/ml) resistance and/or streptomycin (1,000 μg/ml) resistance were determined by using a modified protocol based on protocols described previously (24, 25). For each strain, independent aliquots containing approximately 103 cells were inoculated into five flasks containing 10 ml of Mueller-Hinton broth and incubated at 37°C and 180 rpm for 16 to 18 h. Cultures were then collected by centrifugation and resuspended in 1 ml of a saline solution, and serial 1:10 dilutions were plated on Mueller-Hinton agar plates plates and Mueller-Hinton agar plates supplemented with 300 μg/ml of rifampin and/or 1,000 μg/ml of streptomycin. Mutation frequencies were then calculated by dividing the median numbers of mutants by the median numbers of total cells. The breakpoint used to define hypermutable strains was a frequency of mutation to rifampin resistance of >2 × 10−7, as recommended previously (24). Additionally, as a control, frequencies of mutation to rifampin and streptomycin resistance were determined with reference strain PAO1, as well as mutant derivatives with mutations in mucA (PAOMA), mutS (PAOMS), and mucA mutS (PAOMSA), which were constructed in another study (submitted for publication). At least three independent mutation frequency experiments (each with five cultures) were performed for the control strains, and the results were expressed as means ± standard deviations.
Complementation of mutator strains with PAO1 wild-type MMR genes.
To explore the genetic basis for the mutator phenotypes, complementation studies were performed with all hypermutable strains. Plasmid pUCPMS harboring PAO1 wild-type mutS, plasmid pUCPML harboring PAO1 wild-type mutL, and plasmid pUCP24, a control cloning vector, were electroporated into the hypermutable isolates as previously described (33). The MICs of gentamicin (a resistance marker for these plasmids) were determined by a conventional broth microdilution method for the hypermutable isolates before electroporation, and depending on the level of resistance, transformants were selected on Luria-Bertani agar plates containing 50 or 250 μg/ml of the antibiotic.
Complementation was demonstrated by reversion of the increased rate of mutation to rifampin or streptomycin resistance in two independent transformant colonies for each strain. Complementation was considered positive if the mutation frequency after pUCPMS or pUCPML transformation decreased ≥2 logs, negative if the mutation frequency decreased <1 log, and partial if the reduction was at least 1 log but was not as high as the wild-type level.
To construct plasmid pUCPML, the PAO1 wild-type mutL gene was obtained from plasmid pJMML by BamHI restriction and was ligated to plasmid pUCP24 opened with the same enzyme. The ligation product was then transformed into Escherichia coli XL-1 Blue made competent by CaCl2, and the resulting transformants were selected on MacConkey agar plates containing 5 μg/ml of gentamicin and confirmed by PCR and sequencing.
PCR amplification and sequencing of MMR genes.
The mutS and mutL genes from all the mutator strains were PCR amplified using primer pairs mutSF1-mutSR1 and mutLF-mutLR, respectively (Table 1). After purification (QIAquick PCR purification kit; Qiagen, Germany), the PCR products were fully sequenced. A BigDye Terminator kit (PE-Applied Biosystems, United States) was used for the sequencing reactions, which were analyzed with an ABI Prism 3100 DNA sequencer (PE-Applied Biosystems, United States). Additionally, the mutS and/or mutL alleles of the nonmutator isolates that preceded and followed the mutator isolates were PCR amplified and sequenced for all patients. In all cases, the mutations detected were confirmed by sequencing a new PCR product. The sequences were compared with the sequences of PAO1 using the BLAST computer program (http://www.ncbi.nlm.nih.gov/BLAST).
Molecular strain typing.
The epidemiological relatedness of the sequential isolates obtained from the patients infected by hypermutable strains was studied by using pulsed-field gel electrophoresis (PFGE). Bacterial DNA embedded in agarose plugs was digested with SpeI. DNA was separated in a contour-clamped homogeneous electric field DRIII apparatus (Bio-Rad, La Jolla, CA) under the following conditions: 6 V/cm2 for 26 h with pulse times of 5 to 40 s. DNA macrorestriction patterns were interpreted by using the criteria established by Tenover et al. (38).
Statistical analysis.
A statistical analysis was conducted using Fischer's exact test, Student's t test, or the Mann-Whitney U test, as appropriate. A P value of <0.05 was considered significant.
RESULTS
Prevalence and nature of CF mutator strains.
In this work we analyzed the collection of P. aeruginosa CF isolates previously characterized by Smith et al. (34). This collection includes 35 longitudinal isolates from one CF patient (patient 1). The complete genomes (6.5 Mb) of the earliest available isolate (isolate 1, collected at 6 months of age) and the latest available isolate (isolate 35, collected at 96 months of age) were sequenced previously, and the mutations in the remaining intermediate isolates were detected and traced by PCR-based genotyping (34). In that work it was shown that isolate 35 (96-month isolate) had a nonsynonymous mutation in mutS (R490L), but hypermutability was not assessed because this strain was rifampin resistant. In order to determine whether this fully sequenced late CF isolate was indeed a mutator, we determined the frequencies of mutation to streptomycin resistance for isolates 1 and 35 and compared the results with the results for the control strains. As shown in Table 2, isolate 35 showed a 2-log-higher mutation frequency than isolate 1. Moreover, the frequencies of mutation to streptomycin resistance for isolates 1 and 35 were very similar to those for the wild-type and mutS-deficient control strains, respectively (Table 2). Furthermore, the increased mutation frequency was restored by complementation with the plasmid harboring the wild-type mutS gene, demonstrating that the first fully sequenced late CF isolate was indeed a mutator and also that the R490L substitution, affecting DNA binding domain IV of MutS (13), results in a nonfunctional MutS protein. As expected, isolates 32 to 34, which also harbored the R490L substitution, showed increased mutation frequencies, whereas the latest isolate that did not harbor the mutation (isolate 31, 60-month isolate) showed wild-type mutation frequencies (not shown).
TABLE 2.
Frequencies of mutation to streptomycin resistance and complementation with pUCPMS for isolates from patient 1
| Strain | Frequency of mutation to streptomycin (1,000 μg/ml) resistance | Complementation with pUCPMSa |
|---|---|---|
| PAO1 | 9.1 × 10−10 ± 2.6 × 10−10 | |
| PAOMAb | 5.6 × 10−10 ± 0.6 × 10−10 | |
| PAOMSb | 1.2 × 10−7 ± 0.7 × 10−7 | + |
| PAOMSAb | 2 × 10−7 ± 0.2 × 10−7 | + |
| 1-0.5 (6 mo) | 9.8 × 10−10 | |
| 1-8 (96 mo) | 7.6 × 10−8 | + |
Complementation was considered positive when the mutation frequency after transformation of pUCPMS was decreased ≥2 logs.
PAOMA is a mucA mutant of PAO1, PAOMS is a mutS mutant of PAO1, and PAOMSA is a mutS mucA mutant of PAO1.
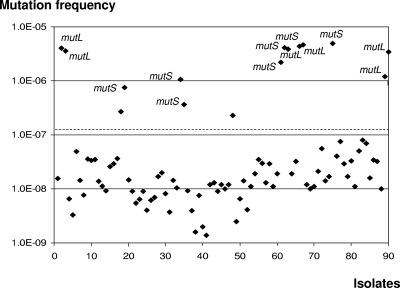
We then continued to characterize the collection of Smith et al. (34), which also included 90 P. aeruginosa clinical isolates obtained longitudinally from sputum samples from 29 additional CF patients (patients 2 to 30). There were at least two clonal isolates from each patient; the earliest isolate from each patient was collected before he or she was 11 years old, and the latest isolate was collected >5 years after the recovery of the earliest isolate. For this set of 90 CF isolates a total of 34 genes and their regulatory regions were fully sequenced by Smith et al. (34). Because all the isolates showed wild-type rifampin susceptibility, the frequencies of mutation to rifampin resistance were assessed for all 90 isolates. As shown in Fig. 1, the analysis of the mutation frequencies revealed that there were 15 (16.7%) hypermutable isolates, based on the defined breakpoint (>2 × 10−7). Mutator isolates were detected for 9 of the 29 patients (31.0%); for 2 of these patients (6.9%) hypermutable strains were detected among the earliest isolates at the onset of lung infection, whereas for the 7 remaining patients (24.1%) mutators emerged during the course of the CRI.
FIG. 1.
Distribution of the frequencies of mutation to rifampin resistance in the 90 P. aeruginosa isolates from CF patients 2 to 30. mutS- or mutL-deficient isolates are indicated. The dashed line indicates the mutation frequency breakpoint used to identify mutator strains (>2 × 10−7).
The genetic relatedness of the sequential isolates was explored by PFGE. In all seven cases in which mutators emerged during the course of the chronic infection, the mutator isolates and the corresponding preceding nonmutator isolates showed the same macrorestriction pattern. For one additional patient (patient 8) the first isolate and the last isolate had mutator phenotypes and produced the same PFGE pattern. Finally, for the single patient in whom nonmutator isolates followed a mutator, the same PFGE pattern was also found. These results suggest that the transition from the nonmutator phenotype to the mutator phenotype (and the other way around) was a consequence of mutational events and not of strain replacement.
The genetic basis for the hypermutability of all the P. aeruginosa mutator CF isolates was investigated by performing complementation studies and by sequencing the MMR system genes. The main results are shown in Table 3, and the complete allelic profiles of MMR system genes are shown in Table S1 in the supplemental material. Of the 15 hypermutable strains, 13 (86.6%) were found to be defective in either mutS (7) or mutL (6). As shown in Table 3, in most cases (11 of the 13 strains), the MMR system-deficient mutator isolates contained directly inactivating mutations (frameshifts or stop codons) in mutS or mutL. As expected, in all cases, sequencing of the nonmutator isolates that preceded or followed the mutator isolates revealed the presence of identical mutS or mutL alleles except for the inactivating mutation. In the two remaining MMR-deficient isolates (22-20 and 21-19.5), missense mutations were responsible for the mutator phenotypes (Table 3). The mutS-deficient isolate 22-20 contained two nonsynonymous substitutions in mutS, A187S and R306C, although the former was also detected in the preceding nonhypermutable isogenic isolates. These results show that the previously undescribed R306C substitution, involving a highly conserved residue located in DNA binding domain IV (13), must be primarily responsible for the inactivation of MutS in this strain. The A187S substitution was also found in the preceding nonhypermutable isogenic isolates of mutS-deficient (4-bp insertion) isolate 13-19 (Table 3). Nevertheless, this polymorphism is apparently frequent in mutator isolates in several other different collections (8, 11, 21). This suggests that it may have a potential effect on MutS activity that should be explored further. The results for patient 21 are particularly interesting. Early nonhypermutable isolates (21-0.5 and 21-10.7) were followed by two sequential mutS-deficient isolates (21-14.3 and 21-15.2) and a final mutL-deficient isolate (21-19.5) (Table 3). All the isolates were found to be isogenic by PFGE, suggesting that there was independent emergence of two different mutator lineages during the chronic infection. Intriguingly, isolate 21-19.5 harbored two missense mutations in mutL, neither of which was present in the preceding isolates, despite the fact that the allelic profiles were otherwise conserved (see Table S1 in the supplemental material). It seems unlikely that acquisition of both missense mutations is required for the inactivation of MutL, since there are hundreds of possible mutations leading to one-step gene inactivation. A more likely explanation is that one of the mutations leading to MutL inactivation was selected first and that the second mutation was selected afterwards in the mutator background as a compensatory mutation to restore a potential deleterious effect of the first mutation. The emergence of two independent mutator genotypes apparently also occurred for patient 8, in which an early weak mutator (not MMR deficient) was followed by a mutS-deficient isogenic lineage (Table 3).
TABLE 3.
Results for mutation frequencies, complementation studies, and sequencing of MMR system genes in the CF mutator isolates
| Isolate | Frequency of mutation to rifampin (300 μg/ml) resistance | Complementationa
|
Sequencing of MMR system genesb | |
|---|---|---|---|---|
| mutS (pUCPMS) | mutL (pUCPML) | |||
| PAO1 | 2.4 × 10−8 ± 1.3 × 10−8 | |||
| PAOMSc | 4.0 × 10−6 ± 1.2 × 10−6 | + | ||
| PAOMAc | 1.1 × 10−8 ± 0.2 × 10−8 | |||
| PAOMSAc | 3.1 × 10−6 ± 1.0 × 10−6 | + | ||
| 2-12.8d | 4.1 × 10−6 | − | + | mutL, 2-bp insertion (CG) after nucleotide 1822 |
| 2-13 | 3.6 × 10−6 | − | + | mutL, 2-bp insertion (CG) after nucleotide 1822 |
| 8-1.1 | 2.7 × 10−7 | − | − | MutS, wild type; MutL, G427S |
| 8-13.2A | 7.6 × 10−7 | ND | ND | mutS, 4-bp deletion (CGAG) at nucleotide 1761; MutL, G427S |
| 13-19.6A | 1.1 × 10−6 | + | − | MutS, A187S; mutS, 4-bp insertion (CGCC) after nucleotide 1600 |
| 13-19.6B | 3.7 × 10−7 | + | − | MutS, A187S; mutS, 4-bp insertion (CGCC) after nucleotide 1600 |
| 17-22.2 | 2.3 × 10−7 | − | − | mutS, wild type; mutL, wild type |
| 21-14.3 | 2.2 × 10−6 | + | − | mutS, 1-bp deletion (T) at nucleotide 2437; mutL, wild type |
| 21-15.2 | 4.1 × 10−6 | + | − | mutS, 1-bp deletion (T) at nucleotide 2437; mutL, wild type |
| 21-19.5 | 3.9 × 10−6 | − | + | mutS, wild type; MutL, T118A, A124V |
| 22-20 | 4.4 × 10−6 | + | − | MutS, A187S, R306C |
| 23-6.2 | 4.6 × 10−6 | − | + | MutL, Q492X |
| 25-21 | 4.9 × 10−6 | + | − | mutS, 1-bp deletion (A) at nucleotide 2428 |
| 30-16.8 | 1.2 × 10−6 | − | + | mutL, 1-bp insertion (C) after nucleotide 100 |
| 30-18.5 | 3.4 × 10−6 | − | + | mutL, 1-bp insertion (C) after nucleotide 100 |
Complementation was considered positive when the mutation frequency after transformation of pUCPMS or pUCPML was decreased ≥2 logs, negative when the mutation frequency was decreased <1 log, and partial if the reduction was 1 log. In one case it was not possible to perform complementation (ND) due to the high level of gentamicin resistance of the clinical strain.
Inactivating mutations are indicated by bold type.
PAOMA is a mucA mutant of PAO1, PAOMS is a mutS mutant of PAO1, and PAOMSA is a mutS mucA mutant of PAO1.
Patient number-age (in years) at the time of isolation.
Hypermutation and genetic adaptation to the airways of CF patients.
In order to analyze the effect of hypermutation during the process of genetic adaptation to the airways of CF patients, we recorded the mutations that accumulated in mutator and nonmutator isolates. Initially, we analyzed the data for patient 1. A total of 68 mutations were detected in the latest isolate (isolate 35, 96 month of age) that were not present in the earliest isolate (isolate 1, 6 months of age) by whole-genome (6.5 Mb) sequencing, and only 8 of these mutations were synonymous mutations (34). Interesting, the vast majority (42 of 68) of the detected mutations were present only in the mutator isolates (isolates 32 to 35) selected during the last 36 months of the 90-month period evaluated. On the other hand, the last nonhypermutable isolate (isolate 31, 60 months of age) had accumulated only 26 of the 68 final mutations in a longer period (54 months, from 6 to 60 months of age). According to these data, the established rate of accumulation of mutations per year of infection was notably higher in the period in which mutators were selected (14.0 mutations per year) than in the preceding period in which only nonmutators were present (5.7 mutations per year).
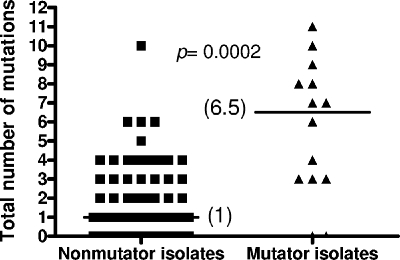
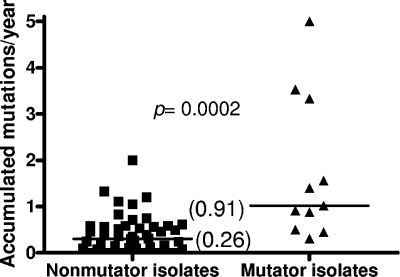
We then analyzed the collection of 90 clinical isolates obtained longitudinally from respiratory samples from 29 additional CF patients (patients 2 to 30) in which 34 genes and their regulatory regions had been fully sequenced (34). We recorded the total number of mutations in the 34 genes, as well as the number of new mutations accumulated per year of infection in the sequential isolates, for each mutator and nonmutator strain. As shown in Fig. 2, there were dramatic differences in the median numbers of all mutations in the 34 genes; nonmutator isolates had a median of only 1 mutation, whereas the median number of mutations for mutators was 6.5 (P = 0.0002). Similarly, the proportion of mutators was dramatically increased for CF isolates containing a growing number of total mutations (Fig. 3). For instance, the proportion of mutators increased from 16.7% when all isolates were considered to 87.5% when only isolates containing at least seven mutations were considered (Fig. 3). Obviously, the total number of mutations in a particular isolate is expected to be influenced by the time frame of the chronic infection (late isolates should have more mutations than early isolates), and therefore the number of mutations should be higher in mutator isolates because they are overrepresented in late isolates. For instance, the mean age of patients at the time of isolation of mutator isolates (15.5 ± 5.8 years) was significantly higher (P = 0.009) than the mean age of patients at the time of isolation of nonmutator isolates (11.3 ± 5.5 years). To normalize the time frame of chronic infection, we analyzed the number of new mutations that accumulated per year of infection for the sequential isolates. As shown in Fig. 4, notable differences were observed in the median numbers of mutations in the 34 genes; nonmutator isolates had a median of only 0.26 mutation per year of infection, whereas the median value for mutators was 0.91 mutation per year (P = 0.0002). Furthermore, these data are expected to underestimate the real rate of accumulation of new mutations in the mutator isolates, since in an important proportion of the cases these isolates were preceded by a nonmutator isolate. Therefore, in the time between the times of acquisition of the isolates there had to be a period when the isolate was nonmutator and a period when the isolate was a mutator. Consistent with this observation, the number of new mutations that accumulated per year of infection by mutators whose preceding isolate was also a mutator (3.33 new mutations per year) was significantly (P = 0.04) higher than the number of new mutations that accumulated per year of infection by the mutator isolates whose preceding isolate was a nonmutator (0.81 new mutation per year). Using the same reasoning, nonmutator isolates whose preceding isolate was a mutator should also have had a higher rate of accumulation of new mutations; indeed, the median rate for these isolates was 0.90 new mutation per year, in contrast to the rate for nonmutator isolates preceded by nonmutator isolates (0.25 new mutation per year).
FIG. 2.
Total numbers of mutations (in the 34 sequenced genes) detected in mutator and nonmutator isolates from CF patients 2 to 30.
FIG. 3.
Percentages of mutators among P. aeruginosa CF isolates containing different numbers of mutations in the 34 sequenced genes.
FIG. 4.
Numbers of new mutations (in the 34 sequenced genes) that accumulated per year of infection in the sequential mutator and nonmutator isolates from CF patients 2 to 30.
Overall, these results showed that mutators dramatically enhance the process of genetic adaptation to the airways of CF patients. Subsequently, we explored whether, in addition to quantitative differences in the accumulation of new mutations, there were also qualitative differences between mutator and nonmutator isolates in the types of new mutations accumulated and in the genes affected. Table 4 shows the distribution of the types of new mutations detected in mutator and nonmutator isolates. Overall, the observed differences were not statistically significant, although nonmutator isolates proportionally tended to have more base substitutions (particularly nonsense mutations and single-nucleotide polymorphisms in intergenic regions) and mutator isolates tended to have more insertions and deletions, specifically due to the contribution of frameshift mutations produced by short (1- to 4-bp) insertions or deletions, as would be expected for MMR-system deficient strains (10).
TABLE 4.
Distribution of the types of new mutations in mutator and nonmutator isolates
| Mutation type | No. of new mutations (%)
|
|
|---|---|---|
| Mutator isolates (n = 15) | Nonmutator isolates (n = 75) | |
| All new mutations | 66 (100) | 102 (100) |
| Base substitutions | 42 (63.6) | 69 (67.6) |
| Synonymous | 1 (1.5) | 1 (1.0) |
| Missense | 37 (56.1) | 52 (51.0) |
| Nonsense | 3 (4.5) | 11 (10.8) |
| Single-nucleotide polymorphisms (intergenic) | 1 (1.5) | 5 (4.9) |
| Insertions or deletions | 24 (36.4) | 33 (32.3) |
| 1- to 4-bp insertions or deletions | 14 (21.2) | 16 (15.7) |
| >4-bp insertions or deletions | 10 (15.2) | 17 (16.7) |
Table 5 shows the distribution of the new mutations among the sequenced genes in mutator and nonmutator isolates. Since a clear link between hypermutation and development of antimicrobial resistance has been well established in previous studies (1, 7, 16, 24), we examined whether the increased numbers of mutated genes in mutator isolates were a consequence of overrepresentation of mutations in genes involved in antimicrobial resistance. As shown in Table 5, the proportion of mutations that affect genes involved in antimicrobial resistance for mutator isolates (35.4%) was not higher than the proportion for nonmutator isolates (38.6%). Furthermore, significant differences were not observed for any of the genes studied, demonstrating that hypermutation has a generalized effect on genetic adaptation not linked to a specific adaptive trait. For instance, of the two genes showing a higher number of mutations in CF isolates, one (mexZ) is involved in antibiotic resistance and the other (lasR) is involved in quorum sensing regulation and virulence (34). As shown in Table 5, mutations in these two genes were increased equally in mutator isolates (0.60 new mutation per mutator isolate versus 0.27 mutations per nonmutator isolate [P = 0.015] for mexZ and 0.53 new mutation per mutator isolate versus 0.19 mutation per nonmutator isolate [P = 0.008] for lasR).
TABLE 5.
Distribution of the new mutations among the sequenced genes in mutator and nonmutator isolates
| Mutations or genea | Mutator isolates (n = 15)
|
Nonmutator isolates (n = 75)
|
||
|---|---|---|---|---|
| No. of mutations (%) | No. of mutations per isolate | No. of mutations (%) | No. of mutations per isolate | |
| All new mutations | 65 (100) | 4.3 | 101 (100) | 1.3 |
| All new mutations in antibiotic resistance genes | 23 (35.4) | 1.5 | 39 (38.6) | 0.52 |
| mexZ (PA2020) | 9 (13.8) | 0.60 | 20 (19.8) | 0.27 |
| lasR (PA1430) | 8 (12.3) | 0.53 | 14 (13.9) | 0.19 |
| PA0313 | 8 (12.3) | 0.53 | 12 (11.9) | 0.16 |
| mexA (PA0425) | 4 (6.2) | 0.27 | 7 (6.9) | 0.09 |
| accC (PA4848) | 3 (4.6) | 0.20 | 6 (5.9) | 0.08 |
| vfr (PA0652) | 2 (3.1) | 0.13 | 5 (5.0) | 0.07 |
| mexS (PA2491) | 1 (1.5) | 0.07 | 7 (6.9) | 0.09 |
| exsA (PA1713) | 3 (4.6) | 0.20 | 6 (5.9) | 0.08 |
| PA0506 | 2 (3.1) | 0.13 | 5 (5.0) | 0.07 |
| wspF (PA3703) | 0 (<1.5) | <0.07 | 4 (4.0) | 0.05 |
| rpoN (PA4462) | 4 (6.2) | 0.27 | 3 (3.0) | 0.04 |
| fleQ (PA1097) | 3 (4.6) | 0.20 | 2 (2.0) | 0.03 |
| mexT (PA2492) | 4 (6.2) | 0.27 | 2 (2.0) | 0.03 |
Only genes for which at least five mutated isolates were available are included. The two synonymous mutations detected (one from each group [mutators and nonmutators]) were excluded from the analysis.
DISCUSSION
Over the last decade, several in vitro and in vivo experiments have shown that mutator phenotypes may confer an evolutionary advantage during bacterial exposure to new, stressful, or fluctuating environments by increasing the rate of appearance of adaptive mutations (3, 5, 35, 37). Adaptation to the host immune system (23), antibiotic treatments (17), or viral parasites (28) has been shown to be accelerated in mutator backgrounds in various experimental models. Mutator strains have also been found in diverse natural bacterial populations with variable prevalence, although they appear to be overrepresented in chronic infections, while they are rather uncommon in acute settings (16, 24, 29, 31). The case of P. aeruginosa is highly illustrative; the prevalence of mutators has been reported to be <1% in acute infections (6), 5 to 10% in environmental and early CF isolates (11), and 30 to 60% in long-term CRI (1, 16, 24). Not surprisingly, our results obtained with the collection of CF isolates of Smith et al. (34), the first collection reported for North America, agree well within these previous findings. In most cases, mutator lineages emerged during CRI from otherwise clonal strains, although in two of the patients (6.9%) mutators were detected in the earliest isolates available. Furthermore, the vast majority of the mutator strains detected, including the first fully sequenced late CF isolate, were found to be defective in the MMR system gene mutS or mutL, also confirming previous observations (21, 25). Moreover, the documented independent selection of mutS and mutL mutants in a single patient provides further clear evidence of positive selection of MMR system-inactivating mutations during CRI.
CRI with P. aeruginosa in CF patients is one of the most illustrative examples of extreme natural bacterial evolution during the course of human infections. Indeed, the genetic properties of the bacterial cells present late in P. aeruginosa infections of CF patients differ greatly from the genetic properties of the cells that initiated the infections many years before the decline in lung function became life threatening (19, 34). Long-term persistence in the airways of CF patients seems to be driven by the selection of multiple, frequently loss-of-function mutations leading to an overall pattern of increased antimicrobial resistance, reduced virulence, and specific metabolic adaptation. Mutational inactivation of the transcriptional regulator lasR can be considered one of the paradigms of such a pattern of evolution, since it is extraordinarily frequent in the CF setting and meets all three criteria mentioned above (2). Several other frequently selected mutations, such as those leading to alginate hyperproduction, the loss of flagellum expression, or a modified lipopolysaccharide, have been demonstrated experimentally to significantly contribute to bacterial adaptation during CRI (27).
Consistent with previous hypotheses and in vitro and in vivo models, we showed that hypermutation has a dramatic effect on the intense evolution of P. aeruginosa during CRI. As shown here, the presence of adaptive traits is not homogeneous among the CF isolates; on the contrary, it is significantly concentrated in MMR system-deficient lineages. Indeed, the presence of mutations in the 34 genes studied was rather infrequent for nonhypermutable isolates (median, one mutation per isolate), whereas it was up to 6.5-fold higher in mutator backgrounds. Moreover, for the first time the predicted amplification of mutator populations by hitchhiking with adaptive mutations (18) is clearly documented in the course of natural human infections, as shown in Fig. 4. The documented differences in the number of mutations that accumulated per year of infection are certainly overwhelming; while sequential nonmutator lineages acquire a median of only 0.25 mutation per year of infection, sequential mutator lineages accumulate over 3 mutations per year, a 13-fold-higher rate. Interestingly, increased accumulation of mutations in mutator isolates was not a consequence of overrepresentation of mutations in genes involved in antimicrobial resistance, the only adaptive trait linked so far to hypermutation in CF patients (7, 24), demonstrating that hypermutation has a generalized effect on P. aeruginosa evolution and adaptation during CRI. Our results are consistent with recent findings for the CF mouse model of chronic colonization, in which P. aeruginosa mutator strains were shown to favor long-term persistence even in the absence of antimicrobial therapy (20). They also support the results of recent studies showing that inactivation of the MMR system favors the emergence of multiple P. aeruginosa phenotypic variants in vitro, including the typical markers of CF lung infection lasR and mucA (mucA22) mutants (14, 22). Nevertheless, it should be noted that while the effect of hypermutation in bacterial evolution during CRI is clear, its implications for disease progression are still uncertain. In a very recent study, the presence of mutators was indeed associated with poorer lung function, although hypermutation could not be demonstrated to be an independent risk factor in this negative outcome (40). It should also be noted that the shortcut to genetic adaptation to the airways of CF patients provided by mutators is not expected to be cost free. While the acquisition of mutations under positive selective pressure should speed up adaptation, it may also lead to accumulation of mutations deleterious for secondary environments. Indeed, adapted mutator lineages recovered from CF mice showed reduced transmissibility in a previous study (20). Moreover, mutator isolates recovered from late CRI in CF patients have recently been shown to have reduced fitness and virulence once they are retrieved from the primary lung environment (9, 21).
Supplementary Material
Acknowledgments
This work was supported by the Ministerio de Educación y Ciencia of Spain (grant SAF2003-02851), by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III through the Spanish Network for Research in Infectious Diseases (grants REIPI C03/14 and RD06/0008), and by the Canadian Cystic Fibrosis Foundation (D.P.S.).
Footnotes
Published ahead of print on 10 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ciofu, O., B. Riis, T. Pressler, H. E. Poulsen, and N. Hoiby. 2005. Ocurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 492276-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Déziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denamur, E., and I. Matic. 2006. Evolution of mutation rates in bacteria. Mol. Microbiol. 60820-827. [DOI] [PubMed] [Google Scholar]
- 4.Gibson, R. L., J. L. Burns, and B. W. Rammsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168918-951. [DOI] [PubMed] [Google Scholar]
- 5.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 2912606-2608. [DOI] [PubMed] [Google Scholar]
- 6.Gutiérrez, O., C. Juan, J. L. Pérez, and A. Oliver. 2004. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob. Agents Chemother. 483573-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrichfreise, B., I. Wiegand, W. Pfister, and B. Wiedemann. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 514062-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogardt, M., S. Schubert, K. Adler, M. Götzfried, and J. Heesemann. 2006. Sequence variability and functional analysis of MutS of hypermutable Pseudomonas aeruginosa cystic fibrosis isolates. Int. J. Med. Microbiol. 296313-320. [DOI] [PubMed] [Google Scholar]
- 9.Hogardt, M., C. Hoboth, S. Schmoldt, C. Henke, L. Bader, and J. Heesemann. 2007. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 19570-80. [DOI] [PubMed] [Google Scholar]
- 10.Horst, J. P., T. H. Wu, and M. G. Marinus. 1999. Escherichia coli mutator genes. Trends Microbiol. 729-36. [DOI] [PubMed] [Google Scholar]
- 11.Kenna, D. T., C. J. Doherty, J. Foweraker, L. Macaskill, V. A. Barcus, and J. R. W. Govan. 2007. Hypermutability in environmental Pseudomonas aeruginosa and in populations causing pulmonary infection in individuals with cystic fibrosis. Microbiology 1531852-1859. [DOI] [PubMed] [Google Scholar]
- 12.Labat, F., O. Pradillon, L. Garry, M. Peuchmaur, B. Fantin, and E. Denamur. 2005. Mutator phenotype confers advantage in Escherichia coli chronic urinary tract infection pathogenesis. FEMS Immunol. Med. Microbiol. 44317-321. [DOI] [PubMed] [Google Scholar]
- 13.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Win, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a GT mismatch. Nature 407711-717. [DOI] [PubMed] [Google Scholar]
- 14.Lujan, A. M., A. J. Moyano, I. Segura, C. E. Argaraña, and A. M. Smania. 2007. Quorum-sensing-deficient (lasR) mutants emerge at high frequency from a Pseudomonas aeruginosa mutS strain. Microbiology 153225-237. [DOI] [PubMed] [Google Scholar]
- 15.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infection associated with cystic fibrosis. Clin. Microbiol. Rev. 15194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macia, M. D., D. Blanquer, B. Togores, J. Sauleda, J. L. Pérez, and A. Oliver. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 493382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macia, M. D., N. Borrell, M. Segura, C. Gómez, J. L. Pérez, and A. Oliver. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathee, K., G. Narasimhan, C. Valdes, X. Qiu, J. M. Matewish, M. Koehrsen, A. Rokas, C. N. Yandava, R. Engels, E. Zeng, R. Olavarietta, M. Doud, R. S. Smith, P. Montgomery, J. R. White, P. A. Godfrey, C. Kodira, B. Birren, J. E. Galagan, and S. Lory. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. USA 1053100-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mena, A., M. D. Maciá, N. Borrell, B. Moya, T. de Francisco, J. L. Pérez, and A. Oliver. 2007. Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J. Bacteriol. 1893665-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montanari, S., A. Oliver, P. Salerno, A. Mena, G. Bertoni, B. Tümmler, L. Cariani, M. Conese, G. Döring, and A. Bragonzi. 2007. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology 1531445-1454. [DOI] [PubMed] [Google Scholar]
- 22.Moyano, A. J., A. M. Lujan, C. E. Argaraña, and A. M. Smania. 2007. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol. Microbiol. 64547-559. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson, A. I., E. Kugelberg, O. G. Berg, and D. I. Andersson. 2004. Experimental adaptation of Salmonella typhimurium to mice. Genetics 1681119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver, A., R. Cantón, P. Campo, F. Baquero, and J. Blázquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2881251-1253. [DOI] [PubMed] [Google Scholar]
- 25.Oliver, A., F. Baquero, and J. Blázquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 431641-1650. [DOI] [PubMed] [Google Scholar]
- 26.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blázquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 484226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, A., A. Mena, and M. D. Maciá. 2007. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections, p. 433-444. In F. Baquero, C. Nombela, G. H. Cassell, and J. A. Gutiérrez (ed.), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC.
- 28.Pal, C., M. D. Maciá, A. Oliver, I. Schachar, and A. Buckling. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 4501079-1081. [DOI] [PubMed] [Google Scholar]
- 29.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclerc. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 1871709-1716. [DOI] [PubMed] [Google Scholar]
- 30.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 39469-72. [DOI] [PubMed] [Google Scholar]
- 31.Román, F., R. Cantón, M. Pérez-Vazquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 421450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Römling, U., B. Fiedler, J. Bobhammer, D. Grothues, J. Greipel, H. Von der Hart, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 1701616-1621. [DOI] [PubMed] [Google Scholar]
- 33.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachack, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sniegowski, P. D., P. J. Gerrish, and R. E. Lenski. 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387703-705. [DOI] [PubMed] [Google Scholar]
- 36.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkmann, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406959-964. [DOI] [PubMed] [Google Scholar]
- 37.Taddei, F., M. Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon, and B. Godelle. 1997. Role of mutator alleles in adaptive evolution. Nature 387700-702. [DOI] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventre, I., A. L. Goodman, I. Vallet-Gey, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waine, D. J., D. Honeybourne, E. G. Smith, J. L. Whitehouse, and C. G. Dowson. 6 August 2008. The association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J. Clin. Microbiol. doi: 10.1128/JCM.00357-08. [DOI] [PMC free article] [PubMed]
- 41.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.