Abstract
Anabaena variabilis grows heterotrophically using fructose, while the close relative Anabaena sp. strain PCC 7120 does not. Introduction of a cluster of genes encoding a putative ABC transporter, herein named frtRABC, into Anabaena sp. strain PCC 7120 on a replicating plasmid allowed that strain to grow in the dark using fructose, indicating that these genes are necessary and sufficient for heterotrophic growth. FrtR, a putative LacI-like regulatory protein, was essential for heterotrophic growth of both cyanobacterial strains. Transcriptional analysis revealed that the transport system was induced by fructose and that in the absence of FrtR, frtA was very highly expressed, with or without fructose. In the frtR mutant, fructose uptake was immediate, in contrast to that in the wild-type strain, which required about 40 min for induction of transport. In the frtR mutant, high-level expression of the fructose transporter resulted in cells that were extremely sensitive to fructose. Even in the presence of the inducer, fructose, expression of frtA was low in the wild-type strain compared to that in the frtR mutant, indicating that FrtR repressed the transporter genes even in the presence of fructose. FrtR bound to the upstream region of frtA, but binding was not visibly altered by fructose, further supporting the hypothesis that fructose has only a modest effect in relieving repression of frtA by FrtR. A. variabilis grew better with increasing concentrations of fructose up to 50 mM, showing increased cell size and heterocyst frequency. Anabaena sp. strain PCC 7120 did not show any of these changes when it was grown with fructose. Thus, although Anabaena sp. strain PCC 7120 could take up fructose and use it in the dark, fructose did not improve growth in the light.
Although the majority of cyanobacteria are obligate photoautotrophs, dependent on sunlight for ATP and on CO2 for carbon, a few well-studied strains can take up and use sugars either only in the light, growing mixotrophically, or in the dark, growing heterotrophically (41). Synechocystis sp. strain PCC 6803 is one of the best-studied strains that can grow in the dark using glucose as the sole carbon source; however, it requires short, regular exposure to light for heterotrophic growth (3). Two well-studied filamentous heterocyst-forming cyanobacterial strains, namely, Nostoc punctiforme ATCC 29133 and Anabaena variabilis ATCC 29413, are capable of true heterotrophic growth in complete darkness (14, 43). The former strain grows on glucose or fructose, while A. variabilis ATCC 29413 can use only fructose (14). In these two heterocyst-forming strains, sugars support not only growth in the dark but also nitrogen fixation, an energetically expensive reaction. N. punctiforme ATCC 29133 is a symbiont of the bryophyte Anthoceros punctatus, and hence, it is likely that its ability to use sugars is essential for its role in symbiosis (49). A. variabilis is not known to be an endosymbiont; however, by morphology, phenotype, and genetics, it is virtually identical to many strains called Anabaena azollae, isolated from the symbiotic association of cyanobacteria with the water fern Azolla (6, 12, 28, 36, 37, 39, 52). Although it was first believed that Anabaena azollae was the symbiont, it was shown later that the true symbiont is different and is probably nonculturable (13, 28). Hence, there is some possibility that A. variabilis also once came from Azolla and that fructose utilization is associated with symbiosis in this strain.
Fructose dramatically affects the physiology of A. variabilis. The cells grow faster, are bigger, and in filaments that have differentiated heterocysts, produce more and larger heterocysts, fixing more nitrogen and producing more hydrogen than do cells grown photoautotrophically (14, 35, 42). [14C]fructose, which is taken up almost immediately by vegetative cells in a filament, is quickly transported in some form to the heterocysts, where the 14C compound accumulates and is metabolized to provide a reductant for nitrogen fixation (14). Although fructose supports nitrogen fixation in whole filaments, isolated heterocysts cannot use fructose as a source of reductant, suggesting either that fructose cannot be transported by heterocysts or that fructose is converted to another compound in the vegetative cell before it moves to the heterocyst (16). For N. punctiforme, a mutant deficient in glucose-6-phosphate dehydrogenase cannot fix nitrogen and cannot grow in the dark with fructose, indicating that the oxidative pentose phosphate pathway is the major pathway for fructose metabolism and is important in heterocysts for nitrogen fixation (43). In fructose-grown filaments, the heterocysts not only are bigger than those in cells grown photoautotrophically but also store more glycogen and are morphologically different (21).
Growth with fructose results in increased respiration and decreased chlorophyll (14, 33, 36, 46). In long-term, dark-grown, fructose-adapted cells, there is an increase in photosystem II, resulting in a decrease in the ratio of photosystem I to photosystem II (23). Cells grown with low CO2 in the presence of fructose do not fix CO2 well because of decreased carbonic anhydrase and decreased ribulose bis-phosphate carboxylase oxygenase (29). The decrease in oxygen production in fructose-grown cells is thought to contribute to a micro-oxic environment that better supports nitrogen fixation (14). Microarray analysis of RNA from the non-nitrogen-fixing unicellular cyanobacterium Synechocystis sp. strain PCC 6803 under conditions of nitrogen starvation shows increased expression of genes important in glycolysis, the oxidative pentose phosphate pathway, and glycogen catabolism and increased activities of glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, two key enzymes of the oxidative pentose phosphate pathway (31). In Synechocystis, transcription of the genes for sugar catabolism is regulated by Hik8 (40), a homolog of a protein (SasA) in Synechococcus that regulates kaiC, which is part of the central oscillator of circadian rhythm (15, 20). In addition, the sigma factor SigE positively regulates three glycolytic genes, four oxidative pentose phosphate genes, and two glycogen metabolism genes (32). Activation of sugar catabolic genes under conditions of nitrogen starvation requires the global nitrogen activator NtcA (30, 32). SigE in Anabaena sp. strain PCC 7120, a strain that cannot use fructose, is not essential for nitrogen fixation but is expressed late in heterocyst differentiation, suggesting that it has a role in heterocyst function (1, 19).
The transport of glucose in Synechocystis is known to occur via a glucose-fructose permease, the product of the glcP transport gene (11, 17, 50). Transport of fructose is toxic to the cells; inactivation of glcP relieves the toxicity but no longer allows the cells to grow using glucose (11, 17, 50). Expression of glcP in Synechococcus sp. strain PCC 7942 resulted in a strain that was capable of glucose transport but also died in the presence of glucose (51). Uptake of fructose in A. variabilis and Nostoc sp. strain ATCC 29150 is constitutive but increases after exposure to fructose (38, 46) and is energy dependent in A. variabilis (46). The Km for fructose uptake is about 160 μM for cells that have not been grown with fructose and about 50 μM for cells pregrown with fructose, and it does not change in the light versus the dark (14, 16). We describe here the genes for fructose transport in A. variabilis, their regulation, and the effect of their expression on growth of the obligately photoautotrophic strain Anabaena sp. strain PCC 7120.
MATERIALS AND METHODS
Strains and growth conditions.
Strains of A. variabilis FD, a derivative of A. variabilis 29413 that can grow at 40°C and supports the growth of bacteriophages better than the parent strain does (9), and Anabaena sp. strain PCC 7120 were maintained on agar-solidified Allen and Arnon (AA) medium (2) supplemented, when appropriate, with 5 mM NH4Cl, 10 mM N-tris (hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES), pH 7.2, 25 to 40 μg ml−1 neomycin sulfate, or 3 μg ml−1 each of spectinomycin and streptomycin. Strains were grown photoautotrophically in liquid cultures in an eightfold dilution of AA medium (AA/8) or in AA/8 supplemented with 5 mM NH4Cl and 10 mM TES, pH 7.2, at 30°C, with illumination at 50 to 80 microeinsteins m−2 s−1. Antibiotics were included as follows (when required): neomycin (3 to 5 μg ml−1) and spectinomycin (0.3 μg ml−1 for liquid). For experiments to measure the growth of strains with fructose, cells were harvested at an optical density at 720 nm (OD720) of 0.2, washed once in AA/8, and resuspended in AA/8 with the indicated concentrations of fructose at an OD720 of 0.02. All growth experiments were performed three times, and a representative graph is presented.
Construction of plasmids and strains.
A neomycin resistance (Nmr) cassette containing a transcriptional terminator was PCR amplified from pRL648 (10), using primers nptTerm-3′and nptTerm-5′, digested with EcoRI, and cloned into the EcoRI site of pUC1819RI to create pBP285. Primer sequences are provided in Table S1 in the supplemental material. The Nmr cassette was used to create pBP299, a plasmid with an insertional mutation in frtR. The bom site of pRL1075, required for conjugation, was inserted into pBP299 to create pBP301. Replacement of the wild-type frtR gene in the chromosome of strain FD with the mutant frtR allele in pBP301 was accomplished by conjugation followed by double recombination (45). The mutant was segregated as described previously and tested by PCR to verify that no wild-type copies of the gene remained (22).
Plasmid pBP289 was created to contain the ava2169 to ava2173 genes from genomic library clone pAAWY3009. This plasmid was used to construct the replicating plasmids pBP291 (containing frtRABC) and pBP292 (containing frtABC without frtR). Plasmids were constructed as described in Table 1. Replicating plasmids pBP291 (containing frtRABC) and pBP292 (containing frtABC without frtR) were conjugated into Anabaena sp. strain PCC 7120, selecting for the antibiotic resistance on the plasmid, and the presence of the plasmid was verified by PCR.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| A. variabilis FD | Anabaena variabilis parent strain | 9 |
| Anabaena sp. strain PCC 7120 | Wild-type strain | |
| Anabaena sp. strain PCC 7120 BP291 | frtRABC expressed from plasmid pBP291 | This work |
| Anabaena sp. strain PCC 7120 BP292 | frtABC expressed from plasmid pBP292 | This work |
| A. variabilis FD BP301 | frtR mutated with insert of Nmr cassette at NaeI sites | This work |
| A. variabilis BP352 | pBP352 integrated into the chromosome | This work |
| A. variabilis BP353 | pBP353 integrated into the chromosome | This work |
| A. variabilis BP356 | pBP356 integrated into the chromosome | This work |
| A. variabilis JU336 | pJU336 integrated into the chromosome | This work |
| A. variabilis JU338 | pJU338 integrated into the chromosome | |
| A. variabilis JU377 | pJU377 integrated into the chromosome | This work |
| Anabaena sp. strain PCC 7120 JU356 | pJU336 integrated into the chromosome | This work |
| Anabaena sp. strain PCC 7120 JU357 | pJU338 integrated into the chromosome | This work |
| Anabaena FD JU353 | pJU336 integrated into the chromosome of A. variabilis FD BP301 | This work |
| Anabaena FD JU355 | pJU338 integrated into the chromosome of A. variabilis FD BP301 | This work |
| Plasmids | ||
| pAAWY3009 | 9.3-kb clone of A. variabilis DNA containing frtRABC; Cmr | JGI sequencing clone |
| pBP285 | Kmr Nmr cassette in a polylinker [C.K.3 (10)] with a transcriptional terminator | This work |
| pBP288 | Cloning vector for integration of transcriptional fusions into the chromosome; Tcr Kmr Nmr Spr Smr Apr | This work |
| pBP289 | 9.8-kb EcoRI fragment of pAAWY3009 (containing Ava2169 to Ava2173 but lacking Ava4074), self-ligated | This work |
| pBP291 | 8.4-kb ScaI/SmaI fragment from pBP289 (containing frtRABC) inserted into the SmaI sites of pRL57 | This work |
| pBP292 | 4.9-kb NaeI/SmaI fragment from pBP289 (containing frtABC) ligated into the SmaI sites of pRL57 | This work |
| pBP299 | 1.1-kb SmaI fragment containing the Nmr cassette of pBP285 inserted into NaeI site of frtR in pHL110 | This work |
| pBP301 | 5-kb BglII fragment of pRL1075 ligated into the BamHI site of pBP299 | This work |
| pBP313 | Cloning vector to overexpress genes from psbA promoter; Kmr Nmr Spr Smr Apr | This work |
| pBP314 | PCR-amplified Tcr cassette of pBR322 inserted into the NdeI/BamHI sites of pET22b | This work |
| pBP328 | PCR-amplified Tcr cassette of pBR322 inserted into the SmaI/SacI sites of pBP313 | This work |
| pBP350 | Replaced BglII/BamHI fragment containing Nmr cassette of pPE20 with PCR-amplified Ω Spr Smr cassette from pBP288 | This work |
| pBP351 | 4.2-kb EcoRI/ScaI fragment of pHL110 (containing Ava2169 to -71) cloned into EcoRV/EcoRI fragment of pBR322 | This work |
| pBP352 | 5-kb SmaI fragment (containing a lacZ Spr Smr cassette) of pBP350 inserted into EcoRV site of pBP351 | This work |
| pBP353 | 5-kb SmaI fragment (containing a lacZ Spr Smr cassette) of pBP350 inserted into ClaI site (blunted) of pBP351 | This work |
| pBP354 | PCR-amplified frtR gene inserted into NdeI/BamHI sites of pBP314 for overexpression of native FrtR | This work |
| pBP356 | PCR-amplified frtR gene inserted into SmaI/SacI sites of pBP313 under the control of psbA promoter | This work |
| pBR322 | Mobilizable plasmid; Apr Tcr | 5 |
| pET22b | T7 expression vector; expression induced by isopropyl-β-d-thiogalactopyranoside | Invitrogen |
| pHL110 | 4.5-kb HindIII frt region of pAAWY3009 inserted into HindIII site of pUC18 | This work |
| pJU336 | PCR-amplified 400-bp frtR promoter fragment inserted into BglII/SmaI sites of pBP288 | This work |
| pJU338 | PCR-amplified 500-bp frtA promoter fragment inserted into BglII/SmaI sites of pBP288 | This work |
| pJU377 | PCR-amplified frtA gene inserted into SmaI/SacI sites of pBP313 under the control of psbA promoter | This work |
| pPE20 | Source of lacZ for transcriptional fusions; Apr Kmr Nmr | 44 |
| pRL1075 | Source of mobilization site, oriT, and sacB gene, which confers sucrose sensitivity; Cmr Emr | 4 |
| pRL57 | S.K5 + L.HEH2 + C.S3; positive selection shuttle (pDU1) cloning vector | 10 |
| pRL648 | Kmr Nmr cassette in a polylinker (C.K.3) | 10 |
| pUC18 | Cloning vector; Apr | 47 |
| pUC1819RI | Cloning vector; Apr | 7 |
The BglII and BamHI Nmr fragment in pPE20 was replaced with a Spr Smr cassette containing a transcriptional terminator (amplified from pBP288, using the Sm Ω L and Sm Ω R primers, engineered with BglII and BamHI sites at the 5′ and 3′ ends, respectively, for cloning) to create pBP350. The Spr Smr version of pPE20, pBP350, was used to create lacZ transcriptional fusions of frtA (pBP352) and frtR (pBP353) at the EcoRV and ClaI sites, respectively. Integration of the transcriptional fusions, pBP352 and pBP353, into the chromosome of FD was accomplished by conjugation of the nonreplicative plasmids, selecting for single recombinants containing the entire plasmid in the chromosome.
Plasmid pBP288 is a 16.7-kb pBR322-based vector that contains (i) a promoterless lacZ gene for assaying promoter activity in vivo; (ii) a 6.5-kb ntcA region of A. variabilis that allows for good homologous recombination; (iii) a 1.1-kb npt gene from pRL648 interrupting ntcA, ensuring only one functional copy of ntcA after recombination; (iv) a 1.0-kb Ω Spr Smr cassette with a transcriptional terminator from pRL277 upstream of and directed away from the lacZ gene; and (v) a Tetr cassette between the BglII and SmaI cloning sites to allow for easy cloning of promoter fragments upstream of the lacZ gene.
Plasmid pBP313 contained the psbA promoter in the BglII/SmaI sites of pBP288. A Tetr gene (PCR amplified from pBR322 by use of primers pBR322-L2 and pBR322-R2) was inserted into the SmaI/SacI sites of pBP313 to create pBP328, a plasmid that destroyed the lacZ gene but gave selection for inserting fragments under the control of the psbA promoter in vivo. The plasmid used to overexpress FrtR in A. variabilis was constructed by PCR amplifying the frtR gene, using primers psbAFrtR-5psbAFruR-5 and psbAFrtR-3′psbAFruR-3′, and inserting it into the SmaI/SacI sites of pBP328 to create pBP356. Additionally, the frtABC coding region (PCR amplified using primers frtABC-L and frtABC-R) was cloned downstream of the psbA promoter on pBP313 to generate pJU377. These plasmids were conjugated into FD by single recombination to yield BP356 and JU377.
A 500-bp frtA promoter fragment (amplified from FD by use of the frtA498A-L and frtA-R10 primers) and a 400-bp frtR promoter fragment (amplified from FD by use of the frtR397-L and frtR-R10 primers) were cloned into the BglII/SmaI sites upstream of lacZ on pBP288 to generate pJU338 and pJU336, respectively. These plasmids were then conjugated, with selection for single recombinants, into FD to generate strains JU338 and JU336, into Anabaena sp. strain PCC 7120 to generate strains JU357 and JU356, and into BP301 to produce strains JU355 and JU353.
The plasmid pBP354, used to overexpress FrtR in Escherichia coli, was constructed by PCR amplification of frtR with NdeI and BamHI sites at the 5′ and 3′ ends, respectively, using primers FrtR-L3 and FrtR-R3, and insertion into the same sites of pBP314. pBP314 was constructed by inserting a Tetr gene [PCR amplified from pBR322 by use of primers Tet(NdeI)-L and Tet(BamHI)-R] into the NdeI/BamHI sites of pET22b (Invitrogen), therefore making it easier to select for insertion of a DNA fragment encoding protein into the vector.
FrtR overexpression and purification and electrophoretic mobility shift assay.
The FrtR protein was purified from E. coli/pBP354, overexpressing FrtR, as inclusion bodies as described by Campbell et al. (8), with the following modifications: cells were lysed by four 30-s rounds of sonication and the protein concentration was adjusted to 1.0 mg ml−1 before renaturation. Electrophoretic mobility shift assay binding reaction mixtures contained 4 mM Tris, pH 8.0, 12 mM HEPES, 12% glycerol, 100 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2, 1.0 μg poly(dI-dC), and 10,000 cpm 32P-end-labeled probe. FrtR was added (100 to 700 ng of protein), and the mixture was incubated for 20 min at 30°C. After the binding reaction, the reaction mixtures were loaded into a 4% polyacrylamide gel with a Tris-glycine buffer (pH 8.0) and were electrophoresed at 40 mA for 25 min. Bands were visualized using a phosphorimager.
RNA isolation and RT-PCR.
RNAs were isolated from 50-ml cultures grown in AA/8, with or without fructose, and subjected to DNase digestion using a Turbo DNA-free kit (Ambion). Reverse transcription-PCR (RT-PCR) was performed as previously described (34), with the following change: 2.5 ng of RNA and 2.5 U of Superscript III (Invitrogen) were added per reaction. Primers were annealed at 58°C. The primers frtA-L/R and frtR-L/R were used to amplify the frtA and frtR transcripts, respectively. RNA from the housekeeping gene rnpB was amplified using rnpB-L/R primers as a control (48).
Microtiter β-galactosidase assays.
Cultures were grown as described above to an OD720 of 0.1 and divided into two equal portions, and 5 mM fructose was added to one portion to induce expression of the frt genes. Two hours after induction, the cultures were adjusted to an OD720 of 0.05, and 700 μl of culture was added to 700 μl of 2× LacZ buffer (120 mM Na2HPO4, 80 mM NaH2PO4, 20 mM KCl, 2 mM MgSO4, 100 mM β-mercaptoethanol). The samples were vortexed for 60 s with 30 μl 0.1% sodium dodecyl sulfate and 60 μl chloroform. The chloroform was removed, and 250 μl of sample was placed in microtiter wells. Eighty microliters of o-nitrophenyl-β-d-galactopyranoside (4 mg ml−1) was added to the wells, and a microtiter plate reader measured the OD420 every 90 s for 1 h. Eight replicates were done for each sample. Excel was used to process the raw data, yielding the rate of the reaction, which was normalized to the OD720 of the culture.
Light micrographs.
Filaments were viewed with a Zeiss epifluorescence microscope and imaged using a Retiga EXi (QImaging) cooled charge-coupled device camera with IP Labs 4.0 software (BD Biosciences). The exposure time was about 0.05 s for bright-field images.
RESULTS
Identification of fructose transport genes.
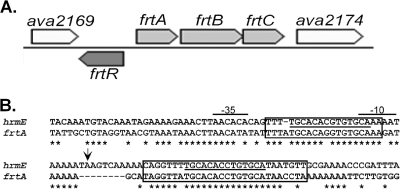
Fructose transport genes were identified as putative ABC-type sugar transport genes present in the genomes of A. variabilis and N. punctiforme, both capable of heterotrophic growth in the dark with fructose as the sole carbon source, but not in the genome of Anabaena sp. strain PCC 7120, an obligate photoautotroph. The organization of the ABC-type transport genes, frtABC, and a lacI-like regulatory gene, frtR, is shown in Fig. 1A. The flanking ava2169 gene is tentatively identified as encoding dihydrouridine synthase and has close homologs in the genomes of Anabaena sp. strain PCC 7120, N. punctiforme, and Nodularia spumigena CCY9414, while the ava2174 gene encodes a putative porin-like protein. There is a close homolog of the ava2174 gene in the genome of N. punctiforme but not in the genomes of Anabaena sp. strain PCC 7120 and N. spumigena. The homolog of FrtR in N. punctiforme (73% identical) is HrmR (Npun02008536), a repressor of its own gene, hrmR, and of hrmE (Npun02008530), which are genes involved in the differentiation of hormogonia in N. punctiforme (8). There is no homolog of hrmE in A. variabilis or in the genomes of any other sequenced cyanobacteria. The homologs of FrtABC in N. punctiforme (Npun02008528, Npun02008527, and Npun02008526 to Npun02006538), with 72 to 85% identities to the proteins in A. variabilis, are downstream of hrmE in the N. punctiforme genome. Downstream of the frtABC homologs in N. punctiforme is a gene (Npun02006539) that appears to encode a glucose transporter. This gene is absent from A. variabilis, consistent with the fact that this strain cannot use glucose as a carbon source; however, there is a homolog (sll0771; named glcP for glucose permease) with 71% identity in Synechocystis sp. strain PCC 6803, a strain that can grow heterotrophically in the dark with glucose.
FIG. 1.
Fructose transport genes. (A) The region of the chromosome of A. variabilis with the fructose transport genes, namely, frtR (ava2170), encoding a putative lacI-like transcriptional regulator, and frtABC (ava2171 to -2173), encoding a putative periplasmic binding component, ATPase component, and transmembrane component, respectively. (B) Alignment of the promoter region of hrmE of N. punctiforme with a conserved region of the frtA promoter region, beginning about 300 bp upstream from the start codon of frtA. The transcription start site of hrmE is indicated by an arrow, and the −10 and −35 regions of the hrmE promoter are labeled. The HrmR binding sites, which are underlined, are shown within boxes that indicate longer conserved palindromic sequences.
The hrmE gene in N. punctiforme is regulated by HrmR, a homolog of FrtR (8). Although there is no similarity between HrmE in N. punctiforme and FrtA in A. variabilis, there is striking similarity in the regulatory regions of these genes. The region of hrmE that contains the transcription start site and the two binding sites for HrmR is well conserved in the region upstream of frtA in A. variabilis (Fig. 1B), and the two binding sites for HrmR are nearly identical between frtR and hrmE. In the regulatory region of frtA, the second HrmR-like binding site is within a larger, 28-bp, almost perfect palindrome (Fig. 1B); however, that larger palindrome is not as well conserved in hrmE. In contrast, in hrmE, the first HrmR binding site is within a 20-bp perfect palindrome (Fig. 1B) that is not as well conserved in frtA. The striking similarity in these regions suggests that hrmE and frtA have similar modes of regulation, which is supported by the similarity of FrtR and HrmR. There are two HrmR binding sites in the promoter region of hrmR, and HrmR binds to this region; thus, it is autoregulatory (8). Since frtA and frtR in A. variabilis are divergent genes (Fig. 1A), they share a regulatory region and therefore share the two HrmR binding sites shown in Fig. 1B.
Function of frtRABC.
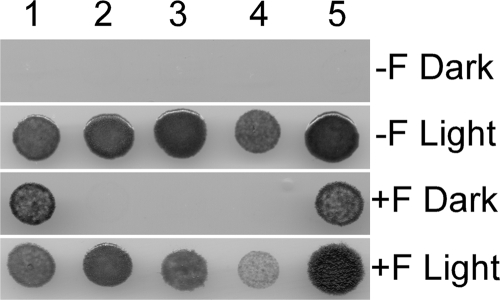
In order to determine whether the frtRABC genes function to transport fructose, we transferred frtRABC or frtABC, lacking frtR, to Anabaena sp. strain PCC 7120, a strain that lacks the frtRABC genes and cannot grow heterotrophically in the dark with sugars. An frtR mutant of A. variabilis was also constructed. The wild-type strain of A. variabilis grew well in the light with or without fructose but grew in the dark only in the presence of fructose (Fig. 2, lane 5). In contrast, Anabaena sp. strain PCC 7120 grew only in the light and could not grow in the dark with fructose (Fig. 2, lane 4) unless the strain also contained the ftrRABC genes of A. variabilis (Fig. 2, lane 1). Thus, the frtRABC genes in Anabaena sp. strain PCC 7120 were sufficient to allow the strain to transport fructose. In Anabaena sp. strain PCC 7120, the only barrier to the utilization of fructose in the dark is the inability of the strain to transport the sugar. The frtR gene was essential for growth in the dark with fructose; neither the frtR mutant of A. variabilis (Fig. 2, lane 3) nor a mutant of Anabaena sp. strain PCC 7120 containing only frtABC, without frtR, was able to grow in the dark in the presence of fructose. These results suggested that FrtR is essential for expression of frtABC and might be an activator; however, this was not consistent with its similarity to the LacI repressor and to HrmR, which is also a repressor, so we explored this further.
FIG. 2.
Growth of strains with or without fructose transport genes. Cells of A. variabilis strain FD or Anabaena sp. strain PCC 7120 with or without ftrRABC genes were grown on BG-11 agar medium with or without 5 mM fructose (F) for 4 days in the light or 7 days in the dark. Lane 1, Anabaena sp. strain PCC 7120 BP291, containing the frtRABC genes; lane 2, Anabaena sp. strain PCC 7120 BP292, containing the frtABC genes; lane 3, A. variabilis BP301 (frtR mutant); lane 4, Anabaena sp. strain PCC 7120; lane 5, A. variabilis strain FD.
Transcription of frtA and frtR.
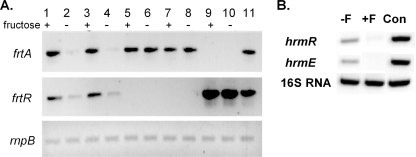
If the frtRABC genes encode a fructose transport system, then they would likely be regulated by fructose. Analysis of transcripts of frtA and frtR in cells grown with or without fructose indicated that in both A. variabilis and Anabaena sp. strain PCC 7120 with frtRABC, transcription of frtA and frtR was induced by fructose, although low levels of transcript were detected for both genes in the absence of fructose. frtA was more strongly induced by fructose than frtR was (see Fig. 4). If FrtR were an activator, then transcription of frtABC would depend on FrtA. However, it is a repressor, since in both the frtR mutant of A. variabilis and in Anabaena sp. strain PCC 7120 with frtABC but not frtR, expression of frtA was constitutive (Fig. 3A, lanes 5 to 8). Further supporting the role of FrtR as a repressor was the finding that in a strain of A. variabilis that overexpressed frtR, there was no expression of frtA, with or without fructose (Fig. 3A, lanes 9 and 10).
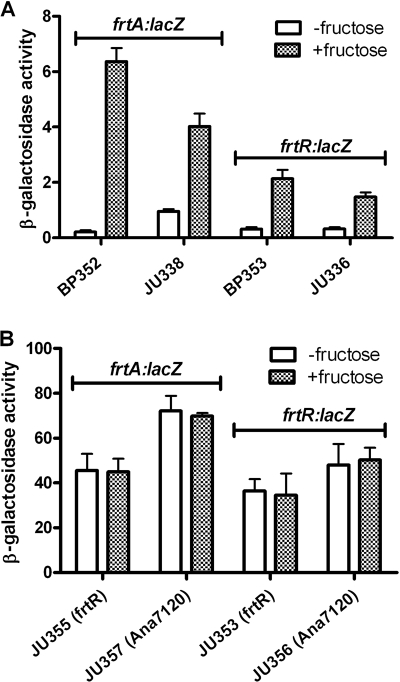
FIG. 4.
Expression of frtA-lacZ and frtR-lacZ fusions. (A) Relative rates of β-galactosidase activity were measured in strains BP352 (lacZ inserted within the frtA gene), JU338 (containing a 500-bp frtA promoter fragment fused to lacZ), BP353 (lacZ inserted within the frtR gene), and JU336 (containing a 400-bp frtR promoter fragment fused to lacZ), grown in AA/8 with or without 5 mM fructose. (B) Relative rates of β-galactosidase activity were measured in strains JU355 (containing a 500-bp frtA promoter fragment fused to lacZ in strain BP301, the frtR mutant), JU357 (containing a 500-bp frtA promoter fragment fused to lacZ in Anabaena sp. strain PCC 7120), JU353 (containing a 400-bp frtR promoter fragment fused to lacZ in strain BP301, the frtR mutant), and JU356 (containing a 400-bp frtR promoter fragment fused to lacZ in Anabaena sp. strain PCC 7120), grown with or without 5 mM fructose.
FIG. 3.
Transcription of frtA, frtR, hrmR, and hrmE. (A) Transcription of frtA and frtR was determined by RT-PCR, using RNAs extracted from A. variabilis strains grown with or without 5 mM fructose for 24 h. Lanes 1 and 2, wild-type A. variabilis; lanes 3 and 4, BP291 (Anabaena sp. strain PCC 7120 with frtRABC); lanes 5 and 6, BP292 (Anabaena sp. strain PCC 7120 with only frtABC); lanes 7 and 8, BP301 (A. variabilis frtR mutant); lanes 9 and 10, BP356 (A. variabilis strain overexpressing frtR); lane 11, positive control (FD DNA). Transcription of rnpB was the control for equal amounts of RNA in each reaction. (B) Transcription of hrmR and hrmE was determined by RT-PCR, using RNAs extracted from N. punctiforme grown in AA/8 without (−F) or with (+F) 5 mM fructose for 24 h. Con, positive control using chromosomal DNA from N. punctiforme. 16S rRNA was the control for equal amounts of RNA in each reaction.
Regulation of hrmR by fructose.
Since the homologue of frtR in N. punctiforme is hrmR, which has been shown to regulate itself and another gene of unknown function, hrmE (8), we used RT-PCR to determine whether these genes were also induced by growth with fructose. Both genes were repressed by fructose (Fig. 3B). Since the results were the reverse of those expected, the entire experiment was done a second time, beginning with new cultures of N. punctiforme grown with or without fructose, but the results were the same. Thus, despite the similarity in the encoded proteins, frtR and hrmE respond differently to fructose.
Expression of frtA-lacZ and frtR-lacZ fusions.
The frtA and frtR promoters were fused to lacZ to measure changes in expression of these genes in response to fructose in the presence and absence of FrtR. Two fusions were created for each gene. In the first type, a promoterless lacZ gene was inserted into frtA and frtR, thus providing not only a normal promoter but also a normal context for the promoter in the chromosome. The second type of fusion placed a 400-bp frtR or 500-bp frtA promoter fragment in front of lacZ, and then the entire construct was integrated into the chromosome. For the first type of fusion, expression of frtA (strain BP352), as measured by β-galactosidase activity, increased about 30-fold with fructose, while expression of frtR (strain BP353) increased about 7-fold with fructose (Fig. 4A). For the second type of fusion, expression of frtA (strain JU338) increased about fourfold with fructose, while expression of frtR (strain JU336) increased about fivefold (Fig. 4A). Even though the promoter fragments used were large and should have had all the necessary cis-acting elements, expression of frtA and ftrR in the BP352 and BP353 strains, in which the fusions were in the normal chromosomal locations, was more stringently controlled than that in strains JU338 and JU336, which had the promoter-lacZ fusion integrated at a different site by single-crossover recombination. This suggests that control of expression of frtA and frtR may depend on additional cis-acting sites that are not within the 400- to 500-bp promoter fragments used in the second type of fusion. Consistent with the results from RT-PCR (Fig. 3), in the presence of fructose the expression of frtA was higher than the expression of frtR, and the expression of frtA was more strongly induced by fructose than the expression of frtR.
To determine the effect of FrtR on expression, the promoter fragment fusions were also constructed in a frtR mutant of A. variabilis (BP301) and in Anabaena sp. strain PCC 7120, which naturally lacks frtR. In the absence of FrtR, frtA expression, as measured by β-galactosidase activity, was about 10-fold higher than that in the wild-type strain (compare strains JU355 and JU338) (note the difference in scale of the y axes in Fig. 4A and B) and was unaffected by growth with fructose (Fig. 4B). In the absence of FrtR, frtR expression, as measured by β-galactosidase activity, was about 25-fold higher than that in strains with FrtR (compare strains JU353 and JU336) and was also unaffected by growth with fructose (Fig. 4B). This indicates that in the presence of fructose, FrtR represses itself more strongly than it represses frtA. For both frtA and frtR, constitutive expression in the absence of FrtR was about 1.5-fold higher in Anabaena sp. strain PCC 7120 than in the frtR mutant of A. variabilis (BP301) (Fig. 4B). The constitutive expression of frtA and frtR in the absence of FrtR provides further evidence that FrtR is a repressor.
In strains with FrtR, expression of frtA and frtR was much more strongly repressed, even in the presence of fructose, than that in strains lacking FrtR, indicating that there was a repression of frtA and frtR by FrtR under all growth conditions tested. In the strains with FrtR in which frtA and frtR were expressed with fructose, frtA was more strongly expressed than frtR. However, in the absence of FrtR, the difference in expression between frtA and frtR was much smaller, suggesting that the lower level of frtR expression in the strains with FrtR was the result of stronger repression of frtR than of frtA in the presence of fructose and not the result of a much stronger promoter for frtA. Together, these results indicated that FrtR repressed expression of both frtA and frtR in the presence or absence of fructose, but the repression was much weaker in the presence of fructose. Furthermore, FrtR repressed frtR more than it repressed frtA in cells grown with fructose.
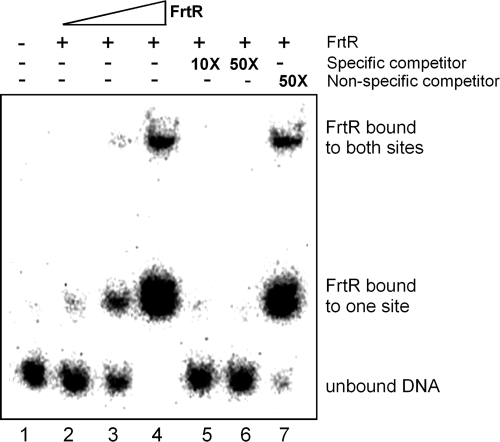
Binding of FrtR to the frtR-frtA promoter region.
Recombinant FrtR was purified from E. coli as inclusion bodies, and the protein was renatured. The protein bound to two sites on a DNA fragment that included the intergenic region between ftrR and frtA (Fig. 5). This region includes the two HrmR-like binding sites shown in Fig. 1. The binding was competed using the same cold DNA fragment but was not competed using an unrelated DNA fragment from the rnpB gene. The addition of fructose to the binding reaction mix had no effect on the mobility shift. This may be due to binding of FrtR to this region even in the presence of fructose. This is evident from the repression of frtA and frtR by FrtR even in the presence of fructose, as shown by the much higher levels of expression of frtA-lacZ and frtR-lacZ in an frtR mutant than in the wild-type strain in the presence of fructose (Fig. 4A and B).
FIG. 5.
Binding of FrtR to the promoter region of frtA. A 32P-labeled 131-bp DNA fragment upstream of frtA was incubated with or without recombinant FrtR protein. Samples in lanes 2 and 3 contained 100 ng and 300 ng of FrtR protein extract, respectively. Samples in lanes 4 to 7 contained 700 ng of FrtR protein extract.
Growth of strains with fructose.
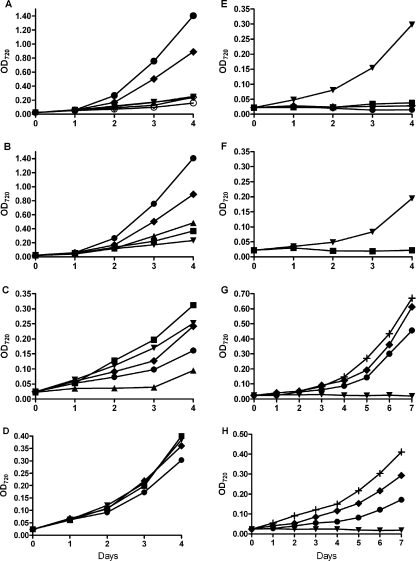
As shown in Fig. 2, neither the frtR mutant of A. variabilis nor the mutant of Anabaena sp. strain PCC 7120 with frtABC but without frtR grew in the dark with fructose, suggesting that FrtR might be an activator. However, the transcription data shown in Fig. 3 and 4 demonstrate that FrtR is a repressor. Furthermore, FrtR repressed frtABC even in the presence of fructose, suggesting that overexpression of these genes may be detrimental to cells. We grew the wild-type strain of A. variabilis and the frtR mutant with various concentrations of fructose in the light. The wild-type strain grew much better with fructose than without it, whereas the frtR mutant did not (Fig. 6A). For the wild-type strain, increasing concentrations of fructose, from 1 to 50 mM, supported increased growth rates, but 200 mM fructose decreased the growth rate to about the same rate as that with 1 mM fructose (Fig. 6B). For the frtR mutant, increasing concentrations of fructose did not increase the growth rate (Fig. 6C; note the difference in scale of the y axis). In fact, concentrations of fructose of >1 mM decreased growth, and 200 mM fructose almost completely inhibited growth. Strain JU377, in which the frtABC genes were overexpressed from the strong psbA promoter in a wild-type frtR+ background, grew poorly even in the absence of fructose and died after exposure to even 1 mM fructose (Fig. 6F). Thus, it appears that overexpression of frtABC in the frtR mutant leads to transport of fructose at levels that are toxic to the cells. In the light, the frtR mutant was apparently able to overcome this toxicity when the concentration of fructose was low, but in the dark, when the cells were dependent on fructose as a carbon source, even 5 mM fructose was toxic (Fig. 2, lane 3), suggesting that metabolism of fructose was involved in the toxic effect of high intracellular concentrations of fructose. Even without fructose, the poor growth of strain JU377, which overexpressed frtABC, suggested that excessive amounts of the transporter itself are deleterious to cell growth.
FIG. 6.
Growth of strains with fructose. The strains indicated for each panel were grown in AA/8 without fructose and then diluted in medium containing the concentrations of fructose indicated by the symbols on day 0. (A) Strains FD (A. variabilis wild type) (solid symbols) and BP301 (frtR mutant) (open symbols). (B) Wild-type strain FD. (C) A. variabilis BP301 (frtR mutant). (D) Anabaena sp. strain PCC 7120 with the frtRABC genes (strain BP291). (E) Anabaena sp. strain PCC 7120 with the frtABC genes (lacking frtR) (strain BP292). (F) Strain JU377, a strain of A. variabilis in which the frtABC genes are constitutively expressed from the strong psbA promoter. (G) Wild-type strain FD grown in the dark. (H) Anabaena sp. strain PCC 7120 with the frtRABC genes (strain BP291) grown in the dark. Fructose concentrations were as follows: ▾, 0 mM; ▪, 1 mM; ⧫, 5 mM; +, 10 mM; •, 50 mM; and ▴, 200 mM.
Anabaena sp. strain PCC 7120 mutant BP291 (with frtRABC) grew in the dark with fructose (Fig. 6H); however, it grew more slowly than A. variabilis FD (Fig. 6G). However, unlike the growth of A. variabilis FD in the light, the growth of BP291 in the light was not enhanced by fructose (Fig. 6D), and 50 mM fructose, which greatly stimulated growth of FD (Fig. 6A), slightly inhibited growth of BP291 (Fig. 6D). Anabaena sp. strain PCC 7120 with frtRABC grew in the light with 5 mM fructose and DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea], but growth was slow, particularly under nitrogen-fixing conditions, and increasing the fructose concentration above 5 mM did not help the growth (data not shown). Thus, although Anabaena sp. strain PCC 7120 mutant BP291 was able to transport and use fructose in the dark, in the light the strain did not use the fructose and grew photoautotrophically. Anabaena sp. strain PCC 7120 mutant BP292 (with frtABC but lacking frtR) did not grow in the dark with 5 mM fructose (Fig. 2, lane 2) and died in the light with as little as 1 mM fructose (Fig. 6E), showing even greater sensitivity to fructose than that of BP301, the A. variabilis strain lacking frtR (Fig. 6C). Unregulated expression of frtABC led to a complete inhibition of growth, suggesting that fructose is toxic to this strain in the light.
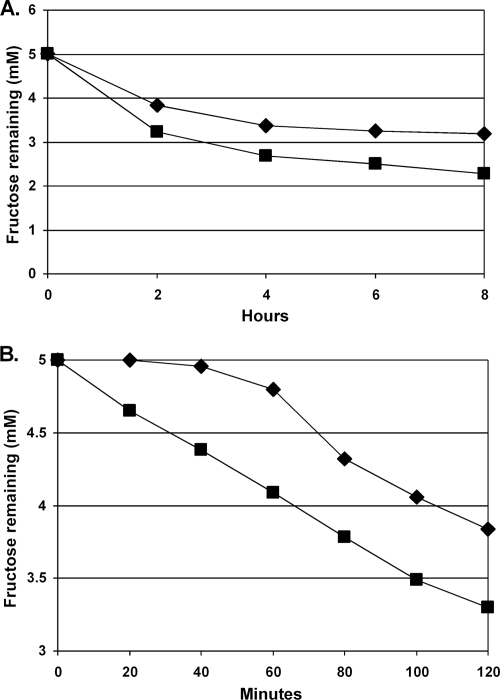
Uptake of fructose in the frtR mutant.
Overexpression of frtABC in BP301 (frtR mutant) might be expected to affect the rate of fructose uptake. We measured fructose uptake by the disappearance of fructose from the medium in the wild-type strain and in BP301 (frtR mutant). In the first 2 hours after the addition of fructose, the rate of uptake was greater in the BP301 mutant, but the initial high rate slowed for both strains about 2 hours after the addition of fructose (Fig. 7A). BP301 continued to take up fructose slightly faster than the wild-type strain for even up to 8 h. In the absence of the repressor, FrtR, high levels of expression of frtABC allowed the uptake of fructose to begin immediately upon its addition, while the wild-type strain showed a lag in uptake of about 40 min (Fig. 7B). This suggests that while frtABC is transcribed at low levels even in the absence of fructose, fructose is required for synthesis of sufficient FrtABC to efficiently transport fructose. In the absence of the repressor (strain BP301), sufficient FrtABC is made to allow the immediate transport of fructose.
FIG. 7.
Fructose transport in wild-type (⧫) and BP301 (frtR mutant) (▪) strains. Cells were grown in AA/8 to an OD720 of 0.250, fructose was added at time zero, and transport was measured as the disappearance of fructose from the medium over time (hours [A] or minutes [B]). Fructose was measured using a fructose assay kit (Sigma-Aldrich).
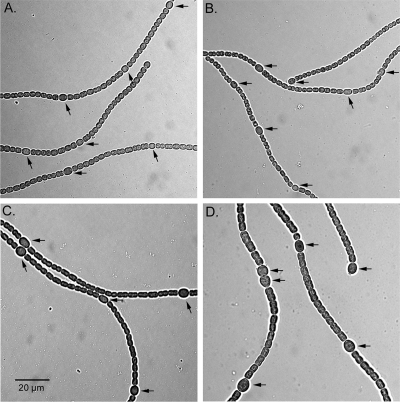
Filaments and heterocysts of fructose-grown cells.
Fructose has been reported to stimulate both the number and size of heterocysts of A. variabilis, consistent with the strain's ability to use fructose as a carbon source in the light (14, 21). Forty-eight hours after nitrogen stepdown, A. variabilis strain FD grown with 50 mM fructose had 11.9% heterocysts, compared to only 7.6% in filaments grown without fructose. In contrast, under the same growth conditions, BP291, the Anabaena sp. strain PCC 7120 mutant with frtRABC, produced 7.1% heterocysts with fructose and 6.6% heterocysts without fructose. Also, in contrast to the increased size of cells in filaments of A. variabilis FD grown with 5 mM fructose, filaments of BP291 showed no increase in size with 5 mM fructose (Fig. 8), and in contrast to the case for A. variabilis, they showed no increase in the rate of nitrogen fixation when grown with fructose (data not shown). These results support the growth data indicating that Anabaena sp. strain PCC 7120 mutant BP291 grown in the light does not use fructose as a carbon source.
FIG. 8.
Light micrographs of filaments of Anabaena sp. strain PCC 7120 with the frtABC genes (strain BP291) grown without (A) or with (B) 5 mM fructose and of filaments of A. variabilis FD grown without (C) or with (D) 5 mM fructose. The size scale is the same for all panels. Heterocysts are indicated by arrows.
DISCUSSION
The normal Frt− phenotype of Anabaena sp. strain PCC 7120 was complemented by the addition of the A. variabilis fructose transport genes, frtRABC. Therefore, the lack of a fructose transport system in Anabaena sp. strain PCC 7120 was the only barrier to heterotrophic growth on fructose in the dark. However, both Anabaena sp. strain PCC 7120 BP292, which constitutively expressed frtABC due to a lack of the repressor FrtR, and A. variabilis strain BP301, a frtR mutant, were unable to grow on fructose in the dark even though they expressed frtABC and transported fructose. These results indicate that the repressor, FrtR, is required for heterotrophic growth on fructose.
The evidence presented here suggests that the lack of FrtR caused excessive fructose uptake via the high-level constitutive expression of the transport genes and that this led to toxicity. Fructose is toxic in two cyanobacterial strains that have glucose transporters, i.e., Synechocystis sp. strain PCC 6714 and Synechocystis sp. strain PCC 6803, and expression of glcP from Synechocystis sp. strain PCC 6803 in the obligate photoautotroph Synechococcus sp. strain PCC 7942 results in glucose sensitivity (17, 51). Our results support this explanation by the requirement for the repressor, FrtR. With the inducer, fructose, expression of frtABC in a wild-type frtR+ background increased about 30-fold. However, in an frtR mutant background, expression was 400-fold higher than that in the wild-type strain. This indicates that in the wild-type strain, under inducing conditions with fructose, frtABC was still highly repressed by FrtR. Furthermore, A. variabilis strain JU377, which overexpressed frtABC in a wild-type frtR+ background, was extremely sensitive to fructose. This indicated that overexpression of the fructose transport genes in the presence of FrtR was sufficient to produce a fructose-sensitive phenotype. Finally, fructose toxicity resulted in impaired phototrophic growth as a function of fructose concentration in strains lacking a functional repressor but not in strains in which fructose uptake was regulated. Together, these findings indicate that fructose uptake must be tightly regulated in order to prevent toxic levels of fructose uptake. The fact that simply overexpressing the fructose transport proteins, even in the absence of fructose, greatly decreased growth suggests that at least part of the problem was the excessive amount of transporters made. However, the addition of fructose to the strains overexpressing the transport proteins resulted in much greater toxicity, which was proportional to the amount of fructose added, indicating that fructose or a metabolic product of fructose was toxic when present in large concentrations in the cell.
For Anabaena sp. strain PCC 7120, which normally cannot take up fructose, the addition of the frtRABC genes of A. variabilis allowed this strain to use fructose, but only in the dark. In contrast to the case for A. variabilis, fructose did not stimulate growth, increase heterocyst frequency, increase cell size, or stimulate nitrogen fixation in Anabaena sp. strain PCC 7120 with the frtRABC genes. Excessive entry of fructose into the Anabaena sp. strain PCC 7120 mutant with the frtABC genes but lacking frtR resulted in death. Expression of glcP in Synechococcus sp. strain PCC 7942 resulted in a glucose sensitivity in that strain (51). Analysis of the carbon catabolic pathways of the two Anabaena strains by use of KEGG, which is based on genome sequences, revealed no obvious differences (18); hence, a more detailed metabolomic analysis of fructose metabolism in the two strains may be necessary to understand the mechanism of fructose toxicity.
Although we demonstrated that the expression of frtABC is induced by fructose, we were unable to show that fructose directly affected the binding activity of FrtR to DNA in vitro. Our data suggest that FrtR remained bound to its target sequence irrespective of the presence or absence of fructose. Thus, either fructose has a low affinity for FrtR or binding of fructose to FrtR has little effect on the affinity of FrtR for DNA. Either condition would make it difficult to detect an FrtR-fructose interaction by our methods. It is also possible that the binding activity of FrtR is modulated by a secondary metabolite of fructose.
In N. punctiforme, specialized motile filaments called hormogonia are important in symbiosis (25, 26, 27). The hrm locus plays an important role in repressing further hormogonium differentiation after a functional symbiosis has been established between N. punctiforme and its host (8). The homologue of frtR in N. punctiforme, hrmR, has been shown to regulate itself and another gene of unknown function, hrmE. The activity of hrmR is modulated by an unidentified hormogonium repressing factor that is present in plant extracts (8). Immediately downstream of hrmE are the homologs of frtABC, namely, hrmB1, hrmB2, hrmT, and hrmP (25). It appears likely that hrmB1-hrmB2-hrmTP, like frtABC, is responsible for fructose transport in N. punctiforme. These genes are induced by the hormogonium repressing factor and are thus thought to be part of the hrm locus (25). The close similarity between frtABC and hrmB1-hrmB2-hrmTP (71 to 85% identity) and the proximity of hrmB1-hrmB2-hrmTP to other genes known to be involved in hormogonium formation suggest that fructose or a metabolite thereof might also be involved in regulating hormogonium differentiation. The fructose could be converted to a signaling metabolite that would then provide the signal to repress hormogonium differentiation and establish a lasting relationship with the plant. HrmR is the regulator of hrmR and hrmE (8), and both of these genes are negatively regulated by fructose (Fig. 3). It seems unlikely that HrmR directly regulates hrmB1-hrmB2-hrmTP because there is not a putative HrmR binding site upstream of hrmB1-hrmB2-hrmTP. A conserved 15-bp regulatory sequence upstream of hrmB1-hrmB2-hrmTP that is not bound by HrmR (26) and is absent in the intergenic region between frtR and frtA in A. variabilis might be the regulatory site for another regulatory protein controlling expression of hrmB1-hrmB2-hrmTP (26).
These data and other reports of sugar toxicity in other cyanobacteria (17, 51), combined with the apparent inability of the photoautotrophic strain Anabaena sp. strain PCC 7120 to use fructose when growing in the light, suggest that strains that are naturally capable of sugar transport and utilization have evolved mechanisms that allow them both to use sugars efficiently and to overcome sugar toxicity. These are of course likely to be metabolically linked processes. N. punctiforme and the free-living organism Anabaena azollae, which is genetically and morphologically very similar to A. variabilis, depend on sugar supplies from a plant when they are in a symbiotic association (26, 39). In the free-living state, these cyanobacteria retain the ability to use sugars and even show, in modified form, some of the characteristics of symbiosis (36, 37), including larger cells, more heterocysts, increased respiration, and increased nitrogen fixation, suggesting that some of the important changes associated with symbiosis are controlled by sugar metabolism in the cyanobacterium rather than by plant-derived factors. A. variabilis and Anabaena sp. strain PCC 7120 are very similar genetically, sharing about 95% nucleotide identity between homologous genes. They share about 5,000 homologous genes, but A. variabilis has about 650 genes that are not present in Anabaena sp. strain PCC 7120, and of these, about 240 have homologs in N. punctiforme (data calculated from information available at the Joint Genome Institute (JGI) integrated microbial genome website) (24). Among these 240 genes, which include the frtRABC genes and their homologs in N. punctiforme, are likely to be other genes that will provide answers to questions concerning how sugars are used by and may modify important physiological characteristics of true heterotrophic strains. Further system-level analysis, comparing transcriptomes, proteomes, and metabolomes for photoautotrophic versus heterotrophic strain growth with and without sugars, should help to provide answers to these interesting questions.
Supplementary Material
Acknowledgments
Support for this research was provided by National Science Foundation grants MCB-0416663 and CHE-610177.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldea, M. R., R. A. Mella-Herrera, and J. W. Golden. 2007. Sigma factor genes sigC, sigE, and sigG are upregulated in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1898392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, M. B., and D. I. Arnon. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, S. L., and L. McIntosh. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 1732761-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 977-84. [DOI] [PubMed] [Google Scholar]
- 5.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heynker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 295-113. [PubMed] [Google Scholar]
- 6.Braun-Howland, E. B., P. Lindblad, S. A. Nierzwicki-Bauer, and B. Bergman. 1988. Dinitrogenase reductase of nitrogenase in the cyanobacterial symbionts of three Azolla species: localization and sequence of appearance during heterocyst differentiation. Planta 176319-332. [DOI] [PubMed] [Google Scholar]
- 7.Brusca, J. S., M. A. Hale, C. D. Carrasco, and J. W. Golden. 1989. Excision of an 11-kilobase-pair DNA element from within the nifD gene in Anabaena variabilis heterocysts. J. Bacteriol. 1714138-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, E. L., F. C. Wong, and J. C. Meeks. 2003. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in the differentiation of hormogonia. Mol. Microbiol. 47573-582. [DOI] [PubMed] [Google Scholar]
- 9.Currier, T. C., and C. P. Wolk. 1979. Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J. Bacteriol. 13988-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68119-138. [DOI] [PubMed] [Google Scholar]
- 11.Flores, E., and G. Schmetterer. 1986. Interaction of fructose with the glucose permease of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 166693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franche, C., and G. Cohen-Bazire. 1987. Evolutionary divergence in the nifHDK gene region among nine symbiotic Anabaena azollae and between Anabaena azollae and some free-living heterocystous cyanobacteria. Symbiosis 3159-178. [Google Scholar]
- 13.Gebhardt, J. S., and S. A. Nierzwicki-Bauer. 1991. Identification of a common cyanobacterial symbiont associated with Azolla spp. through molecular and morphological characterization of free-living and symbiotic cyanobacteria. Appl. Environ. Microbiol. 572141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haury, J. F., and H. Spiller. 1981. Fructose uptake and influence on growth of and nitrogen fixation by Anabaena variabilis. J. Bacteriol. 147227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki, H., S. B. Williams, Y. Kitayama, M. Ishiura, S. S. Golden, and T. Kondo. 2000. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101223-233. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, B. 1990. Fructose utilization by the cyanobacterium Anabaena variabilis studied using whole filaments and isolated heterocysts. Arch. Microbiol. 15492-98. [Google Scholar]
- 17.Joset, F., T. Buchou, C.-C. Zhang, and R. Jeanjean. 1988. Physiological and genetic analysis of the glucose-fructose permeation system in two Synechocystis species. Arch. Microbiol. 149417-421. [Google Scholar]
- 18.Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, and Y. Yamanishi. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36D480-D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khudyakov, I. Y., and J. W. Golden. 2001. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J. Bacteriol. 1836667-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo, T., C. A. Strayer, R. D. Kulkarni, W. Taylor, M. Ishiura, S. S. Golden, and C. H. Johnson. 1993. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA 905672-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang, N. J., J. M. Krupp, and A. L. Koller. 1987. Morphological and ultrastructural changes in vegetative cells and heterocysts of Anabaena variabilis grown with fructose. J. Bacteriol. 169920-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons, E. M., and T. Thiel. 1995. Characterization of nifB, nifS, and nifU genes in the cyanobacterium Anabaena variabilis: NifB is required for the vanadium-dependent nitrogenase. J. Bacteriol. 1771570-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mannan, R. M., and H. B. Pakrasi. 1993. Dark heterotrophic growth conditions result in an increase in the content of photosystem II units in the filamentous cyanobacterium Anabaena variabilis ATCC 29413. Plant Physiol. 103971-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeks, J. C. 2006. Molecular mechanisms in the nitrogen fixing Nostoc-bryophyte symbiosis, p. 165-196. In J. Overmann (ed.), Molecular basis of symbiosis. Springer, Dordrecht, The Netherlands. [DOI] [PubMed]
- 26.Meeks, J. C. 2006. Molecular mechanisms in the nitrogen-fixing Nostoc-bryophyte symbiosis. Prog. Mol. Subcell. Biol. 41165-196. [DOI] [PubMed] [Google Scholar]
- 27.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 6694-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meeks, J. C., C. M. Joseph, and R. Haselkorn. 1988. Organization of the nif genes in cyanobacteria in symbiotic association with Azolla and Anthoceros. Arch. Microbiol. 15061-71. [DOI] [PubMed] [Google Scholar]
- 29.Nieva, M., and E. F. Valiente. 1996. Inorganic carbon transport and fixation in cells of Anabaena variabilis adapted to mixotrophic conditions. Plant Cell Physiol. 371-7. [Google Scholar]
- 30.Osanai, T., M. Azuma, and K. Tanaka. 2007. Sugar catabolism regulated by light- and nitrogen-status in the cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 6508-514. [DOI] [PubMed] [Google Scholar]
- 31.Osanai, T., S. Imamura, M. Asayama, M. Shirai, I. Suzuki, N. Murata, and K. Tanaka. 2006. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res. 13185-195. [DOI] [PubMed] [Google Scholar]
- 32.Osanai, T., Y. Kanesaki, T. Nakano, H. Takahashi, M. Asayama, M. Shirai, M. Kanehisa, I. Suzuki, N. Murata, and K. Tanaka. 2005. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J. Biol. Chem. 28030653-30659. [DOI] [PubMed] [Google Scholar]
- 33.Pils, D., C. Wilken, A. Valladares, E. Flores, and G. Schmetterer. 2004. Respiratory terminal oxidases in the facultative chemoheterotrophic and dinitrogen fixing cyanobacterium Anabaena variabilis strain ATCC 29413: characterization of the cox2 locus. Biochim. Biophys. Acta 165932-45. [DOI] [PubMed] [Google Scholar]
- 34.Pratte, B. S., and T. Thiel. 2006. High-affinity vanadate transport system in the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 188464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, P. M., H. Spiller, S. L. Albrecht, and K. T. Shanmugam. 1996. Photodissimilation of fructose to H2 and CO2 by a dinitrogen-fixing cyanobacterium, Anabaena variabilis. Appl. Environ. Microbiol. 621220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozen, A., H. Arad, M. Schönfeld, and E. Tel-Or. 1986. Fructose supports glycogen accumulation, heterocysts differentiation, N2 fixation and growth of the isolated cyanobiont Anabaena azollae. Arch. Microbiol. 145187-190. [Google Scholar]
- 37.Rozen, A., M. Schönfeld, and E. Tel-Or. 1988. Fructose-enhanced development and growth of the N2-fixing cyanobiont Anabaena azollae. Z. Naturforsch. 43c408-412. [Google Scholar]
- 38.Schmetterer, G., and E. Flores. 1988. Uptake of fructose by the cyanobacterium Nostoc sp. ATCC 29150. Biochim. Biophys. Acta 94233-37. [Google Scholar]
- 39.Shi, D.-J., and D. Hall. 1988. The Azolla-Anabaena association: historical perspective, symbiosis and energy metabolism. Bot. Rev. 54353-386. [Google Scholar]
- 40.Singh, A. K., and L. A. Sherman. 2005. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp. strain PCC 6803 and its requirement for heterotrophic growth. J. Bacteriol. 1872368-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, A. J. 1983. Modes of cyanobacterial carbon metabolism, p. 47-85. In N. G. Carr and B. A. Whitton (ed.), The biology of the cyanobacteria. University of California Press, Berkeley, CA.
- 42.Spiller, H., G. Bookjans, and K. T. Shanmugam. 1983. Regulation of hydrogenase activity in vegetative cells of Anabaena variabilis. J. Bacteriol. 155129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Summers, M. L., J. G. Wallis, E. L. Campbell, and J. C. Meeks. 1995. Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 1776184-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel, T., E. M. Lyons, J. C. Erker, and A. Ernst. 1995. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 929358-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiel, T., and C. P. Wolk. 1987. Conjugal transfer of plasmids to cyanobacteria. Methods Enzymol. 153232-243. [DOI] [PubMed] [Google Scholar]
- 46.Valiente, E. F., M. Nieva, M. C. Avendano, and E. S. Maeso. 1992. Uptake and utilization of fructose by Anabaena variabilis ATCC 29413. Effect on respiration and photosynthesis. Plant Cell Physiol. 33307-313. [Google Scholar]
- 47.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19259-268. [DOI] [PubMed] [Google Scholar]
- 48.Vioque, A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 253471-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, F. C., and J. C. Meeks. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148315-323. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, C.-C., M.-C. Durand, R. Jeanjean, and F. Joset. 1989. Molecular and genetic analysis of the fructose-glucose transport system in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 31221-1229. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, C. C., R. Jeanjean, and F. Joset. 1998. Obligate phototrophy in cyanobacteria: more than a lack of sugar transport. FEMS Microbiol. Lett. 161285-292. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman, W. J. H., B. Rosen, and T. A. Lumpkin. 1989. Enzymatic, lectin, and morphological characterization and classification of presumptive cyanobionts from Azolla Lam. New Phytol. 113497-503. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.