Abstract
A novel thermostable arylesterase, a 35-kDa monomeric enzyme, was purified from the thermoacidophilic archaeon Sulfolobus solfataricus P1. The optimum temperature and pH were 94°C and 7.0, respectively. The enzyme displayed remarkable thermostability: it retained 52% of its activity after 50 h of incubation at 90°C. In addition, the purified enzyme showed high stability against denaturing agents, including various detergents, urea, and organic solvents. The enzyme has broad substrate specificity besides showing an arylesterase activity toward aromatic esters: it exhibits not only carboxylesterase activity toward tributyrin and p-nitrophenyl esters containing unsubstituted fatty acids from butyrate (C4) to palmitate (C16), but also paraoxonase activity toward organophosphates such as p-nitrophenylphosphate, paraoxon, and methylparaoxon. The kcat/Km ratios of the enzyme for phenyl acetate and paraoxon, the two most preferable substrates among all tested, were 30.6 and 119.4 s−1·μM−1, respectively. The arylesterase gene consists of 918 bp corresponding to 306 amino acid residues. The deduced amino acid sequence shares 34% identity with that of arylesterase from Acinetobacter sp. strain ADP1. Furthermore, we successfully expressed active recombinant S. solfataricus arylesterase in Escherichia coli. Together, our results show that the enzyme is a serine esterase belonging to the A-esterases and contains a catalytic triad composed of Ser156, Asp251, and His281 in the active site.
Sulfolobus solfataricus P1 (DSM1616), an extreme thermoacidophilic archaeon, inhabits sulfur-rich volcanic hot springs in a high-temperature and low-pH environment (10). Enzymes from thermoacidophilic archaea have attracted considerable attention among researchers because of the significance of their evolutionary phylogeny, the structural and functional stability of their enzymes despite their extreme environments, and the broad applications in biotechnology and industry available due to their special properties (23, 44).
Esterases (ester hydrolases) (EC 3.1.1.x) are widely distributed in various organisms, including animals, plants, and microorganisms, and catalyze the hydrolysis and formation of ester bonds (43). Many of the esterases have wide substrate specificity, leading to the assumption that they have evolved to enable access to carbon sources or to be involved in catabolic pathways. Archaeal esterases also display a nonspecific substrate spectrum, hydrolyzing aliphatic esters, aromatic esters, and triacylglycerols. Esterases were initially classified into three groups on the basis of substrate specificity and sensitivity to various inhibitors. However, the enzymes exhibit overlapping substrate specificity (2, 6), leading to problems in identification and classification (3, 31). Arylesterases, or A-esterases, are not inhibited by organophosphates such as paraoxon, whereas carboxylesterases, or B-esterases (serine esterases), are inhibited by organophosphates, and C-esterases are inhibited by organophosphates and eserine.
Bacterial lipolytic enzymes, including carboxylesterases (EC 3.1.1.1) and lipases (EC 3.1.1.3), have been shown to usually possess a typical Ser-Asp-His catalytic triad and the general motif Gly-x-Ser-x-Gly (GXSXG) around the active-site serine (5, 8), although the esterase from Streptomyces scabies contains a catalytic Ser-His dyad instead of the common Ser-Asp-His triad (49). In relation to arylesterases, it was reported that the extracellular arylesterase from Vibrio mimicus has the Ser-Asp-His catalytic triad (46) but contains a GDSLS motif not identical to the active-site serine-containing consensus motif (GXSXG) found in a variety of lipases and esterases (19). The arylesterase from Agrobacterium radiobacter also contains the Ser-Asp-His catalytic triad and a GDSLT motif (41). On the other hand, the arylesterase from Acinetobacter sp. strain ADP1 contains the typical catalytic triad, Ser-Asp-His, and the general motif, GXSXG (24).
Arylesterases (EC 3.1.1.2) showing paraoxonase activity were less understood than other esterases, especially carboxylesterases and lipases. Until recently, most of the arylesterases and/or paraoxonases reported were from mammalian sources. The mammalian arylesterases/paraoxonases play important roles in the detoxification of the organophosphoric compounds as well as in the prevention of artherosclerosis (1). The enzymes are also used as diagnostic indicators for liver cirrhosis and carcinoma (25) and as oxidative stress markers for patients with cardiac syndrome X (20). A few arylesterases from insect pests displaying resistance against organophosphate insecticides such as paraoxon and chlorpyrifos-oxon were reported (17, 50). There were also a few reports on bacterial arylesterases (26, 29) and no reports regarding the paraoxonase activity. Bacterial arylesterases may be used for similar industrial and environmental purposes, especially considering the fact that they have broad substrate specificities, but little is known about their genetics and biochemistry. In addition, no arylesterase showing paraoxonase activity from archaea was reported, except for two phosphotriesterases having paraoxonase activity (34, 38).
In this study, we report the purification and characterization of a thermostable arylesterase (accession number FM205057) from S. solfataricus P1 (DSM1616), the isolation of the gene, and the expression of the recombinant enzyme in Escherichia coli. The purified arylesterase showed remarkable stability against high temperature and denaturants such as sodium dodecyl sulfate (SDS), urea, and detergents and contains a catalytic triad composed of Ser156, Asp251, and His281 in its active site. To our knowledge, this is the first arylesterase displaying a paraoxonase activity isolated and identified so far from archaea.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
S. solfataricus P1 (DSM1616) was purchased from American Type Culture Collection (ATCC). S. solfataricus P1 (DSM1616) was aerobically grown at 75°C and pH 4.0 in a 5-liter fermentor with moderate stirring (120 rpm). The medium containing 0.2% (wt/vol) yeast extract, and the mineral base was used according to the description of ATCC medium formulations. Cells were harvested by centrifugation (4,000 × g) for 45 min at 4°C. These cells were stored at −20°C. E. coli BL21(DE3) was used as a host for gene expression. E. coli BL21(DE3) cells containing the recombinant DNA were routinely grown at 37°C on Luria-Bertani (LB) medium with ampicillin (100 μg/ml). The vectors utilized were pGEM-T vector system I for sequencing of PCR products and pET-11d for expression.
Enzyme purification.
All purification steps were performed at 4°C. The fractions containing arylesterase activity were preferentially traced by the formation of a clear zone on the tributyrin-emulsified agar plates as described below. A 40-g batch of frozen cells was thawed and resuspended in 10 mM sodium phosphate buffer (pH 7.0) (buffer A) to make a 20% homogeneous solution. The cells were disrupted by passing them three times through a chilled French pressure cell (Thermo Spectronic Inc.) at a pressure of 1,200 lb/in2. Cell debris was removed by centrifugation (100,000 × g) for 75 min at 4°C. The supernatant after centrifugation was collected as cell extract. Next, 2% (wt/vol) protamine sulfate in buffer A was slowly added with gentle stirring to the cell extract in an ice bath to a final concentration of 0.4% to remove nucleic acids, and then the solution was left in ice for 1 h as it was. The solution was centrifuged at 40,000 × g for 50 min. The supernatant was applied to a column of DEAE-cellulose (5.5 by 30 cm) equilibrated with buffer A and eluted with 400 ml of a linear gradient of 0 to 0.15 M NaCl in buffer A. The fractions containing arylesterase activity were pooled. The salt concentration of the pooled enzyme solution was adjusted to 2 M with powdered NaCl and loaded onto a butyl-Sepharose column (3.0 by 15 cm) equilibrated with buffer A containing 2 M NaCl. The solution was then washed with buffer A and subsequently eluted with a series of step gradients of 150 ml of buffer A containing 30%, 40%, 50%, 60%, and 70% (vol/vol) ethylene glycol, respectively. The fractions containing arylesterase activity were collected. The collected solution was dialyzed twice to remove ethylene glycol against 5 liters of buffer A for 18 h. The dialyzed protein solution was applied to a Q-Sepharose column (3.0 by 15 cm) equilibrated with buffer A and eluted with 300 ml of a linear gradient of 0 to 0.2 M NaCl in buffer A. The fractions containing arylesterase activity were collected. The collected solution was dialyzed twice against 5 liters of buffer A for 18 h to remove NaCl. The dialyzed protein solution was applied to a hydroxyapatite column (2.0 by 10 cm) equilibrated with buffer A and eluted with a linear 160-ml gradient of 10 to 100 mM sodium phosphate buffer (pH 7.0). The fractions containing arylesterase activity were collected and used as purified enzyme. The protein concentration was determined by the method of Bradford (9) using bovine serum albumin as a standard.
Enzyme assays.
The standard arylesterase assay was carried out using phenyl acetate (PA) as a substrate according to the modified method of Aviram et al. (7). The reaction was allowed to proceed for 1 or 2 min, and the initial rate of hydrolysis was determined by measuring the amount of phenol released from PA at 270 nm at 60°C using a spectrophotometer (Shimatzu UV2401PC) fitted with the constant-temperature controlled cell holder. The substrate solution was prepared by mixing 0.1 ml of a methanolic solution of PA (5 mM) and 0.8 ml of prewarmed 100 mM sodium phosphate buffer (pH 7.0) in cells held in the cell holder at 60°C or at a desired temperature. After 1 min, 0.1 ml enzyme solution (8 μg/ml) was added to this substrate solution. Immediately after incubation for 1 min, the reaction was started and measured. The same buffer solution without the enzyme was used in a blank. Nonenzymatic hydrolysis of PA was less than 0.1% during the initial 1 or 2 min. Each determination was performed at least in triplicate. One enzyme unit is defined as the amount of enzyme releasing 1 μmol of phenol per min from PA at 60°C.
For the determination of substrate specificity, the following substrates were tested: aromatic esters such as PA and α-naphthyl acetate (NA); p-nitrophenyl (PNP) esters such as PNP-butyrate (C4), PNP-caproate (C6), PNP-caprylate (C8), PNP-caprate (C10), PNP-laurate (C12), and PNP-palmitate (C16); and organophosphates such as PNP-phosphate, paraoxon, and methylparaoxon. Arylesterase activity was determined by using aromatic esters such as PA and α-NA as substrates. The measurement of the arylesterase activity using PA was performed as described previously. However, arylesterase activity using α-NA was determined by measuring the amount of α-naphthol released from α-NA at 600 nm according to the modified method of Morana et al. (35). The assay mixture contained 0.8 ml of 100 mM sodium phosphate buffer (pH 7.0), 0.05 ml of α-NA dissolved in methanol, and 0.1 ml enzyme solution (8 μg/ml); the reaction mixture was incubated for 5 min at 60°C. The reaction was determined after the addition of 0.05 ml Fast Blue BB-SDS solution (10 mg/ml Fast Blue BB in 20% [wt/vol] SDS) followed by incubation for another 5 min at 37°C. One unit of arylesterase is defined as the amount of the enzyme required to liberate 1 μmol of α-naphthol per min. Carboxylesterase activity was assessed by measuring the initial rate of hydrolysis of PNP esters containing unsubstituted fatty acids from butyrate (C4) to palmitate (C16) to yield p-nitrophenol at 405 nm and 60°C (45). The basal reaction mixture included 0.1 ml of freshly prepared and prewarmed PNP ester solution, 0.1 ml enzyme solution (8 μg/ml), and 0.8 ml of prewarmed 100 mM sodium phosphate buffer (pH 7.0) containing sodium taurocholate (2 mg/ml) and gum arabic (1 mg/ml) as emulsifiers at 60°C. One unit of carboxylesterase is defined as the amount of the enzyme required to liberate 1 μmol of p-nitrophenol per min. Paraoxonase activity was determined by measuring the initial rate of hydrolysis of organophosphates such as PNP-phosphate, paraoxon, and methylparaoxon to yield p-nitrophenol at 412 nm at 60°C by modifying the method of Aviram et al. (7). The basal reaction mixture contained 0.1 ml of organophosphate solution (1 mM), 0.1 ml enzyme solution (8 μg/ml), and 0.8 ml of 100 mM sodium phosphate buffer (pH 7.0). One unit of paraoxonase is defined as the amount of the enzyme required to liberate 1 μmol of p-nitrophenol per min. The extinction coefficients of p-nitrophenol were determined prior to the measurements under every condition. Activities for all substrates were determined similarly. The examination of the substrate specificity was performed by using PA (0.01 to 1 mM), α-NA (0.01 to 1 mM), PNP-butyrate (0.02 to 5 mM), PNP-caproate (0.01 to 1 mM), PNP-caprylrate (0.01 to 1 mM), PNP-caprate (0.01 to 1 mM), PNP-laurate (0.02 to 2 mM), PNP-palmitate (0.02 to 5 mM), PNP-phosphate (0.2 to 5 mM), paraoxon (0.001 to 1 mM), and methyl paraoxon (0.2 to 5 mM) as substrates. The extent of nonspecific hydrolysis varied with substrates, but it was typically less than 5% during the initial 1 or 2 min. Each determination was performed at least in triplicate.
Activity staining.
Activity staining was used preferentially for tracing arylesterase activity during the purification procedure. The staining was carried out on tributyrin-emulsified agar plates containing 1.5% (wt/vol) agar powder, 0.01% (wt/vol) rhodamine B, and 1% (vol/vol) tributyrin in buffer A as described previously (22). The presence of active arylesterase in the sample was evidenced from the formation of a clear zone on a turbid tributyrin-emulsified agar plate at 75°C. Tricaprylin, trimyristin, tripalmitin, and triolein were also used in the same way for the preferential detection of the substrate specificity.
Another activity staining method (35), which is useful in identifying the presence of active arylesterase, was employed after SDS-polyacrylamide gel electrophoresis (SDS-PAGE). A renaturation procedure was carried out to recover the enzyme activity in a gel. The gel was washed twice in 2-propanol-50 mM Tris-HCl (pH 7.0) (1:4, vol/vol) for 15 min, subsequently rinsed three times in 50 mM Tris-HCl (pH 7.0) buffer after each 15 min, and finally rinsed with water. The gel was stained in the dark at 37°C by immersing it in a solution consisting of 100 ml of 50 mM sodium phosphate buffer (pH 7.5) and 50 mg of Fast Blue BB plus 1 ml of acetone solution containing 10 mg α-NA until a dark-gray band appeared on the gel. The stained gel was rinsed with water and fixed in 3% (vol/vol) acetic acid.
Identification and determination of the molecular weight of the enzyme.
Native PAGE (28) was performed using a 10% (wt/vol) gel to assess the purity of the purified arylesterase. The molecular weight of the purified enzyme was determined on a 12.5% SDS-polyacrylamide gel (28) with and without mercaptoethanol in the sample buffer using low-molecular-weight standards (Amersham Biosciences). Gels were stained with silver (36). The molecular weight of the native protein was also determined by high-pressure liquid chromatography (HPLC) (GPC column [Protein-PakTM 300SW, 7.8 by 300 mm; Waters]) using molecular weight standards (Sigma MW-GF-200 kit).
Effect of pH and temperature on enzyme activity.
The optimum pH and temperature for the arylesterase activity were determined using PA. The effect of pH on the arylesterase activity was examined at 60°C, with the pH in the range of 3.0 to 10.0. The following buffers (100 mM) were used: sodium citrate (pH 3.0 to 6.0), sodium phosphate (pH 6.0 to 8.0), Tris-HCl (pH 8.0 to 9.0), and sodium bicarbonate (pH 9.0 to 10.0). The desired pH of each buffer was adjusted at 60°C. The effect of temperature on the arylesterase activity was investigated at the optimum pH (pH 7.0) in the temperature range from 30°C to 98°C at 2°C intervals near the optimum temperature.
Examination of stability against temperature and chemicals.
The thermal stability of the purified arylesterase was examined using the standard arylesterase assay after incubation of the enzyme (8 μg/ml) for designated time periods (0 to 120 h) at three different temperatures (50°C, 70°C, and 90°C). The stability of the arylesterase against several compounds {the detergents Tween 20, Tween 80, Triton X-100, Lubrol, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and SDS; the water-miscible organic solvents methanol, ethanol, 2-propanol, acetonitrile, and dimethyl sulfoxide; urea; and NaCl} was investigated by measuring the residual activity using the standard arylesterase assay after incubation of the enzyme (8 μg/ml) with each compound for 60 min at 30°C and 70°C. Blank samples were prepared by incubation with the buffer solution instead of the enzyme under the same conditions. Control experiments were performed in the absence of each compound under the same conditions. Each measurement was carried out with one or two different concentrations of the compounds: 1% and 5% (wt/vol) detergents, 50% and 90% (vol/vol) organic solvents, 4 M and 8 M urea, and 2 M NaCl.
Analysis of N-terminal sequence.
The analysis of the N-terminal amino acid sequence was carried out using an automatic amino acid sequencer (Hewlett-Packard protein sequencer HP241) according to the manufacturer's instructions.
Effect of inhibitors and divalent cations on enzyme activity.
The inhibitory effect of chemical modifiers that are specific to particular amino acids (such as pyridoxal 5′-phosphate [PLP] to Lys, phenylglyoxal [PGO] to Arg, HgCl2 and p-chloromercuribenzoate [PCMB] to Cys, diethyl pyrocarbonate [DPC] to His, and diisopropylfluorophosphate [DFP] and phenylmethylsulfonyl fluoride [PMSF] to Ser) on the purified arylesterase was examined. Enzyme activity was measured using the standard arylesterase assay after one to three different concentrations (0.05, 0.5, and 5 mM) of each inhibitor were added to the enzyme (8 μg/ml) and then incubated for 30 min at 30°C. In addition, 0.5 mM and 5 mM paraoxon or eserine, which are known as indicators for the classification of esterases, were also examined under the same conditions as described above. To investigate the effect of divalent cations on arylesterase activity, 5 mM of each divalent cation (CaCl2, CuSO4, FeSO4, MnCl2, MgCl2, and ZnSO4) was separately added to the enzyme (8 μg/ml) and incubated for 30 min at 30°C. In order to clarify whether divalent cations are required for the reaction, 10 mM EDTA was added to the enzyme (8 μg/ml) and incubated for 60 min at 75°C. The residual activities were measured using the standard arylesterase assay after incubation.
Identification of the sequence of the S. solfataricus arylesterase gene.
Routine DNA manipulations were performed by standard methods (42). Two oligonucleotide primers were designed to identify the sequence of the S. solfataricus arylesterase gene. The forward primer (5′-TAYTAYAAMGCNAAYAUHAUH-3′, where Y is C or T; M is T or C; H is A, C, or T; and N is A, T, G, or C) was deduced from the N-terminal amino acid sequence YYKANII of the purified protein. The reverse primer (5′-NGGRTCRTAYTCNGCNGTNAC-3′, where R is A or G; Y is C or T; and N is A, T, G, or C) was derived from the consensus sequence VTAEYDP obtained from the multiple alignments of archaeal esterase genes. A DNA fragment, part of the arylesterase gene, was obtained by PCR using these primers and S. solfataricus genomic DNA as a template, and the PCR product was then sequenced. The PCR conditions were as follows: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 60 s, 50°C for 30 s, and 72°C for 60 s; and finally 1 cycle of 72°C for 7 min. The PCR products were separated by electrophoresis in a 0.7% agarose gel, and the DNA fragments were purified using the gel purification kit. The complete base sequence of the S. solfataricus arylesterase gene from this sequenced fragment was obtained by PCR using the DNA Walking SpeedUp Premix kit (Seegene) according to the protocol from manufacturer with reference to the method of Park et al. (37). This kit was used to obtain unknown sequences that lie adjacent to the known sequence of the PCR product obtained above. This PCR technique was employed to directly amplify unknown sequences using six target-specific primers (N-terminal side, TSP1 [5′-GGA TAA GAT TGC CAC ATT-3′], TSP2 [5′-GAC ATC AAT TTG CCA GTA G-3′], and TSP3 [5′-CCT CAG GCT ATA GTC GTT-3′]; C-terminal side, TSP1 [5′-AAT GTG GCA ATC TTA TCC-3′], TSP2 [5′-GAT AGC ATC TTC AGT CTC-3′], and TSP3 [5′-GCC TTT GCC AAC AAT TCA-3′]) in known sequence and DNA walking annealing control primers and a universal primer in unknown sequence, which were designed by manufacturer to capture unknown target sites with high specificity. The DNA fragments used to obtain unknown sequences were cloned into pGEM-T vector system I and sequenced.
Construction of S. solfataricus arylesterase plasmids.
For the expression of recombinant arylesterase, two primers were constructed using the information from the complete coding sequence of the arylesterase gene obtained previously and considering the insertion of the gene into the NcoI site of the expression vector pET-11d. The constructed primers were 5′-AATCCATGGAAATGCCGTTGGATCCTGA-3′ (forward) and 5′-CGCCCATGGTTACTTAAAAATGTCCTTAAGTA-3′ (reverse), where the underlined letters indicate the initiation codon in the forward primer and the termination codon in the reverse primer. The S. solfataricus arylesterase gene containing an NcoI site at both ends was synthesized by PCR using S. solfataricus genomic DNA as a template. The PCR product was digested with NcoI and then inserted into the expression vector, pET-11d, digested with the same enzyme to construct the recombinant DNA plasmid, pSsoAre. In addition, this pSsoAre was used as a template DNA to create mutations (Ser156Ala, Asp251Asn, His281Asn, Cys105Ser, Cys107Ser, and Cys129Ser) in the expected catalytic triad, Ser-Asp-His, and in all three cysteines of the S. solfataricus arylesterase gene by site-directed mutagenesis. The mutagenic oligonucleotide primers were designed, and PCR based on the QuikChange II site-directed mutagenesis kit for the construction of S. solfataricus arylesterase mutant plasmids (pSsoAreSer156Ala, pSsoAreAsp251Asn, pSsoAreHis281Asn, pSsoAreCys105Ser, pSsoAreCys107Ser, and pSsoAreCys129Ser) was carried out using pSsoAre as template DNA and forward and reverse primers with lengths of 29 to 41 bases (Table 1) mutated at the desired sites (Ser156Ala, Asp251Asn, His281Asn, Cys105Ser, Cys107Ser, and Cys129Ser) of the S. solfataricus arylesterase gene according to the guidelines and protocol recommended by Stratagene Co. The PCR conditions were as follows: 1 cycle of 30 s at 95°C followed by 12 cycles of 30 s at 95°C, 1 min at 55°C, and 7 min at 68°C. The products were digested with 1 μl of DpnI (10 U/μl) at 37°C for 1 h to digest the parental template DNA, and then the PCR products were transformed into XL1-Blue supercompetent cells to obtain the mutant plasmids pSsoAreSer156Ala, pSsoAreAsp251Asn, pSsoAreHis281Asn, pSsoAreCys105Ser, pSsoAreCys107Ser, and pSsoAreCys129Ser. Finally, insert S. solfataricus arylesterase genes from these mutant plasmids were sequenced to confirm the correct mutations.
TABLE 1.
Complementary mutagenic primers for the generation of mutant arylesterases
| Primera | Sequenceb |
|---|---|
| Ser156Ala-f | 5′-TGCCACATTTGGAATAGCTGCTGGAGGTAATTTG-3′ |
| Ser156Ala-r | 5′-CAAATTACCTCCAGCAGCTATTCCAAATGTGGCA-3′ |
| Asp251Asn-f | 5′-GTTACTGCAGAGTACAATCCTTTGAGGGATC-3′ |
| Asp251Asn-r | 5′-GATCCCTCAAAGGATTGTACTCTGCAGTAAC-3′ |
| His281Asn-f | 5′-AGAGTAAATGGCAATGTTAATGCATTTTTGGGTTCACC-3′ |
| His281Asn-r | 5′-GGTGAACCCAAAAATGCATTAACATTGCCATTTACTCT-3′ |
| Cys105Ser-f | 5′-CAAGAATTCTCTCAAACTCCAGCGAGTGCACAGTCATATCG-3′ |
| Cys105Ser-r | 5′-CGATATGACTGTGCACTCGCTGGAGTTTGAGAGAATTCTTG-3′ |
| Cys107Ser-f | 5′-CAAACTCCTGCGAGAGCACAGTCATATCG-3′ |
| Cys107Ser-r | 5′-CGATATGACTGTGCTCTCGCAGGAGTTTG-3′ |
| Cys129Ser-f | 5′-CAACCGCAGTCTATGATAGTTTCAATGCTATAGTG-3′ |
| Cys129Ser-r | 5′-CACTATAGCATTGAAACTATCATAGACTGCGGTTG-3′ |
f and r, forward and reverse primers, respectively.
Boldface indicates mutated nucleotide residues.
Expression and purification of recombinant and mutant arylesterases.
The recombinant DNA plasmid, pSsoAre, and the mutant plasmids, pSsoAreSer156Ala, pSsoAreAsp251Asn, pSsoAreHis281Asn, pSsoAreCys105Ser, pSsoAreCys107Ser, and pSsoAreCys129Ser, were transformed into E. coli BL21(DE3) competent cells for expression and cultivated in 1 liter of LB broth supplemented with ampicillin (100 mg/liter) at 37°C. The expression vector pET-11d was used as a control. When the A600 of the culture reached 0.5, expression was induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Recombinant and mutant arylesterases were subsequently purified as follows. IPTG-induced BL21(DE3) cells that had been harvested from a 1-liter culture were suspended in 50 ml of buffer A containing 0.5 M NaCl. The cell extract was obtained by sonication for 5 min on ice and then heated two times for 30 min at 75°C. After each heating, centrifugation was performed at 10,000 × g for 15 min to remove the denatured proteins from E. coli, and the supernatant was applied to a butyl-Sepharose column (3.0 by 15 cm) that was equilibrated with buffer A containing 2 M NaCl. The expressed arylesterase and the mutant arylesterases were eluted with 40% (vol/vol) ethylene glycol. The fractions containing arylesterase activity were collected and dialyzed twice against 5 liters of buffer A for 18 h to remove ethylene glycol. The dialyzed protein solution was used as purified enzyme solution.
RESULTS
Purification and physical properties of S. solfataricus arylesterase.
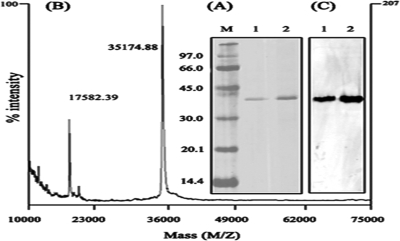
The purification procedure reported here yielded electrophoretically homogeneous arylesterase in five steps starting from cell extracts of S. solfataricus P1 (DSM1616) (Table 2). The arylesterase was purified 858.5-fold, with a yield of 5.6%. The enzyme showed a specific activity of 5,580 units/mg for the hydrolysis of PA at 60°C and pH 7.0. The homogeneity of the enzyme was confirmed on an SDS-polyacrylamide gel with a single band of approximately 37 kDa (Fig. 1A, lane 1). The molecular weight of this enzyme, determined by using and HPLC sizing column, was almost identical to the molecular weight obtained by SDS-PAGE and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analyses (Fig. 1B). These results indicate that the enzyme is monomeric. In addition, SDS-PAGE followed by activity staining using α-NA showed that the purified enzyme has an esterase activity (Fig. 1C, lane 1). This result indicates that heating the sample buffer containing the purified enzyme, 1% β-mercaptoethanol, and 1% SDS for 1 min in boiling water before performing a usual SDS-PAGE does not appear to significantly affect the enzyme activity (see below).
TABLE 2.
Purification of the arylesterase from Sulfolobus solfataricus P1
| Step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 6,120 | 39,700 | 6.5 | 100 | 1 |
| Protamine sulfate | 3,710 | 28,650 | 7.7 | 72.2 | 1.2 |
| DEAE-cellulose | 236.9 | 10,300 | 43.5 | 25.9 | 6.7 |
| Butyl-Sepharose | 18.9 | 5,872 | 310.7 | 14.8 | 47.8 |
| Q-Sepharose | 1.7 | 3,830 | 2,252.9 | 9.6 | 346.6 |
| Hydroxyapatite | 0.4 | 2,232 | 5,580.0 | 5.6 | 858.5 |
FIG. 1.
Electrophoretic and MS analyses of S. solfataricus arylesterase and recombinant S. solfataricus arylesterase. (A and C) Samples were loaded on an SDS-polyacrylamide gel in duplicate, and the gel was cut following the run. One portion was stained with silver (A), and the other was stained by the activity staining method using the chromogenic substrate α-NA after renaturation (C). M, molecular weight standards; lanes 1, purified S. solfataricus arylesterase; lanes 2, purified recombinant S. solfataricus arylesterase. (B) MALDI-TOF MS spectrum of the purified S. solfataricus arylesterase.
Catalytic properties of S. solfataricus arylesterase.
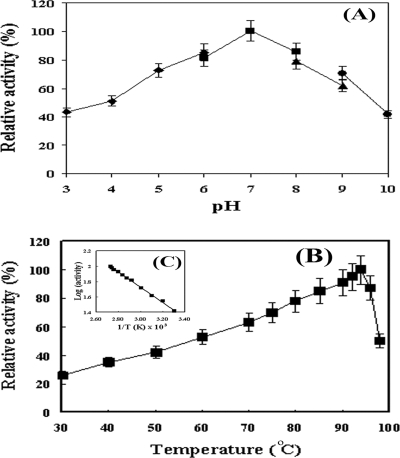
The optimum pH and temperature of the arylesterase activity were determined by standard arylesterase assay using PA as a substrate. The arylesterase displayed the maximum activity at pH 7.0 (Fig. 2A) and at 94°C (Fig. 2B). The activation energy calculated from the Arrhenius plot was 34.9 kJ/mol in the range of 30 to 94°C (Fig. 2C). The enzyme, however, was rapidly inactivated at above 94°C.
FIG. 2.
Effects of pH and temperature on the activity of the purified S. solfataricus arylesterase. The effects of pH and temperature on the activity of the purified arylesterase were determined spectrophotometrically using PA as a substrate. (A and B) The enzyme activity was examined at 60°C and the indicated pHs (A) or at pH 7.0 and the indicated temperatures (B). The pH was obtained using 100 mM sodium citrate buffer (♦) for the pH range of 5.0 to 6.0 100 mM sodium phosphate buffer (▪) for the pH range of 6.0 to 8.0, 100 mM Tris-HCl buffer (▴) for the pH range of 8.0 to 9.0, and 100 mM sodium bicarbonate buffer (•) for the pH range of 9.0 to 11.0. (C) Arrhenius plot of values from panel B. Error bars indicate standard deviations.
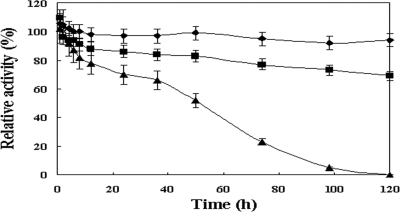
The thermostability of the arylesterase was evaluated using the standard arylesterase assay at three different temperatures (50°C, 70°C, and 90°C) with increasing incubation time up to 120 h (Fig. 3). Most of the enzyme activity was maintained for 5 days at 50°C. Although the enzyme activity gradually decreased with time at higher temperatures, approximately 70% of the enzyme activity remained after 5 days at 70°C, and 52% of it was still preserved after 50 h at 90°C. However, at 90°C after 5 days, the enzyme was completely inactivated.
FIG. 3.
Stability of the purified S. solfataricus arylesterase at different temperatures. The residual activity was determined after incubation of the enzyme for the indicated times at 50°C (♦), 70°C (▪), or 90°C (▴). The activity measurement was carried out using the standard enzyme assay. Error bars indicate standard deviations.
The effects of detergents, organic solvents, and urea on the arylesterase activity were investigated using the standard enzyme assay. Although the enzyme activity was gradually decreased with the increase of the concentration of each compound and with the increase of the incubation temperature, on the whole the enzyme displayed considerable stability against these compounds (Table 3). Incubation with Tween 20, Lubrol, or CHAPS for 60 min, regardless of concentrations (1% and 5%) and temperatures (30°C and 70°C), had little effect on the enzyme; instead, the enzyme was activated by incubation with 1% of these detergents, which might be due to partial unfolding of the enzyme. However, after incubation for 60 min with Triton X-100, the enzyme activity was remarkably decreased with increased temperature and concentration. In the case of incubation with 1% SDS, about half of the activity was retained even after incubation for 60 min at 70°C. However, the addition of 5% SDS completely eliminated the enzyme activity at 70°C. The denaturation effects of the organic solvents tested on the enzyme activity were not significant at 30°C, but at 70°C approximately half of the enzyme activity was still preserved after incubation with 90% organic solvents. Furthermore, the enzyme was not significantly inactivated by 8 M urea at 70°C. These data clearly showed that the purified arylesterase possesses remarkable stability against chemicals that can otherwise affect protein folding. However, we cannot rule out the possibility that the unfolded enzyme in 8 M urea was refolded in the assay buffer upon dilution.
TABLE 3.
Effects of various compounds on stability of the arylesterase from S. solfataricus P1 at 30°C and 70°C
| Compound | Concn | % Residual activity (mean ± SD) after incubation for 60 min ata:
|
|
|---|---|---|---|
| 30°C | 70°C | ||
| None | 100 | 100 | |
| Detergents | |||
| Tween 20 | 1% | 167 ± 13 | 109 ± 5 |
| 5% | 93 ± 1 | 92 ± 1 | |
| Tween 80 | 1% | 81 ± 7 | 66 ± 5 |
| 5% | 56 ± 4 | 21 ± 4 | |
| Triton X-100 | 1% | 95 ± 2 | 41 ± 3 |
| 5% | 14 ± 4 | 11 ± 2 | |
| Lubrol | 1% | 114 ± 7 | 102 ± 2 |
| 5% | 92 ± 7 | 91 ± 5 | |
| CHAPS | 1% | 187 ± 8 | 182 ± 6 |
| 5% | 110 ± 3 | 109 ± 3 | |
| SDS | 1% | 68 ± 3 | 47 ± 1 |
| 5% | 7 ± 2 | 0 | |
| Organic solvents | |||
| Methanol | 50% | 106 ± 4 | 76 ± 4 |
| 90% | 96 ± 1 | 48 ± 2 | |
| Ethanol | 50% | 115 ± 2 | 78 ± 2 |
| 90% | 98 ± 2 | 63 ± 4 | |
| 2-Propanol | 50% | 108 ± 3 | 86 ± 6 |
| 90% | 92 ± 7 | 55 ± 5 | |
| Acetonitrile | 50% | 92 ± 3 | 61 ± 3 |
| 90% | 86 ± 1 | 43 ± 1 | |
| Dimethyl sulfoxide | 50% | 121 ± 5 | 97 ± 3 |
| 90% | 110 ± 2 | 71 ± 6 | |
| Other | |||
| Urea | 4 M | 102 ± 7 | 94 ± 5 |
| 8 M | 91 ± 4 | 88 ± 4 | |
| NaCl | 2 M | 80 ± 11 | 76 ± 6 |
The enzyme was incubated at 30°C and 70°C in 100 mM sodium phosphate buffer (pH 7.0) with different concentrations of compounds. After 60 min, aliquots were removed from each incubation mixture and assayed for residual activity using the standard arylesterase assay.
The substrate specificity of the purified arylesterase was investigated using three kinds of substrates: aromatic esters, PNP esters, and organophosphates. As shown in Table 4, the resulting specificity constants (kcat/Km) for the aromatic esters PA and α-NA were 30.6 s−1·μM−1 for PA and 15.5 s−1·μM−1 for α-NA, indicating that PA was a better substrate than α-NA. Among the PNP esters tested, the highest specificity constant (15.1 s−1·μM−1) was obtained with PNP-caproate (C6), indicating that PNP-caproate (C6) was the best substrate. The specificity constants for organophosphates such as PNP-phosphate, paraoxon, and methylparaoxon are shown in Table 4. After summing up all of these specificity constants toward the substrates tested, paraoxon was determined to be the best substrate for the purified arylesterase. From these results, it was determined that the purified arylesterase had broad substrate specificity, enabling the enzyme to hydrolyze the PNP esters and organophosphates as well as aromatic esters as true substrates. Therefore, the purified arylesterase showed not only carboxylesterase activity but also paroxonase activity. In addition, the arylesterase also displayed tributyrinase activity, because it was preferentially identified on the tributyrin-emulsified agar plates during the purification procedure, but it showed no hydrolytic activity toward other triacylglycerols examined.
TABLE 4.
Kinetic parameters for hydrolysis of various substrates
| Substrate | Mean ± SD
|
||
|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1·μM−1) | |
| Aromatic esters | |||
| PA | 17 ± 1 | 521 ± 21 | 30.6 ± 8 |
| α-NA | 29 ± 1 | 450 ± 26 | 15.5 ± 6 |
| PNP esters | |||
| Butyrate (C4) | 224 ± 12 | 285 ± 11 | 1.3 ± 0.5 |
| Caproate (C6) | 31 ± 4 | 467 ± 22 | 15.1 ± 1 |
| Caprylate (C8) | 28 ± 2 | 410 ± 13 | 14.6 ± 2 |
| Caprate (C10) | 81 ± 3 | 341 ± 16 | 4.2 ± 0.5 |
| Laurate (C12) | 122 ± 5 | 264 ± 21 | 2.2 ± 0.2 |
| Palmitate (C16) | 276 ± 9 | 265 ± 12 | 0.9 ± 0.1 |
| Organophosphates | |||
| PNP-phosphate | 234 ± 14 | 269 ± 18 | 1.2 ± 0.2 |
| Paraoxon | 5 ± 0.5 | 597 ± 15 | 119.4 ± 0.3 |
| Methyl paraoxon | 246 ± 18 | 342 ± 31 | 1.4 ± 0.2 |
To investigate the amino acids involved in the catalytic mechanism, the inhibitory effects of various chemical modifiers to specific amino acids on the enzyme were measured using the standard arylesterase assay after incubation for 30 min at 30°C. As shown in Table 5, the enzyme was completely inhibited by 5 mM DFP or 5 mM HgCl2. PMSF (5 mM), PCMB (5 mM), and DPC (5 mM) severely inhibited the enzyme activity to 77%, 77%, and 94%, respectively. However, PLP, PGO, paraoxon, and eserine had no significant effect on the enzyme activity. These results suggested that Ser, Cys, and His were located at or near the active site and were closely related to the catalytic activity of the arylesterase and also that the enzyme might contain a “catalytic triad” consisting of Ser, His, and Asp. In addition, because the enzyme was not inhibited by paraoxon, the purified enzyme was confirmed to be in the A-esterase group. Moreover, paraoxon was found to be used as a substrate of this enzyme. The enzyme activity was not affected by incubation with 5 mM of each divalent cation for 30 min at 30°C (data not shown) or by incubation with 10 mM EDTA for 60 min at 75°C (Table 5). These results suggested that the purified arylesterase does not require any divalent cation for its activity.
TABLE 5.
Effects of various inhibitors on activity of the arylesterase from S. solfataricus P1
| Inhibitor | Concn (mM) | % Residual activity (mean ± SD) after incubation for 30 mina |
|---|---|---|
| Control | 0 | 100 |
| PLP | 5 | 110 ± 8 |
| PGO | 5 | 97 ± 5 |
| PMSF | 0.5 | 56 ± 3 |
| 5 | 23 ± 1 | |
| DFP | 0.05 | 43 ± 3 |
| 0.5 | 31 ± 2 | |
| 5 | 0 | |
| PCMB | 0.5 | 62 ± 5 |
| 5 | 23 ± 11 | |
| HgCl2 | 0.5 | 12 ± 3 |
| 5 | 0 | |
| DPC | 0.5 | 50 ± 3 |
| 5 | 6 ± 2 | |
| Paraoxon | 0.5 | 111 ± 5 |
| 5 | 103 ± 7 | |
| Eserine | 0.5 | 96 ± 7 |
| 5 | 92 ± 5 | |
| EDTA | 10 | 92 ± 6 |
The enzyme was incubated at 30°C (or at 75°C for EDTA) in 100 mM sodium phosphate buffer (pH 7.0) with different concentrations of inhibitors. After 30 min, aliquots were removed from each incubation mixture and assayed for residual activity using the standard arylesterase assay.
Cloning of the S. solfataricus arylesterase gene.
The homogeneous arylesterase purified from S. solfataricus P1 was subjected to automatic Edman degradation, and the following 30 residues were positively identified: PLDPEVRNFLQVYYKANIIDFTKYQFQEIR. The forward primer of a 21-mer oligonucleotide mixture was designed based on the amino acid sequence from residue 13 to residue 19 (underlined). The reverse primer was derived from the consensus sequence VTAEYDP obtained from an alignment of archaeal esterase genes. A 717-bp DNA fragment, part of the arylesterase gene, was obtained by PCR with these primers using S. solfataricus genomic DNA as a template. The rest of the arylesterase gene was cloned by a PCR-based DNA walking procedure. The complete arylesterase gene revealed an open reading frame of 918 bp encoding a protein of 306 amino acids with a molecular mass of 34.5 kDa (Fig. 4). The calculated molecular mass is similar to those determined by SDS/PAGE, HPLC, and MALDI-TOF MS analyses as described above. The predominant amino acids are hydrophobic amino acids, such as Ala, Ile, Leu, and Val, which correspond to 33% of the 306 amino acid residues.
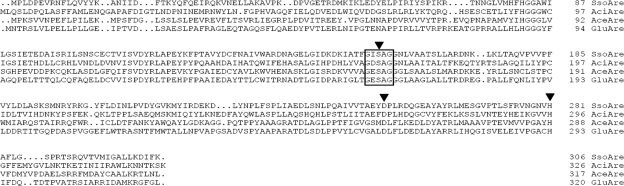
FIG. 4.
Multiple-sequence alignment of S. solfataricus arylesterase (SsoAre), Acinetobacter sp. strain ADP1 arylesterase (AciAre), Acetobacter pasteurianus arylesterase (AceAre), and Gluconobacter oxydans 621H arylesterase (GluAre). The catalytic triad (Ser156, Asp251, and His281) in the S. solfataricus arylesterase as deduced from the alignment is indicated by an arrowhead. The consensus sequence motif, Gly-x-Ser-x-Gly, around the active site Ser156 is boxed.
Amino acid sequence comparison.
The deduced amino acid sequence from the S. solfataricus arylesterase gene showed 34%, 33%, and 29% identity with those from Acinetobacter sp. strain ADP1, Acetobacter pasteurianus, and Gluconobacter oxydans 621H, respectively (Fig. 4). Besides these arylesterases, BLAST searches showed that most of the proteins with higher sequence identity than the Acinetobacter arylesterase were esterases and/or lipases or alpha/beta hydrolases. There were no other arylesterases showing high sequence identity. The amino acid sequence of the S. solfataricus arylesterase was the most similar to that of a putative esterase/lipase from Sulfolobus acidocaldarius DSM 639 (15), with 100% identity, and showed high similarities to that of a carboxylesterase composed of 251 amino acids (72% identity) and to that of a putative lipase composed of 311 amino acids (43% identity) from S. solfataricus P2 (47) from the BLAST search. In addition, sequence alignment revealed that the S. solfataricus arylesterase is a serine esterase containing the typical catalytic triad composed of Ser-Asp-His and the consensus sequence (Gly-x-Ser-x-Gly) around the active-site serine 156 (Fig. 4).
Purification and characterization of recombinant S. solfataricus arylesterase.
For further studies, the S. solfataricus arylesterase gene was expressed in E. coli and some properties of the expressed enzyme were investigated after purification. The recombinant S. solfataricus arylesterase was purified to electrophoretic homogeneity from E. coli extracts using two heat treatments and butyl-Sepharose column chromatography, resulting in a yield of approximately 23% (1.5 mg) for the purified protein (data not shown). The recombinant S. solfataricus arylesterase showed a specific activity of 3,550 units/mg for the hydrolysis of PA at 60°C and pH 7.0 (data not shown). The homogeneity of the purified recombinant S. solfataricus arylesterase was confirmed on SDS-polyacrylamide gels, and the molecular mass of the recombinant S. solfataricus arylesterase was the same as that of native S. solfataricus arylesterase (Fig. 1A, lane 2). In addition, the purified recombinant S. solfataricus arylesterase was stained by the activity staining method using α-NA after SDS-PAGE (Fig. 1C, lane 2), clearly showing that the recombinant protein has an esterase activity. As shown in Fig. S1 in the supplemental material, the effects of pH and temperature on the recombinant S. solfataricus arylesterase were almost the same as those on native S. solfataricus arylesterase (Fig. 2). Moreover, as shown in Fig. S2 in the supplemental material, the thermostability of the recombinant S. solfataricus arylesterase at the three different temperatures (50, 70, and 90°C) with increasing incubation time up to 120 h was also nearly the same as that of native S. solfataricus arylesterase (Fig. 3). These results indicate that the active S. solfataricus arylesterase gene can be expressed in E. coli and that the properties of the expressed enzyme were also the same as those of the native S. solfataricus enzyme. As a consequence, the recombinant S. solfataricus arylesterase was confirmed to be an S. solfataricus arylesterase.
Examination of amino acid residues related to the catalytic activity of the S. solfataricus arylesterase.
Table 5 demonstrates that modification of Ser, Cys, or His with a specific chemical modifier resulted in the loss of enzyme activity. The inhibition of the enzyme by chemical modifications of Ser and His residues can be expected from the sequence alignment data. As shown in Fig. 4, the S. solfataricus arylesterase was presumed to contain the catalytic triad composed of Ser156, Asp251, and His281. Hence, these amino acid residues were replaced by site-directed mutagenesis to give Ser156Ala, Asp251Asn, and His281Asn. As shown in Table 6, the Ser156Ala mutant and the Asp251Asn mutant showed little activity, and the His281Asn mutant was almost fivefold less activity than the wild type. These results strongly suggest that Ser156, Asp251, and His281 compose a catalytic triad. In addition, the chemical modification of Cys residues also caused a significant loss of the enzyme activity (Table 5). There are three cysteine residues (Cys105, Cys107, and Cys129) in native S. solfataricus arylesterase. To determine which of the three is crucial for the activity, we replaced each Cys with Ser by using site-directed mutagenesis. The purified recombinant proteins behaved identically to the wild type on SDS-PAGE (data not shown). However, the specific activities of the three mutants were significantly lower than that of the wild type, as shown in Table 6. The Cys129Ser mutant showed a 28-fold-lower activity, whereas the Cys105Ser and Cys107Ser mutants were 3- to 4-fold less active than the wild type. These results suggest that all three cysteines are required for the full activity, although Cys129 is the most crucial residue.
TABLE 6.
Specific activities of native and mutant S. solfataricus arylesterases
| Arylesterase | Sp act (U/mg) | Relative activity (%) |
|---|---|---|
| Native | 3,048.33 | 100 |
| Ser156Ala | 0.37 | 0.012 |
| Asp251Asn | 18.84 | 0.6 |
| His281Asn | 591.08 | 19.4 |
| Cys105Ser | 731.52 | 24.0 |
| Cys107Ser | 938.43 | 30.8 |
| Cys129Ser | 109.17 | 3.6 |
DISCUSSION
The present work reports the purification and characterization of a thermostable arylesterase displaying highly active paraoxonase activity from S. solfataricus P1 (DSM1616) and its expression in E. coli; this is, to our knowledge, the only arylesterase so far isolated from archaea. This result was supported by (i) the lack of inhibition by paraoxon, (ii) the substrate specificity toward paraoxon, (iii) the lack of a requirement for divalent cations, and (iv) the high stability against temperature and denaturants. These observations strongly indicate that the enzyme purified in this study is a thermostable A-esterase showing paraoxonase activity and is the first isolated and identified from archaea.
The purified arylesterase displays remarkable thermostability, like other enzymes from thermophilic archaea (4, 40). At 90°C, 52% of the enzyme activity remained after 50 h (Fig. 3). In contrast, the activity of the same enzyme from A. radiobacter is completely eliminated after 10 min at 75°C (41). In addition to showing high thermostability, the purified arylesterase also displays high stability against several protein-denaturing compounds, like other enzymes from thermophilic archaea (12, 13, 16, 21), although it is not feasible to compare it with other arylesterases from the same thermophilic archaea due to a lack of reports on the other arylesterases. At 70°C the enzyme can withstand, to a great extent, 8 M urea, 5% Tween 20, 5% Lubrol, 5% CHAPS, 1% SDS, 90% of different water-miscible organic solvents, and 2 M NaCl (Table 3). The high stability of the arylesterase against urea, some detergents, and organic solvents suggests that strong hydrophobic interactions make up the stable core of the enzyme and also that the enzyme may have a high surface hydrophobicity (12, 13, 16). It is also speculated that the proximity of hydrophobic substrates to this arylesterase may be facilitated by the addition of detergents. Similarly, an enzyme presumed to possess a high surface hydrophobicity may be stabilized by most detergents. However, particularly 5% Triton X-100 significantly inactivated the enzyme. This inhibitory effect of Triton X-100 on the enzyme activity could be deduced from reports on the action of Triton X-100 as an inhibitor toward the arylesterases from baker's yeast (48), greenbug (Schizaphis graminum) (50), and Flavobacterium spp. (11). We assume that this inhibitory effect of Triton X-100 might be derived from the structure of aromatic ring in the Triton X-100, which might compete with substrate. In addition, 2 M NaCl does not significantly affect enzyme stability, suggesting that ionic interactions are not involved in maintaining the native conformation of the enzyme and that the enzyme is monomeric. Thus, the high stability of the enzyme against several compounds, along with its thermostability, makes it a very attractive enzyme for future application in industry as well as in biochemistry.
From the examination of substrate specificity, the arylesterase from S. solfataricus displayed paraoxonase activity, and the kcat/Km ratios of the enzyme toward PA and paraoxon were 30.6 s−1·μM−1 and 119.4 s−1·μM−1, respectively. All arylesterases do not necessarily show paraoxonase activity, and not all paraoxonases display arylesterase activity (3, 31). The arylesterase from the organophosphate-resistant clone of the greenbug, S. graminum (50), does not hydrolyze paraoxon. However, the arylesterase from the Flavobacterium spp. is able to hydrolyze paraoxon efficiently as a substrate (11). The arylesterase from mammalian (human, rabbit, mouse, etc.) sera was initially called paraoxonase, whose function is to hydrolyze organophosphates such as paraoxon and physiologically protect low-density lipoprotein against oxidation (30). Recently, it was discovered that there are three paraoxonase (PON) genes closely aligned on human chromosome 7 and mouse chromosome 6 (39). Intensive research on possible functions of the PON family of enzymes shows that PON1 involves both paraoxonase and arylesterase activities. However, other members of the PON family (PON2 and PON3) from human and rabbit sources completely lack paraoxonase activity and have very little, if any, arylesterase activity (18). The paraoxonase from Russell's viper venom does not show arylesterase activity (33).
Concerning the specific chemical modification of the enzyme, we have found that Ser, His, and Cys are involved in the catalytic activity. The result that Ser and His are crucial in the activity of the enzyme leads us to deduce that S. solfataricus arylesterase may contain a catalytic triad composed of Ser, His, and Asp in its active site. The arylesterase from Acinetobacter sp. strain ADP1 (24) or V. mimicus (14) is a serine esterase containing a typical Ser-Asp-His catalytic triad in its active site as well. Judging from the alignment of the S. solfataricus P1 arylesterase amino acid sequence with those from Acinetobacter sp. strain ADP1, Acetobacter pasteurianus, and Gluconobacter oxydans 621H (Fig. 4), the residues that comprise the catalytic triad were suggested to be Ser156, Asp251, and His281, equivalent to Ser166, Asp266, and His296 in Acinetobacter sp. strain ADP1 arylesterase. This presumption was proven to be in good agreement with the result of mutation of Ser156, Asp251, and His281 by site-directed mutagenesis (Table 6). In addition, in the case of the arylesterase from A. radiobacter (41) or baker's yeast (48), a sulfhydryl group and a serine residue play important roles in its activity. However, the arylesterase from Pseudomonas fluorescens shows no inhibition by serine hydrolase inhibitors such as PMSF and DFP (26). The arylesterase from Triatoma infestans (17) or the greenbug S. graminum (50) requires Cys in its catalytic mechanism. In this study we showed by chemical modification (Table 5) and site-directed mutagenesis (Table 6) that the enzyme activity of S. solfataricus arylesterase also depended on Cys residues. Hence, in order to see where the three cysteines are located within the enzyme, we compared the primary structure of S. solfataricus arylesterase with that of Archaoglobus fulgidus carboxylesterase (311 amino acid residues), for which a crystal structure was reported and which shows 30% sequence identity (32). The catalytic triad (Ser160, Asp255, and His285) of A. fulgidus carboxylesterase is fully conserved in S. solfataricus arylesterase (Ser156, Asp251, and His281). Among the three cysteines in S. solfataricus arylesterase, Cys105 and Cys107 are replaced by Ser in A. fulgidus carboxylesterase, whereas Cys129 is conserved. However, it is not easy to explain how modification of cysteines causes enzyme inactivation, since none of the cysteines in S. solfataricus arylesterase constitutes the active site. Inspection of the available three-dimensional structure of A. fulgidus carboxylesterase reveals that the three cysteines in S. solfataricus arylesterase are distant from the active site. This excludes the possibility that replacement of Cys with Ser imposed a steric hindrance to the active site. Considering the fact that all three cysteines are buried within the hydrophobic interior of the enzyme, we speculate that the enhanced polarity of Ser caused a conformational change around the Cys residues which propagated through the backbone to the active site. We expect that the crystal structure of S. solfataricus arylesterase will resolve the issue in the future.
The result (Table 5) that the purified arylesterase/paraoxonase does not require any divalent cation, resulting from the lack of an effect of EDTA on its activity, is the same result as for the arylesterase from A. radiobacter (41) and the arylesterase/paraoxonase from the avian liver (39) but is different from that for the arylesterase from Triatoma infestans (17) (requiring Co2+ and Mn2+), the arylesterases/paraoxonases from the greenbug S. graminum (50) and mammals (27) (requiring Ca2+), and the paraoxonase from Russell's viper venom (33) (requiring Ca2+). In addition, archaeal phosphotriesterases showing paraoxonase activity require divalent cations such as Cd2+, Co2+, Mg2+, Ni2+, and Zn2+ (34, 38).
In conclusion, this is a novel arylesterase, displaying paraoxonase activity, from the thermoacidophilic archaeon S. solfataricus P1. The enzyme has broad substrate specificity and exhibits remarkable stability against high temperature and several protein-denaturing compounds. These features of the enzyme make it an excellent model system to explore the relationship between its structure and properties under nonconventional conditions and to facilitate further research leading to biotechnological and industrial uses.
Supplementary Material
Acknowledgments
This study was supported by a research grant from Kangwon National University.
We thank June M. Kwak and Julie Keating (University of Maryland at College Park) and Sanghwa Han (Kangwon National University) for their critical readings of the manuscript.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aharoni, A., L. Gaidukov, S. Yagur, L. Toker, I. Silman, and D. S. Tawfik. 2004. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. USA 101482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge, W. N. 1953. Serum esterases. I. Two types of esterase (A and B) hydrolysing p-nitrophenyl acetate, propionate and butyrate, and a method for their determination. Biochem. J. 53110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge, W. N. 1993. The esterases: perspectives and problems. Chem. Biol. Interact. 875-13. [DOI] [PubMed] [Google Scholar]
- 4.Arcari, P., L. Masullo, M. Masullo, F. Catanzano, and V. Bocchini. 2000. A NAD(P)H oxidase isolated from the archaeon Sulfolobus solfataricus is not homologous with another NADH oxidase present in the same microorganism. Biochemical characterization of the enzyme and cloning of the encoding gene. J. Biol. Chem. 275895-900. [DOI] [PubMed] [Google Scholar]
- 5.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343177-183. [PMC free article] [PubMed] [Google Scholar]
- 6.Augustinsson, K. B. 1961. Multiple forms of esterase in vertebrate blood plasma. Ann. N. Y. Acad. Sci. 94844-860. [Google Scholar]
- 7.Aviram, M. S., Billecke, R. Sorenson, C. Bisgaier, R. Newton, M. Rosenblat, J. Erogul, C. Hsu, C. Dunlop, and B. N. La Du. 1998. Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: selective action of human paraoxonase allozymes Q and R. Arterioscler. Thromb. Vasc. Biol. 181617-1624. [DOI] [PubMed] [Google Scholar]
- 8.Bornscheuer, U. T. 2002. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2673-81. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 10.Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 8454-68. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K. A. 1980. Phosphotriesterases of Flavobacterium sp. Soil Biol. Biochem. 12105-112. [Google Scholar]
- 12.Burlini, N., P. Magnani, A. Villa, F. Macchi, P. Tortora, and A. Guerritore. 1992. A heat-stable serine proteinase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Biochim. Biophys. Acta 1122283-292. [DOI] [PubMed] [Google Scholar]
- 13.Cacciapuoti, G., M. Porcelli, C. Bertoldo, M. De Rosa, and V. Zappia. 1994. Purification and characterization of extremely thermophilic and thermostable 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus. Purine nucleoside phosphorylase activity and evidence for intersubunit disulfide bonds. J. Biol. Chem. 26924762-24769. [PubMed] [Google Scholar]
- 14.Chang, R. C., J. C. Chen, and J. F. Shaw. 1996. Site-directed mutagenesis of a novel serine arylesterase from Vibrio mimicus identifies residues essential for catalysis. Biochem. Biophys. Res. Commun. 221477-483. [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., K. Brugger, M. Skovgaard, P. Redder, Q. She, E. Torarinsson, B. Greve, M. Awayez, A. Zibat, H. P. Klenk, and R. A. Garrett. 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 1874992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo, S., S. D'Auria, P. Fusi, L. Zecca, C. A. Raia, and P. Tortora. 1992. Purification and characterization of a thermostable carboxypeptidase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Eur. J. Biochem. 206349-357. [DOI] [PubMed] [Google Scholar]
- 17.de Malkenson, N. C., E. J. Wood, and E. N. Zerba. 1984. Isolation and characterization of an esterase of Triatoma infestans with a critical role in the degradation of organophosphorus esters. Insect Biochem. Mol. Biol. 14481-486. [Google Scholar]
- 18.Draganov, D. I., P. L. Stetson, C. E. Watson, S. S. Billecke, and B. N. La Du. 2000. Rabbit serum paraoxonase 3 (PON3) is a high density lipoprotein-associated lactonase and protects low density lipoprotein against oxidation. J. Biol. Chem. 27533435-33442. [DOI] [PubMed] [Google Scholar]
- 19.Feller, G., M. Thiry, and C. Gerday. 1991. Nucleotide sequence of the lipase gene lip2 from the antarctic psychrotroph Moraxella TA144 and site-specific mutagenesis of the conserved serine and histidine residues. DNA Cell Biol. 10381-388. [DOI] [PubMed] [Google Scholar]
- 20.Gur, M., A. Yildiz, R. Demirbag, R. Yilmaz, M. Aslan, I. Ozdogru, and O. Erel. 2007. Paraoxonase and arylesterase activities in patients with cardiac syndrome X, and their relationship with oxidative stress markers. Coron. Artery Dis. 1889-95. [DOI] [PubMed] [Google Scholar]
- 21.Hotta, Y., S. Ezaki, H., Atomi, and T. Imanaka. 2002. Extremely stable and versatile carboxylesterase from a hyperthermophilic archaeon. Appl. Environ. Microbiol. 683925-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeger, K. E., A. Steinbuchel, and D. Jendrossek. 1995. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: bacterial lipases hydrolyze poly(omega-hydroxyalkanoates). Appl. Environ. Microbiol. 613113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaeger, K. E., S. Ransac, B. W. Dijkstra, C. Colson, M. van Heuvel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiol. Rev. 1529-63. [DOI] [PubMed] [Google Scholar]
- 24.Jones, R. M., L. S., Collier, E. L. Neidle, and P. A. Williams. 1999. areABC genes determine the catabolism of aryl esters in Acinetobacter sp. strain ADP1. J. Bacteriol. 1814568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai, H., S. Yomoda, and Y. Inoue. 1991. ELISA using monoclonal antibody to human serum arylesterase. Clin. Chim. Acta 202219-225. [DOI] [PubMed] [Google Scholar]
- 26.Kim, K. K., K. Y. Hwang, K. D. Choi, J. H. Kang, O. J. Yoo, and S. W. Suh. 1993. Crystallization and preliminary X-ray crystallographic analysis of arylesterase from Pseudomonas fluorescens. Proteins 15213-215. [DOI] [PubMed] [Google Scholar]
- 27.Kuo, C. L., and B. N. La Du. 1998. Calcium binding by human and rabbit serum paraoxonases. Structural stability and enzymatic activity. Drug Metab. Dispos. 26653-660. [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J. W., D. Verger, P. D. Carr, H. Yang, and D. L. Ollis. 2000. Expression, purification and preliminary crystallographic studies of a hyperthermophilic esterase from Archaeoglobus fulgidus. Acta Crystallogr. D 56900-901. [DOI] [PubMed] [Google Scholar]
- 30.Mackness, M. I., B. Mackness, P. N. Durrington, P. W. Connelly, and R. A. Hegele. 1996. Paraoxonase: biochemistry, genetics and relationship to plasma lipoproteins. Curr. Opin. Lipidol. 769-76. [DOI] [PubMed] [Google Scholar]
- 31.Mackness, M. I., H. M. Thompson, A. R. Hardy, and C. H. Walker. 1987. Distinction between ‘A’-esterases and arylesterases. Implications for esterase classification. Biochem. J. 245293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandrich, L., L. Merone, M. Pezzullo, L. Cipolla, F. Nicotra, M. Rossi, and G. Manco. 2005. Role of the N terminus in enzyme activity, stability and specificity in thermophilic esterases belonging to the HSL family. J. Mol. Biol. 345501-512. [DOI] [PubMed] [Google Scholar]
- 33.Mende, T. J., and M. Moreno. 1975. A heat stable paraoxonase (O,O-diethyl O-p-nitrophenyl phosphate O-p-nitrophenyl hydrolase) from Russell's viper venom. Biochemistry 143913-3916. [DOI] [PubMed] [Google Scholar]
- 34.Merone, L., L. Mandrich, M. Rossi, and G. Manco. 2005. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: cloning, overexpression and properties. Extremophiles 9297-305. [DOI] [PubMed] [Google Scholar]
- 35.Morana, A., N. Di Prizito, V., Aurilia, M. Rossi, and R. Cannio. 2002. A carboxylesterase from the hyperthermophilic archaeon Sulfolobus solfataricus: cloning of the gene, characterization of the protein. Gene 283107-115. [DOI] [PubMed] [Google Scholar]
- 36.Ohsawa, K., and N. Ebata. 1983. Silver stain for detecting 10-femtogram quantities of protein after polyacrylamide gel electrophoresis. Anal. Biochem. 135409-415. [DOI] [PubMed] [Google Scholar]
- 37.Park, Y. J., S. Y. Choi, and H. B. Lee. 2006. A carboxylesterase from the thermoacidophilic archaeon Sulfolobus solfataricus P1; purification, characterization, and expression. Biochim. Biophys. Acta 1760820-828. [DOI] [PubMed] [Google Scholar]
- 38.Porzio, E., L. Merone, L. Mandrich, M. Rossi, and G. Manco. 2007. A new phosphotriesterase from Sulfolobus acidocaldarius and its comparison with the homologue from Sulfolobus solfataricus. Biochimie 89625-636. [DOI] [PubMed] [Google Scholar]
- 39.Primo-Parmo, S. L., R. C. Sorenson, J. Teiber, and B. N. La Du. 1996. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics 33498-507. [DOI] [PubMed] [Google Scholar]
- 40.Rolfsmeier, M., and P. Blum. 1995. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 177482-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai, Y., K. Ayukawa, H. Yurimoto, K. Yamamoto, and N. Kato. 1998. A novel arylesterase active toward 7-aminocephalosporanic acid from Agrobacterium radiobacter IFO 12607: purification and characterization. J. Ferment. Bioeng. 8558-62. [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Satoh, T., and M. Hosokawa. 1998. The mammalian carboxylesterases: from molecules to functions. Annu. Rev. Pharmacol. Toxicol. 38257-288. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer, S., C. Barkowski, and G. Fuchs. 1986. Carbon assimilation by the autotrophic thermophilic archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 146301-308. [Google Scholar]
- 45.Schmidt-Dannert, C., H. Sztajer, W., Stocklein, U. Menge, and R. D. Schmid. 1994. Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenulatus. Biochim. Biophys. Acta 121443-53. [DOI] [PubMed] [Google Scholar]
- 46.Shaw, J. F., R. C. Chang, K. H. Chuang, Y. T. Yen, Y. J. Wang, and F. G. Wang. 1994. Nucleotide sequence of a novel arylesterase gene from Vibro mimicus and characterization of the enzyme expressed in Escherichia coli. Biochem. J. 298675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 987835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toshimitsu, N., H. Hamada, and M. Kojima. 1986. Purification and some properties of an esterase from yeast. J. Ferment. Technol. 64459-462. [Google Scholar]
- 49.Wei, Y., J. L. Schottel, U. Derewenda, L. Swenson, S. Patkar, and Z. S. Derewenda. 1995. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat. Struct. Biol. 2218-223. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, K. Y., and F. He. 2000. Elevated esterases exhibiting arylesterase-like characteristics in an organophosphate-resistant clone of the greenbug, Schizaphis graminum (Homoptera:Aphididae). Pestic. Biochem. Physiol. 67155-167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.