Abstract
The rumen bacterium Ruminococcus albus binds to and degrades crystalline cellulosic substrates via a unique cellulose degradation system. A unique family of carbohydrate-binding modules (CBM37), located at the C terminus of different glycoside hydrolases, appears to be responsible both for anchoring these enzymes to the bacterial cell surface and for substrate binding.
Ruminococcus albus is widely recognized as one of the specialist cellulose-degrading bacteria resident in the rumena and gastrointestinal tracts of herbivores. Most isolates have been shown to utilize cellulose, xylan, and cellobiose as carbon sources and found to produce a wide range of enzymatic activities, including β-glucosidase, β-xylosidase, α-galactosidase, α-arabinosidase, cellulase, polygalacturonase, and β-1,4-xylanase activities (2, 3, 5). Interestingly, many of these glycoside hydrolases bear a recently described family 37 carbohydrate-binding module (CBM37), which appears to be exclusive to R. albus. These ∼100-residue modules were first identified at the C-terminal ends of an exoglucanase (Cel48A) and a processive endocellulase (Cel9B) from R. albus strain 8 (1) and appear to be nondiscriminatory in their carbohydrate-binding properties, recognizing a variety of polysaccharides, including cellulose, xylan, chitin, and lichenan (21). This breadth of adhesive properties makes the CBM37 family unique among the CBM families known to date. A preliminary examination of the draft genome sequence for R. albus strain 8 suggests that CBM37 modules, which are grouped into three major subtypes, are present in numerous R. albus polysaccharide-degrading enzymes and other nonenzymatic proteins from this bacterium (http://blast.jcvi.org/rumenomics/index.cgi).
Previous studies have shown that effective cellulose hydrolysis by R. albus strains is conditional on the provision of micromolar concentrations of phenylacetic and phenylpropionic acids (9, 17, 18). These compounds appear to influence capsule formation by the bacterium, and cellulase activity is retained as high-molecular-mass complexes on the bacterial cell surface. In the absence of phenylacetic and phenylpropionic acids, the adhesion of the bacterium to cellulose (and its hydrolysis) is negatively affected. Additionally, cellulase activity is secreted into the culture medium and, by size exclusion chromatography, is shown to be present in a form suggesting that there is no aggregation of activity into larger, multiprotein complexes (17). Although it was long believed that these characteristics were attributable to a cellulosomal mode of enzyme organization, the identification of CBM37 modules (rather than dockerins) in these two key enzymes suggests that the CBM37 modules might play some role(s) in protein retention to the bacterial cell surface. In the present study, we present evidence to validate this hypothesis, and we propose that an additional function for the CBM37 family is the attachment of the parent protein to the bacterium's cell surface.
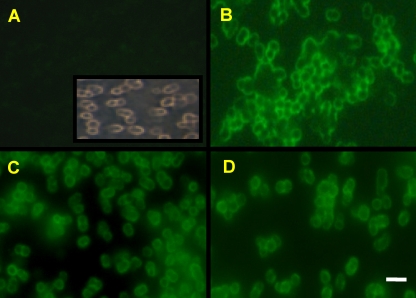
Three different CBM37 modules from R. albus, Cel5G (C-terminal module of AAT48117), Cel9C (AAT48118), and Cel48A (AAR01217), were used in this study, and they map to different branches within the major subgroup of CBM37 modules (see Fig. S1 in the supplemental material). The three CBM37s were cloned and fused to the C-terminal end of a recombinant maltose-binding protein (the resulting fusion proteins are hereinafter referred to as MBP-CBM5G, MBP-CBM9C, and MBP-CBM48A, respectively) (Table 1) and expressed in Escherichia coli as described earlier (21). MBP was required for solubility of the fusion partner and also served as a recognition tag. As a control, the MBP was fused to the catalytic module of Cel5G (hereinafter referred to as MBP-CD5G). Cellobiose-grown cells of R. albus 8 were harvested by centrifugation and washed using previously described procedures (1). Aliquots of the cell suspension were mixed with any one of the four different fusion proteins followed by mouse anti-MBP antibody and fluorescein isothiocyanate-conjugated goat anti-mouse antibody, according to the method described by Orgad et al. (12). The cells were then examined by fluorescence microscopy (Fig. 1). All three MBP-CBM fusion proteins could attach to the cell surface; however, the MBP-CD5G fusion protein did not. These results suggest that the attachment of the recombinant protein to the surface of R. albus 8 cells is mediated via the CBM37 module.
TABLE 1.
Oligonucleotide primers and plasmids used in this study
| MBP fusion proteina | Primer name | Nucleotide sequence | Comment |
|---|---|---|---|
| MBP-CD5G | F-GH5G-CD | ATATGAATTCGCAACATCAGCAGTGAATGACACC | Catalytic domain of Cel5G (without CBM37) |
| R-GH5G-CD | CCCCAAGCTTTTAAGGTGTTTCGGGATCAATGATTATC | ||
| MBP-CBM5G | F-Cel5G-CBM | ATATGAATTCGCTATAAATGTTATGGCGAAAGATG | CBM37 of Cel5G |
| R-Cel5G-CBM | CCCCAAGCTTTTACTTTACAGTGATAGTCACAGCG | ||
| MBP-CBM48A | F-GH48A-CBM | ATATGAATTCGATGATAAGACTTATCCTACCAAC | CBM37 of Cel48A |
| R-GH48A-CBM | CCCCAAGCTTTTAAACTGTAACGTTAACTACAGA | ||
| MBP-CBM9C | F-GH9C-CBM | ATATGAATTCGATCGTTTCGGCGGTTCGAATCCTG | CBM37 of Cel9C |
| R-GH9C-CBM | CCCCAAGCTTTTACTTTATAGTAACAGTACAAGCACG |
Forward (F-) and reverse (R-) primers contain EcoRI and HindIII cleavage sites, respectively (underlined).
FIG. 1.
Binding of different CBM37 fusion proteins to R. albus 8 cells by fluorescence microscopy. Log-phase R. albus cells interacted with the MBP control (MBP-CD5G lacking CBM37) (A), MBP-CBM5G (B), MBP-CBM48A (C), and MBP-CBM9C (D). All test proteins were expressed as fusion proteins with MBP at the N terminus. The labeled cells were subjected to interaction with mouse anti-MBP antibody followed by fluorescein isothiocyanate-conjugated donkey anti-mouse antibody and then visualized by fluorescence microscopy. The inset in panel A shows a phase-contrast micrograph of the R. albus 8 cells used for these studies. Bar = 2 μm.
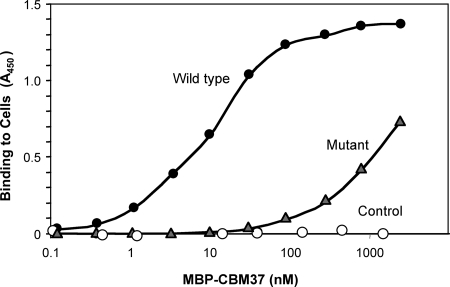
Next, we employed several mutant strains of R. albus 8 isolated by selective enrichment for defective adhesion to cellulose. Proteomic analysis of these mutant strains showed that they possess significantly less than wild-type amounts of the cell-associated CBM37-bearing enzymes Cel9B and Cel48A and are also defective in terms of cellulose hydrolysis kinetics compared to the wild-type strain (1). Following cultivation using cellobiose-containing medium (the same as that used for the wild-type strain), all three mutant strains appeared to be unable to bind the MBP-CBM fusion proteins, because no fluorescent label was detectable by microscopy (data not shown). To further validate this negative result, a cell-based enzyme-linked immunosorbent assay (ELISA) was employed, essentially as described by Rosok et al. (15), using lyophilized and resuspended bacterial cells (R. albus 8 or its ADM-2 mutant). The results, presented in Fig. 2, confirmed that the MBP-CBM5G recombinant protein can attach to the cell surface of wild-type R. albus 8 cells but cannot bind to the cell surface of the mutant strains. These findings suggest that the mutants all lack a key feature on the cell surface that is necessary for the attachment of R. albus proteins via the CBM37 module.
FIG. 2.
ELISA of the binding of CBM37 to the whole cells of R. albus 8. ELISA plates, containing attached R. albus 8 or its ADM-2 mutant, were reacted with the test protein, MBP-CBM37 from Cel5G (MBP-CBM5G), at the indicated final concentrations and labeled with mouse anti-MBP-horseradish peroxidase antibodies. MBP was used as the control.
Previous research showed that cell-free enzyme extracts are often not as effective as the intact cells in cellulose degradation, demonstrating that essential factors that may be missing in the extracellular medium are present in the bacterium (20). In an earlier report (13), transmission electron microscopic visualization of ruthenium red-labeled R. albus cells indicated an extensive “coat” layer, described as a compact mat of polysaccharide fibers external to the cell wall. This polysaccharide coat, or “glycocalyx,” was considered in subsequent works to mediate adhesion of the cells to cellulose (14, 19). In a later work, it was shown that most of the cellulases and xylanases in R. albus SY3 were associated with the capsular and cell wall fraction but were severely reduced on the surfaces of an adhesion-defective mutant (7). Two enzymes critical to effective solubilization by R. albus 8 (Cel48A and Cel9B) were previously demonstrated to be cell-associated proteins and to bind strongly to cellulose (1), presumably via the C-terminal CBM37 modules each enzyme bears. The results of the present study demonstrate that the same module serves to secure the enzymes to the cell surface. Consequently, we propose that CBM37 may function as a shuttle to convey the parent enzyme between the bacterial cell surface and the polysaccharide substrate.
Preliminary scanning electron microscopy of cationized ferritin-treated R. albus 8 versus its adherence-defective mutants clearly revealed a protuberance-laden surface in the wild-type cells as opposed to a smooth surface in the mutant ADM-2 (see Fig. S2 in the supplemental material), but it is as yet unclear whether the CBM37-binding component relates directly to this finding. Preliminary work (not shown) has also demonstrated that a polysaccharide-containing cell extract of R. albus 8 (obtained by a combination of lysosyme and DNase, followed by proteinase K treatments) is highly inhibitory to CBM37 binding to the bacterial cell surface. In contrast, similar extracts derived from the R. albus adherence-defective mutants or from Ruminococcus flavefaciens failed to inhibit the binding. Further work to identify the suspected cell wall carbohydrate component is currently being pursued.
In conclusion, R. albus cellulases are indeed known to be released into the medium during growth (16) and are subsequently bound to the cellulose fibers, yet cellulose digestion is facilitated by the proximity of the cells to the cellulose fibers (6). The model proposed here suggests that the CBM37 acts as a shuttle which transfers the appended enzymes from the bacterial surface to the plant cell wall. Another alternative might be that the CBM37 has two separate carbohydrate-binding sites, as shown previously for other CBMs (4). In this case, one site would bind to the plant cell wall and the other to the bacterial polysaccharide capsule. It remains to be seen, however, whether one or more of the remaining CBM37-bearing nonenzymatic proteins produced by this bacterium might play some role in the binding of the cells to the substrate. Previous research has already demonstrated that more than one mechanism is involved with the adhesion of R. albus to the substrate (8, 10, 11), but the localization of the glycanases at the interface appears to be mediated largely by CBM37.
Supplementary Material
Acknowledgments
We appreciate the assistance of Vered Lavie (Tel Aviv University) with the fluorescence microscopy experiments.
This research was supported by the Israel Science Foundation (grants 422/05 and 159/07); by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel; and by funds provided to M.M. from the National Research Initiative Competitive Grants Program of the USDA (99-35206-8688). Sequencing of the R. albus genome was accomplished with funds provided by grant 00-52100-9618 from the USDA Initiative for Future Agriculture and Food Systems. N.A. received support from research grant US-3106-99C of the United States-Israel Binational Agricultural Research and Development Fund (BARD).
E.A.B. holds the Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Devillard, E., D. E. Goodheart, S. K. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greve, L. C., J. M. Labavitch, and R. E. Hungate. 1984. α-l-Arabinofuranosidase from Ruminococcus albus 8: purification and possible role in hydrolysis of alfalfa cell wall. Appl. Environ. Microbiol. 471135-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greve, L. C., J. M. Labavitch, R. J. Stack, and R. E. Hungate. 1984. Muralytic activities of Ruminococcus albus 8. Appl. Environ. Microbiol. 471141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henshaw, J. L., D. N. Bolam, V. M. Pires, M. Czjzek, B. Henrissat, L. M. Ferreira, C. M. Fontes, and H. J. Gilbert. 2004. The family 6 carbohydrate binding module CmCBM6-2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 27921552-21559. [DOI] [PubMed] [Google Scholar]
- 5.Hungate, R. E., and R. J. Stack. 1982. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl. Environ. Microbiol. 4479-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leatherwood, J. M. 1969. Cellulase complex of Ruminococcus and a new mechanism for cellulose degradation. Adv. Chem. Ser. 9553-59. [Google Scholar]
- 7.Miron, J., J. Jacobovitch, E. A. Bayer, R. Lamed, M. Morrison, and D. Ben-Ghedalia. 2001. Subcellular distribution of glycanases and related components in Ruminococcus albus SY3 and their role in cell adhesion to cellulose. J. Appl. Microbiol. 91677-685. [DOI] [PubMed] [Google Scholar]
- 8.Miron, J., D. Ben-Ghedalia, and M. Morrison. 2001. Invited review: adhesion mechanisms of rumen cellulolytic bacteria. J. Dairy Sci. 841294-1309. [DOI] [PubMed] [Google Scholar]
- 9.Morrison, M., R. I. Mackie, and A. Kistner. 1990. 3-Phenylpropanoic acid improves the affinity of Ruminococcus albus for cellulose in continuous culture. Appl. Environ. Microbiol. 563220-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison, M., and J. Miron. 2000. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol. Lett. 185109-115. [DOI] [PubMed] [Google Scholar]
- 11.Mosoni, P., and B. Gaillard-Martinie. 2001. Characterization of a spontaneous adhesion-defective mutant of Ruminococcus albus strain 20. Arch. Microbiol. 17652-61. [DOI] [PubMed] [Google Scholar]
- 12.Orgad, S., N. Goldfinger, G. Cohen, V. Rotter, and B. Solomon. 2005. Single chain antibody against the common epitope of mutant p53 restores wild-type activity to mutant p53 protein. FEBS Lett. 5795609-5615. [DOI] [PubMed] [Google Scholar]
- 13.Patterson, H., R. Irvin, J. W. Costerton, and K. J. Cheng. 1975. Ultrastructure and adhesion properties of Ruminococcus albus. J. Bacteriol. 122278-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pell, A. N., and P. Schofield. 1993. Computerized monitoring of gas production to measure forage digestion in vitro. J. Dairy Sci. 761063-1073. [DOI] [PubMed] [Google Scholar]
- 15.Rosok, M. J., M. R. Stebbins, K. Connelly, M. E. Lostrom, and A. W. Siadak. 1990. Generation and characterization of murine antiflagellum monoclonal antibodies that are protective against lethal challenge with Pseudomonas aeruginosa. Infect. Immun. 583819-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith, W., R. I. Yu, and R. E. Hungate. 1973. Factors affecting cellulolysis by Ruminococcus albus. J. Bacteriol. 114729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stack, R. J., and R. E. Hungate. 1984. Effect of the 3-phenylpropanoic acid on capsule and cellulases of Ruminococcus albus 8. Appl. Environ. Microbiol. 48218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stack, R. J., R. E. Hungate, and W. P. Opsahl. 1983. Phenylacetic acid stimulation of cellulose digestion by Ruminococcus albus 8. Appl. Environ. Microbiol. 46539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weimer, P. J., N. P. Price, O. Kroukamp, L. M. Joubert, G. M. Wolfaardt, and W. H. Van Zyl. 2006. Studies of the extracellular glycocalyx of the anaerobic cellulolytic bacterium Ruminococcus albus 7. Appl. Environ. Microbiol. 727559-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood, T. M., C. A. Wilson, and C. S. Stewart. 1982. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem. J. 205129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu, Q., M. Morrison, E. A. Bayer, N. Atamna, and R. Lamed. 2004. A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 56611-16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.