Abstract
RNase R is a processive 3′-5′ exoribonuclease with a high degree of conservation in prokaryotes. Although some bacteria possess additional hydrolytic 3′-5′ exoribonucleases such as RNase II, RNase R was found to be the only predicted one in the facultative intracellular pathogen Legionella pneumophila. This provided a unique opportunity to study the role of RNase R in the absence of an additional RNase with similar enzymatic activity. We investigated the role of RNase R in the biology of Legionella pneumophila under various conditions and performed gene expression profiling using microarrays. At optimal growth temperature, the loss of RNase R had no major consequence on bacterial growth and had a moderate impact on normal gene regulation. However, at a lower temperature, the loss of RNase R had a significant impact on bacterial growth and resulted in the accumulation of structured RNA degradation products. Concurrently, gene regulation was affected and specifically resulted in an increased expression of the competence regulon. Loss of the exoribonuclease activity of RNase R was sufficient to induce competence development, a genetically programmed process normally triggered as a response to environmental stimuli. The temperature-dependent expression of competence genes in the rnr mutant was found to be independent of previously identified competence regulators in Legionella pneumophila. We suggest that a physiological role of RNase R is to eliminate structured RNA molecules that are stabilized by low temperature, which in turn may affect regulatory networks, compromising adaptation to cold and thus resulting in decreased viability.
RNA molecules provide the dynamic link between DNA-encoded information and protein synthesis. The unstable, labile nature of RNA is critical, as it allows a rapid adjustment of specific protein expression levels in response to environmental changes. In bacteria, RNA degradation occurs through an initial endoribonuclease cleavage followed by 3′-5′ exoribonuclease digestion (6, 14). In Escherichia coli, the latter reaction involves three processive 3′-5′ exoribonucleases: PNPase, RNase R, and RNase II. PNPase is a phosphorolytic enzyme and part of the degradosome, a multiprotein complex also involving the RNA helicase RhlB, an enolase, and the endoribonuclease RNase E (27). RNase II and RNase R are hydrolytic exoribonucleases that share similar domain architectures and belong to the RNase II family of RNases (18, 23, 38). In contrast to RNase II, RNase R is able to degrade structured RNA in vitro without the aid of helicase activity (11) and leads to 2nt as the end product (1, 11). However, the in vivo function of RNase R is poorly understood. The enzyme has been reported to play a role in rRNA quality control and the decay of mRNA fragments with extensive secondary structures (10, 12). RNase R is emerging as an important component in adaptation to stress. In the ribosome rescue system, RNase R associates with SmpB and the tmRNA (21) and participates in the degradation of the mRNA on which tmRNA-dependent ribosome rescue occurs (29). RNase R also participates in the maturation of the tmRNA itself in Escherichia coli (5), as well as its processing in Caulobacter crescentus and Pseudomonas syringae (19, 26). In E. coli, RNase R is induced twofold in stationary phase (2), and the elevation of RNase R activity in response to cold shock suggested a role of RNase R in adaptation to decreased temperatures (5, 7). The absence of RNase R caused impaired growth at low temperature (5). Consistent with the notion of the involvement of RNase in the degradation of RNA being stabilized by low temperature, RNase R has been found as part of the degradosome in the cold-adapted strain Pseudomonas syringae Lz4W (25). RNase R is also essential for growth at low temperatures for Pseudomonas syringae, Pseudomonas putida, and Aeromonas hydrophila (16, 17, 26). In the reduced-genome intracellular pathogen Mycoplasma genitalium, RNase R appears to be the only exoribonuclease (22) and is also thought to be essential, since the gene could not be interrupted (20). Studies of RNase R in additional species may result in a better understanding of its role in RNA metabolism. Unlike most members of the gammaproteobacterium class, the facultative intracellular pathogen Legionella pneumophila had only one hydrolytic RNase predicted, and by sequence comparison it resembled RNase R. Accordingly, we presumed it was an RNase R-like enzyme. Thus, L. pneumophila may provide another opportunity to study the function of RNase R in the absence of the enzymatically related RNase II.
We report here the characterization of an RNase R mutant of L. pneumophila. We found that RNase R is not essential for the growth of L. pneumophila at optimal temperature as a free-living bacterium or as an intracellular pathogen of mammalian and protozoan host cells. However, the absence of RNase R caused impaired growth at low temperatures that was associated with an accumulation of unprocessed RNA and perturbations of gene regulation. In particular, the lack of the exoribonuclease activity of RNase R at 30°C resulted in derepression of the competence regulon independently of the previously described competence regulators comR and proQ (34). We suggest a role of RNase R in the disposal of RNA molecules stabilized by low temperatures which otherwise would have deleterious impacts on gene regulatory networks.
MATERIALS AND METHODS
Strains and plasmids constructions.
L. pneumophila strains used in this study are derivatives of the strain JR32, a restriction-deficient and streptomycin-resistant derivative of the Philadelphia-1 isolate (32). L. pneumophila strains were grown in liquid medium ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) or on solid-medium ACES-buffered charcoal yeast extract (CYE) plates. Chloramphenicol, kanamycin, gentamicin, and hygromycin were used, respectively, at 5 μg/ml, 10 μg/ml, 50 μg/ml, and 100 μg/ml. Mutants with deletions of comEA and comEC derived from the rnr mutant LELA559C were generated by natural transformation with PCR products made of a kanamycin resistance cassette and the flanking regions (1 kb each) of the gene to be deleted. The PCR products were synthesized by long-flanking-homology PCR (24). The replacement of resistance markers of Tn903dIIlacZ in competent mutants was obtained by natural transformation with plasmid pXDC44, pXDC46, or pXDC57 and the selection of the corresponding marker. Plasmids pXDC44, pXDC46, and pXDC57 were made by cloning a PCR product of Tn903dIIlacZ into pGEM-T Easy (Promega Corporation, Madison, WI), creating pGEM-Tn903. The replacement cassettes conferring resistance to chloramphenicol, gentamicin, and hygromycin were amplified by PCR and cloned into EcoRV and SmaI sites of pGEM-Tn903. Plasmid pXDC18 is a pMMB207C derivative on which the chloramphenicol resistance cassette was removed by digesting with DraI and was replaced with a gentamicin resistance cassette obtained by PCR from pJN105. Plasmids for complementation of rnr, comEA, and comEC were obtained by cloning the corresponding genes under the control of Ptac in the polylinker of pMMB207C or pXDC18. JR32 strains harboring the comEA-lacZ fusions in the chromosome were constructed by allelic exchange using pLAW344 (33). The comEA regions (1 kb) to be fused to lacZ, a 1-kb region downstream of comEA, and a lacZ-aptII cassette were amplified by PCR. The three PCR products were assembled (comEA::lacZ-aptII-downstream comEA) by fusion PCR using the Phusion DNA polymerase (Finnzymes), and the PCR product was cloned into pLAW344. The resulting plasmids was transformed in JR32, and kanamycin-resistant and chloramphenicol-resistant colonies were selected. Transformants were then counterselected on CYE plates with kanamycin and 3% sucrose. Genomic DNA was extracted from the JR32 strains harboring the comEA-lacZ fusions and used to transform the rnr mutant LELA559G.
Axenic growth and determination of doubling time.
Bacterial strains were grown overnight in 3 ml of AYE broth on a rotary shaker. The next morning, stationary cultures were diluted 1:50 in prewarmed media and grown to mid-exponential phase (optical density [OD] = 0.5 to 0.7). Cultures were then diluted to an initial OD of 0.1, 0.05, or 0.025 in 100 μl in a 96-well plate and placed inside a temperature-controlled plate reader. The plate was submitted to intermittent shaking (100 rpm, 60 s every 5 min), and growth was monitored by measuring absorbance at 600 nm every 10 min. Doubling times were determined from the exponential sections of the growth curves.
Intracellular growth in THP-1-derived macrophages and Acanthamoeba castellanii.
THP-1 cells were routinely maintained in RPMI 1640, 2 mM glutamine, 10% fetal calf serum at 37°C under 5% CO2. For Legionella infection, THP-1 cells were incubated overnight in a 24-well microplate (1 × 106 cells/well) in medium containing 10 ng/ml phorbol 12-myristate 13-acetate (PMA). After the overnight stimulation, cells were washed and allowed to stabilize in medium without PMA for 2 days. Cells were then infected with L. pneumophila from stationary AYE cultures at a multiplicity of infection (MOI) of 1:1,000 (bacteria per cell). Infections were synchronized by spinning the bacteria at 650 × g for 10 min onto the adhered host cells. The supernatant of each well was sampled every 24 h, and numbers of CFU were determined by spotting serial dilutions on CYE plates. A. castellanii (ATCC 30234) was grown as adherent cells in proteose peptone-yeast extract-glucose medium (PYG) medium at 30°C. For Legionella infection, A. castellanii cells were resuspended in PYG, seeded in a 24-well microplate (1 × 106 cells/well), and incubated at 30°C for 1 hour or until cells were adherent. The PYG was aspirated and the wells were washed twice with 1 ml of warm (30°C) acetyl (Ac) buffer. L. pneumophila, resuspended in Ac buffer, was added to the wells at a MOI of 1:1,000. Infections were synchronized by spinning the bacteria at 650 × g for 10 min onto the adhered host cells. The plate was incubated for 1 hour at 30°C, and then the Ac buffer was carefully aspirated and replaced with 1 ml of warm Ac buffer. The supernatant of each well was sampled every 24 h, and numbers of CFU were determined by spotting serial dilutions on CYE plates.
Microarray analysis.
The microarray consisted of 2,997 unique 70-mer oligonucleotides representing all predicted L. pneumophila open reading frames spotted in duplicate. Total RNA from bacteria grown on CYE plate as in a transformation experiment (see “Competence assay” below) was isolated using the RNeasy kit (Qiagen Inc, Valencia, CA). RNA samples (20 μg) were reverse transcribed with Superscript II Plus RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA), random primers, and a mixture of deoxynucleotide triphosphates containing aminoallyl-modified dUTP (Sigma-Aldrich, St Louis, MO). RNA templates were removed by alkaline hydrolysis and aminoallyl-cDNAs were purified using the QiaQuick PCR purification kit (Qiagen Inc, Valencia, CA) and coupled with Alexa Fluor 546 or 647 dyes (Invitrogen, Carlsbad, CA). UV cross-linked slides were prehybridized in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 0.1 mg/ml bovine serum albumin for 45 min at 37°C. Slides were then rinsed in 0.1× SSC and dried. Labeled cDNA was added to hybridization solution (40% formamide, 5× SSC, 0.1% SDS, 10 mM Tris-HCl, pH 7.5, 1 mg/ml human placental DNA) and hybridized to microarrays in hybridization chambers (GraceBio Inc, Bend, OR) at 37°C overnight with rotation. Slides were washed twice with 5× SSC plus 0.1% SDS and 0.1× SSC plus 0.1% SDS, rinsed quickly with 0.01× SSC, and dried with high-pressure air by use of a dust remover. Slides were scanned at a 5-μm resolution using a ScanArray express microarray scanner (Perkin Elmer Inc, Wellesley, MA), and spot analysis was performed using Pro Scan Array Express software (Perkin Elmer Inc., Wellesley, MA). Raw signal intensities were corrected for dye labeling effects within and between all slides by use of normalize.lowess() R-function implemented in the Bioconductor affy microarray analysis package. The resulting gene expression data were analyzed using the Spotfire DecisionSite for Functional Genomics software suite (Spotfire).
RNA electrophoresis and Northern blot analysis.
Total RNA from about 109 bacteria grown on a CYE plate were extracted with 1 ml of Trizol reagent (Invitrogen) according to the manufacturer's instructions. Northern blots were performed using either a denaturing TBE-urea 6% acrylamide gel or a formaldehyde-1.5% agarose gel. One to 10 micrograms of total RNA in denaturing buffer was loaded per lane and run in TBE buffer (acrylamide gel) or MOPS (morpholinepropanesulfonic acid) buffer (agarose gel). RNAs were transferred to a nylon membrane (Immobilon-NY+; Millipore Corp., Billerica, MA) either by electrophoretic transfer in TBE buffer (acrylamide gel) or by capillary transfer in 10× SSC (agarose gel). RNAs were cross-linked to the membrane by UV irradiation. Membranes were hybridized at 42°C with 5′-biotinylated oligonucleotide probes (5 nM) in ULTRAhyb ultrasensitive hybridization buffer (Ambion, Austin, TX) and then washed according to the instructions of the ULTRAhyb manufacturer. Membranes were developed using horseradish peroxidase-conjugated streptavidin and enhanced luminol substrate (chemiluminescent nucleic acid detection module; Pierce, Rockford, IL) and Biomax films (Kodak).
Determination of the comEA mRNA half-life.
Wild-type JR32 and the rnr mutant (LELA559) from a CYE plate were inoculated in 30 ml of AYE broth at an OD of 0.05 and grown at 30°C to an OD of 0.7 to 0.8 (about 18 to 20 h of growth). Transcription was arrested by the addition of rifampin (100 μg/ml final), and aliquots of 1 ml were removed at 0, 5, 10, and 15 min after the addition of rifampin. The 1-ml aliquots were added to 500 μl of ice-cold AYE, the bacteria were pelleted by a 30-s centrifugation at 20,000 × g at 4°C, the medium was quickly aspirated, and the pellet was resuspended in 1 ml of Trizol (Invitrogen). Once all aliquots were collected, RNAs were extracted according to the manufacturer's instructions. Traces of genomic DNA were removed by a DNase I treatment, and RNAs were purified by acid phenol-chloroform extraction (5:1) at a pH of 4.5 and ethanol precipitation. RNAs (2 μg) were reverse transcribed and used as the template for real-time PCR with power SYBR green (Applied Biosystems). Data were analyzed with the ΔΔCT method (where CT is the threshold cycle) using 16S rRNA as a reference. Amounts of mRNA were expressed relative to the amount at time zero (t = 0). Data were fit to a first-order exponential decay with the Qtiplot software.
Competence assay.
Competence assays on CYE plates were performed as described by Sexton and Vogel (34). Briefly, one colony was used to produce a circular patch of about 6 to 8 mm in diameter on a CYE plate. One microgram of genomic DNA, previously extracted from the gentamicin-resistant strain JC51 (9) or from the hygromycin-resistant strain KS79 dotB-hygro, was spotted onto the patch, and the plate was incubated for 2 days at 30°C or 37°C. The patch was then resuspended in 1 ml of sterile water, and serial dilutions were plated onto CYE plates with or without a selection agent. The transformation frequency represents the ratio of total CFU determined from plating on selective media by the total CFU determined from plating on nonselective media.
Beta-galactosidase assay.
Beta-galactosidase assays were performed on bacteria grown as described for the transformation assays. One colony was used to produce a circular patch of about 6 to 8 mm in diameter on a CYE plate. After 24 h of growth at 30°C or 37°C, the patch was resuspended in 10 ml of Z buffer and the bacterial suspension was assayed for beta-galactosidase activity as described previously (37).
Immunoblotting.
Proteins from SDS-polyacrylamide gels were electrophoretically transferred onto nitrocellulose sheets (Schleicher and Schuell). Sheets were analyzed by Western blotting with rabbit polyclonal antibody raised against E. coli RNase R (a gift from Murray Deutcher, University of Miami Miller School of Medicine) and an anti-rabbit peroxidase conjugate (Pierce) as the secondary antibody. Nitrocellulose sheets were revealed with a chemiluminescence detection system (Pierce) and Biomax films (Kodak).
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (15) and are accessible through GEO series accession number GSE12796 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12796).
RESULTS
Identification and characterization of an L. pneumophila RNase R mutant.
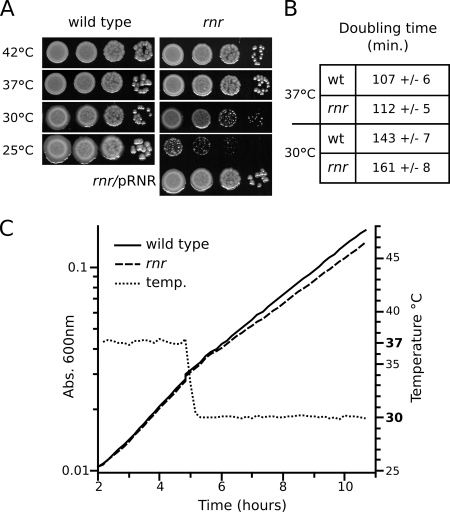
We identified an L. pneumophila rnr mutant from a collection of Tn903dIIlacZ insertions of the Philadelphia-1-derived laboratory strain JR32 (37). The Tn903dIIlacZ insertion has been mapped at nucleotide (nt) 513 of the RNase R coding sequence and does not produce a translational fusion with lacZ. Since RNase R has been reported for several species to be required for growth at low temperatures (5, 16, 17, 26), we analyzed the growth of an L. pneumophila rnr strain under axenic conditions on solid rich medium at optimal and suboptimal growth temperatures. The RNase R mutant did not show significant growth defects at optimal (37°C) or elevated (42°C) growth temperatures (Fig. 1A). In contrast, the rnr mutant was slightly impaired for growth at 30°C and more dramatically impaired at 25°C. The growth defect at low temperature could be suppressed by the plasmid-borne expression of RNase R (Fig. 1A). Growth in liquid medium at 30°C provided similar results, with the doubling time of the RNase R mutant showing a significant increase from that of the parent strain, JR32 (Fig. 1B). A rapid temperature downshift from 37°C to 30°C was found to cause the same growth defect (Fig. 1C). These results are consistent with previous reports for other species and suggest a role of RNase R in processes that are required for adaptation to decreased growth temperature. We also investigated the ability of the rnr mutant to multiply intracellularly in the amoeba Acanthamoeba castellanii and in THP-1 monocytes. Both phagocytic cells were infected with MOI of 1:1,000 (bacteria per cell), and L. pneumophila replication was monitored by enumerating CFU every 24 h. No observable growth defect of the rnr strain was detected in either amoebal or macrophage host cells (data not shown). The infection of A. castellanii was performed at 30°C, and even though the rnr mutant has a mild growth defect at 30°C in rich medium, this defect could not be detected by counting CFU. We conclude that RNase R is dispensable for the intracellular multiplication of L. pneumophila in both human and protozoan hosts. Overall, these results are consistent with previous reports for other species and reinforce the role of RNase R in adaptation to decreased growth temperature.
FIG. 1.
RNase R is required for growth of Legionella pneumophila at low temperature. (A) Serial dilutions of wild-type and rnr mutant Legionella pneumophila were plated on CYE plates and incubated to obtain visible colonies at 42°C (3 days), 37°C (3 days), 30°C (4 days), or 25°C (7 days). (B) Determination of doubling times of wild-type (wt) and rnr mutant strains of Legionella pneumophila in AYE broth at 37°C (n = 4) and 30°C (n = 5). (C) Effect of temperature (temp.) downshift on growth of wild-type and rnr mutant strains of Legionella pneumophila in AYE medium. Abs., absorbance.
Intermediate degradation products of structured RNAs accumulate in the rnr mutant.
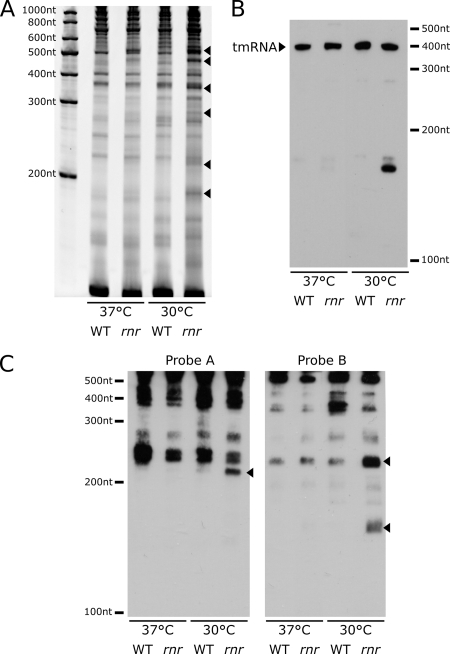
The loss of any RNase activity is expected to affect RNA metabolism. Given the ability of RNase R to degrade structured RNAs, this should primarily affect the metabolism of stable RNAs. We chose to analyze the effect of a loss of RNase at 30°C because at this temperature the growth defect is still mild and it is thus possible to study the rnr mutant and the wild-type parent under similar growth conditions. We extracted total RNA from the rnr mutant and wild-type strains grown at 30°C and 37°C and analyzed them by denaturing polyacrylamide electrophoresis followed by ethidium bromide staining. Compared to the what was seen for the wild-type strain, equal amounts of total RNAs extracted from the rnr mutant grown at 30°C consistently revealed an overall stronger staining intensity at low molecular weights (Fig. 2A). In addition, multiple distinct bands, ranging from about 190 nt to 500 nt, appeared to accumulate in the rnr mutant only when it was grown at 30°C. These bands were either not present or less visible in the wild type grown at either 30°C or 37°C. It has been previously reported that in E. coli RNase R may be involved in the maturation of the small noncoding and highly structured ribonucleotide tmRNA (5). Therefore, we analyzed the extracted RNA and detected the tmRNA by Northern blotting. The abundance of the tmRNA was not affected by temperature or by the presence or absence of RNase R (Fig. 2B). In some experiments we observed a band corresponding to the pre-tmRNA accumulating in the rnr mutant at 30°C and 37°C, but this band was faint and not always detectable. However, very consistently, an additional band at about 150 nt appeared in the rnr mutant at 30°C. Very likely, this represents a partially processed form of the tmRNA. This tmRNA degradation product does not accumulate in an rnr mutant at 37°C, where it might be less stable and processed by other RNases. The small subunit of the ribosome is known to be another target of RNase R. We therefore looked for degradation products of the 16S RNA in the rnr mutant. Northern blots with two different oligonucleotide probes located on the first half of the 16S RNA sequence showed that some fragments of the 16S RNA accumulate in the rnr mutant at 30°C (Fig. 2C). In conclusion, this analysis shows that the rnr mutant is defective for complete processing of the highly structured 16S RNA and tmRNA which accumulates in the rnr mutant at 30°C. In addition, other unidentified RNA products appeared to accumulate in the absence of RNase R at 30°C. Taken together, these results demonstrate a critical role of RNase R in the disposal of structured RNAs and RNA fragments which are stabilized by low temperatures.
FIG. 2.
RNase R is required for efficient degradation of structured RNA at low temperature. (A) Ethidium bromide staining of total RNA separated using denaturing polyacrylamide electrophoresis from Legionella pneumophila strains grown on plates at 30°C and 37°C. (B and C) Total RNA samples were also analyzed by Northern blotting to detect the tmRNA (B) and the 16S subunit of the ribosome (6S RNA probe A and probe B) (C). Arrowheads indicate RNA degradation products accumulating in the rnr mutant. WT, wild type.
Growth at moderate temperature induces changes in the gene expression profile of wild-type and rnr mutant Legionella strains.
The accumulation of undegraded structured RNAs at low temperature can affect bacterial physiology and could be responsible for the decreased viability of the rnr mutant at low temperatures. To evaluate the consequence of rnr disruption on global gene expression, we performed microarray analysis and gene expression profiling on the wild type and the isogenic rnr mutant. Total RNA was isolated from wild-type strain JR32 and an isogenic rnr mutant grown at 30°C or 37°C. Reverse-transcribed RNA samples were used to hybridize a custom-made 70-mer oligonucleotide array containing duplicates of all predicted open reading frames of L. pneumophila Philadelphia-1 (see Materials and Methods). The changes in gene expression at 30°C compared to 37°C are summarized in Table 1. Growth of the wild type and the rnr mutant strain at 30°C resulted in significant increases in the steady-state levels of mRNA of many genes homologous to the E. coli cold shock regulon. These include genes for the major cold shock protein and RNA chaperone CspA (lpg2121 and lpg1205) and the DEAD-box RNA helicase DpbA (lpg2122), which has high activity toward a small hairpin of the 23S ribosomal subunit (36). The increase of cspA (lpg2121) and dpbA mRNA appeared to be more pronounced in the rnr mutant than in the JR32 parent strain, which provides evidence that the rnr mutant is not impaired for the induction of genes required for adaptation to cold. Other genes encoding ribosome-associated proteins appeared as a large class of upregulated genes in both the wild-type and the RNase R-deficient strains. These include genes for four ribosomal proteins (RimM, S3, L18, and L24), the ribosome-interacting proteins of the translocon (SecY and SecE), and the elongation factor EF4 (formerly LepA). EF4 has recently been shown to back-translocate the ribosome in defective tRNA translocation reactions, giving a second chance to translocate the tRNA correctly (28). This activity would be particularly relevant in the event of the depletion of tRNAs. Interestingly, several tRNA genes were downregulated during growth at 30°C, and this downregulation was consistently more pronounced in the RNase R mutant. Low tRNA availability induces ribosome stalling and subsequent tmRNA-mediated tagging (30) and might explain the previous observation that cells lacking RNase R consistently displayed higher levels and a somewhat different pattern of tmRNA-dependent protein tagging (21). Unexpectedly, we also observed that the gene encoding the primary DNA polymerase Pol III, which has DNA repair activity, is also more expressed at 30°C than at 37°C in both the wild type and the rnr mutant strain. Comparative analysis of the gene expression profiles of the RNase R-deficient strain versus the wild type revealed additional genes specifically induced in the rnr mutant strain (Table 2). We found that another gene involved in DNA repair and replication, encoding the single-strand binding protein Ssb, is induced at low temperature specifically in the rnr mutant. Moreover, several other genes that are involved in the metabolism and/or uptake of exogenous DNA were specifically induced in the RNase R-defective strain when it was grown at 30°C. For example, the gene encoding the small periplasmic competence protein ComEA is the most strongly upregulated gene in the RNase R-deficient strain. This gene seems to be upregulated by lower temperature in both the wild type and the rnr mutant, but its expression level is five times higher in the rnr mutant strain than in the wild type at 30°C. In addition, genes encoding components of type IV pili such as PilT and the putative pilin and prepilin Lpg1870 and Lpg0632 are also upregulated. lpg0632 is the first gene of a putative operon encoding six components of type IV pili which are all, to a lesser extent, upregulated in the RNase R-deficient strain (see Table S1 in the supplemental material). Interestingly, this operon is followed by the gene encoding the channel protein ComEC, an essential component of the DNA uptake machinery in bacteria. In conclusion, the growth of L. pneumophila at lower temperature seems to induce a cold shock-like response in both the wild type and the rnr mutant, providing evidence that the reduced growth of the rnr mutant at 30°C is not caused by repression of genes required for adaptation to cold. However, growth at 30°C of the rnr strain was accompanied by the unanticipated expression of additional genes involved in DNA metabolism and DNA uptake.
TABLE 1.
List of genes differentially regulated in response to temperature in the rnr mutant of L. pneumophilaa
| Name; function | Gene | Fold change (30°C/37°C) forb:
|
|
|---|---|---|---|
| rnr mutant | Wild type | ||
| lpg2121; cold shock protein CspA | 5.03 | 2.38 | |
| lpg1397; beta-ketoacyl-acyl carrier protein synthase II | fabF1 | 4.69 | 2.20 |
| lpg0810; competence protein ComEA | comEA | 3.48 | 3.45 |
| lpg1872; elongation factor EF4 (elongation factor LepA) | lepA | 3.30 | 2.64 |
| lpg0910; enhanced entry protein EnhA | enhA | 3.29 | 1.37 |
| lpg0397; 16S rRNA processing protein RimM | rimM | 3.20 | 2.62 |
| lpg0316; preprotein translocase, SecE subunit | secE | 3.18 | 2.08 |
| lpg0335; 30S ribosomal protein S3; RpsC | 2.99 | 2.11 | |
| lpg0977; single-strand DNA binding protein (Ssb) | ssb | 2.95 | 0.59 |
| lpg2429; malonate decarboxylase | mdcG | 2.88 | 0.75 |
| lpg2870; putative permease | 2.77 | 1.37 | |
| lpg0425; ferrochelatase HemH | hemH | 2.77 | 1.54 |
| lpg2122; ATP-dependent RNA helicase DeaD | dbpA | 2.75 | 1.71 |
| lpg2294; hydrolase (haloacid dehalogenase family) | 2.72 | 2.11 | |
| lpg1336; enhanced entry protein EnhA | enhA | 2.69 | 0.77 |
| lpg0753; UDP-N-acetylglucosamine-2-epimerase | neuC | 2.69 | 1.57 |
| lpg0229; heme oxygenase | 2.65 | 3.00 | |
| lpg1400; DNA Pol III, delta prime subunit | holB | 2.64 | 3.51 |
| lpg0751; CMP-N-acetylneuraminic acid synthetase | neuA | 2.64 | 2.08 |
| lpg2472; hydrogenase expression/formation protein HypD | hypD | 2.62 | 2.73 |
| lpg0349; preprotein translocase SecY | secY | 2.62 | 2.56 |
| lpg2041; radical SAM domain protein | 2.60 | 1.16 | |
| lpg1205; cold shock domain family protein CspA | 2.60 | 2.63 | |
| lpg2473; hydrogenase expression/formation protein HypC | hypC | 2.60 | 2.33 |
| lpg1420; cytidylate kinase | cmk | 2.59 | 2.37 |
| lpg0345; 50S ribosomal protein L18 | rplR | 2.58 | 1.98 |
| lpg0853; transcriptional regulator FleQ (FlaK) | fleQ | 2.56 | 2.23 |
| lpg1997; type IV pilin, putative | 2.54 | 1.84 | |
| lpg1915; Tfp pilus assembly protein, type IV pilin | pilE | 2.52 | 1.57 |
| lpg0340; 50S ribosomal protein L24 | rplX | 2.51 | 2.62 |
| lpg1600; tRNA-Ser | 0.30 | 0.54 | |
| lpg0234; SidE | sidE | 0.30 | 0.46 |
| lpg2191; global stress protein GspA | 0.30 | 0.39 | |
| lpg1836; coiled-coil domain protein | 0.30 | 0.43 | |
| lpg1929; tRNA-Phe | 0.29 | 0.47 | |
| lpg1601; tRNA-Arg | 0.27 | 0.33 | |
| lpg0915; cell division transmembrane protein FtsL | 0.26 | 0.35 | |
| lpg1545; DNA binding protein, putative | 0.26 | 0.48 | |
| lpg1878; tRNA-Glu | 0.26 | 0.33 | |
| lpg1330; tRNA-Met | 0.24 | 0.32 | |
| lpg1857; tRNA-Val | 0.23 | 0.45 | |
| lpg2844; proteophosphoglycan, membrane associated | 0.22 | 0.36 | |
| lpg1865; tRNA-Arg | 0.19 | 0.31 | |
| lpg2292; tRNA-Gly | 0.18 | 0.25 | |
| lpg0881; tRNA-Ser | 0.16 | 0.35 | |
| lpg2775; tRNA-Met | 0.16 | 0.31 | |
| lpg1185; tRNA-Ser | 0.15 | 0.30 | |
| lpg1272; tRNA-Arg | 0.14 | 0.18 | |
Genes of unknown function and putative open reading frames were omitted from the list.
Listing is limited to genes showing a change of >2.5-fold or <0.3-fold in the rnr mutant.
TABLE 2.
List of genes differentially regulated in the rnr mutant of L. pneumophila compared to the wild type at 30°Ca
| Name; function | Gene | Fold change (rnr mutant/wild type) atb:
|
|
|---|---|---|---|
| 30°C | 37°C | ||
| lpg0810; competence protein ComEA | comEA | 5.23 | 5.18 |
| lpg0242; D-3-phosphoglycerate dehydrogenase | serA | 4.53 | 3.14 |
| lpg1397; beta-ketoacyl-acyl carrier protein synthase II | fabF1 | 4.46 | 2.09 |
| lpg2013; twitching motility protein PilT | pilT | 4.28 | 3.81 |
| lpg0127; acetyl-coenzyme A synthetase | acsB | 4.00 | 1.72 |
| lpg0063; phospho-2-dehydro-3-deoxyheptonate aldolase | aroH | 3.81 | 4.40 |
| lpg2569; transcription regulator, putative | 3.67 | 4.65 | |
| lpg1340; flagellin | fliC | 3.51 | 1.66 |
| lpg0938; lipoprotein signal peptidase | lspA | 3.25 | 2.52 |
| lpg1370; DNA binding protein Fis | fis | 3.21 | 3.93 |
| lpg0066; carbamoyl phosphate synthase large chain | 2.73 | 3.23 | |
| lpg0952 phosphoesterase (serine protease) | ychK | 2.71 | 2.32 |
| lpg0910; enhanced entry protein EnhA | enhA | 2.65 | 1.12 |
| lpg1219; flagellar hook protein FlgE | flgE | 2.54 | 2.07 |
| lpg2863; pteridine reductase 1 | 2.53 | 2.20 | |
| lpg0397; 16S rRNA-processing protein RimM | rimM | 2.51 | 2.06 |
| lpg1655; class 4 metalloprotease | lasB | 2.49 | 2.39 |
| lpg1218; flagellar basal body rod modification protein FlgD | flgD | 2.45 | 2.54 |
| lpg2837; lysophospholipase A (GDSL motif lipase/hydrolase) | 2.42 | 2.25 | |
| lpg2159; CrtF (enzyme-methylating tetracycline) | 2.40 | 2.25 | |
| lpg0245; NAD-glutamate dehydrogenase | 2.38 | 1.25 | |
| lpg1572; TolA colicin import membrane protein | tolA | 2.36 | 2.71 |
| lpg2974; phosphatidylserine decarboxylase | psd | 2.31 | 1.75 |
| lpg0804; choloylglycine hydrolase (penicillin acylase) | 2.29 | 3.69 | |
| lpg1336; enhanced entry protein EnhA | enhA | 2.27 | 0.65 |
| lpg1889; lipase (triacyglycerol lipase) | 2.25 | 2.62 | |
| lpg0632; type IV prepilin | 2.25 | 1.97 | |
| lpg1870; transmembrane protein (fimbrial, pilin) | 2.25 | 1.74 | |
| lpg1649; myoinositol catabolism protein IolE | iolE | 2.22 | 2.63 |
| lpg2355; Glu-tRNA amidotransferase subunit A | 2.17 | 2.46 | |
| lpg0322; DNA-directed RNA polymerase beta subunit rpoB | 2.16 | 1.42 | |
| lpg0786; cell cycle protein MesJ | mesJ | 2.16 | 2.71 |
| lpg2252; glutamine synthetase | 2.15 | 2.64 | |
| lpg2584; SidF, inhibitor of growth family, member 3 | sidF | 2.10 | 2.01 |
| lpg1052; ATP synthase, putative | 2.10 | 1.05 | |
| lpg2251; succinate semialdehyde dehydrogenase | 2.09 | 2.15 | |
| lpg2426; malonate decarboxylase alpha subunit | mdcA | 2.07 | 2.10 |
| lpg1053; FoF1-type ATP synthase epsilon subunit | 2.07 | 2.11 | |
| lpg0977; single-strand DNA binding protein Ssb | ssb | 2.07 | 0.41 |
| lpg2468; sulfhydrogenase delta subunit | 2.06 | 1.33 | |
| lpg2732; (two-component) response regulator | 2.05 | 3.04 | |
| lpg2222; TPR repeat protein, protein-protein interaction | 2.04 | 1.00 | |
| lpg1093; cation efflux transporter ATPase | pacL | 2.02 | 1.87 |
| lpg1097; polyhydroxyalkanoic synthase | phbC | 2.01 | 1.67 |
| lpg1206; sigma 54 modulation protein YhbH | 2.00 | 1.07 | |
| lpg0093; RNA methyltransferase | 0.30 | 0.98 | |
| lpg0094; ribose-5-phosphate isomerase A | rpiA | 0.26 | 0.41 |
| lpg0092; exoribonuclease R | rnr | 0.26 | 0.57 |
Changes of the corresponding genes in the rnr mutant compared to the wild type at 37°C are also displayed. Genes of unknown function and putative open reading frames were omitted from the list.
Listing is limited to genes showing a change of >2.0-fold or <0.3-fold at 30°C.
The gene expression profile of RNase R mutant shows similarities to the gene expression profiles of constitutively competent Legionella mutants.
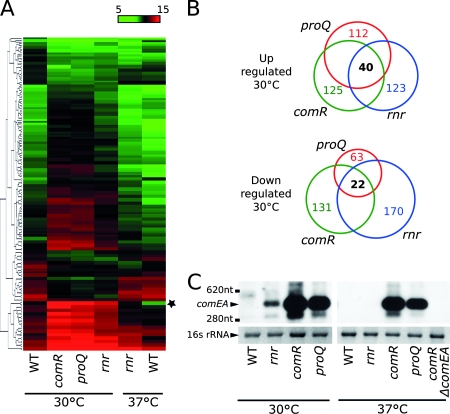
Two L. pneumophila mutants have been described previously to be constitutively competent for DNA uptake and genetic transformation. These two mutants, the comR and proQ mutant strains, were retrieved from a collection of Tn903dIIlacZ mutants (37), and their gene expression profiles were analyzed upon growth at 30°C. The two strains showed a significant overlap of more than a hundred differentially regulated genes (more/less than twofold expressed; P < 0.005). As expected, many of these genes have functions related to competence (see Table S2 in the supplemental material). We then compared the expression of this subset of genes to the expression profile of the rnr mutant grown at 30°C and at 37°C. The gene expression profile of the rnr strain grown at 30°C has great similarities to the gene expression profile of the competent comR and proQ mutant strains. However, upon growth at 37°C, the specific gene expression profile of the rnr strain diverged from that exhibited by the comR, proQ, and rnr strains at 30°C and shared more similarities to the noncompetent JR32 parent strain (Fig. 3A). We found that all three mutant strains shared a set of 62 differentially expressed genes, representing about 20% of all genes differentially expressed in the rnr mutant (Fig. 3B). We performed Northern blot analysis on the comEA gene, which is the most upregulated gene in all three mutants. The comEA transcript was not detectable for wild-type strain JR32 but was detected at 30°C for the rnr mutant and at even higher amounts for the comR and proQ mutants (Fig. 3C). At 37°C, the abundance of the comEA transcript seemed to be unaffected in the comR and proQ strains, whereas consistently with the microarray data it became undetectable in the rnr mutant. In conclusion, RNase R directly or indirectly represses a competence-related regulon in a temperature-dependent manner.
FIG. 3.
The rnr mutant shows an expression profile similar to those of the competent comR and proQ mutants. (A) Heat diagram of spot intensities of differentially regulated genes in the comR and proQ mutants (log2R <> 1; P < 0.001). The star indicates the comEA gene. (B) Venn diagram showing differentially expressed genes (log2R <> 1; P < 0.005) in the comR, proQ, and rnr mutants at 30°C. (C) Northern blot analysis of the comEA transcript in JR32 (wild type) and the rnr, comR, and proQ mutants. WT, wild type.
The L. pneumophila rnr mutant is competent for genetic transformation.
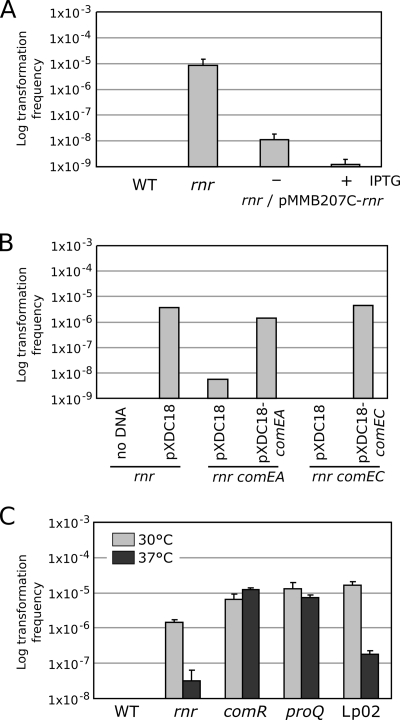
The gene expression profiling of the rnr mutant strongly suggested that it would be competent for genetic transformation. To test the ability of the rnr mutant to take up and integrate exogenous DNA, we grew the rnr mutant on CYE plates together with 1 μg of genomic DNA extracted from the gentamicin-resistant JR32-derived strain JC51. After 48 h of coincubation at 30°C, the transformation frequency was determined. The rnr mutant was competent with transformants appearing at a frequency of about 1 × 10−5 per μg of genomic DNA (Fig. 4A), similar to the transformation frequency observed for the comR and proQ mutant strains. The competence phenotype of the rnr mutation could be decreased by the expression of RNase R under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptac promoter (Fig. 4A). Even in the absence of IPTG, the competence phenotype of the rnr mutation could be decreased, presumably due to promoter leakiness resulting in a low expression level of RNase R. To confirm that the transformants appearing in the competence assays were the results of DNA uptake, we deleted the comEA and comEC genes, encoding the periplasmic DNA binding protein ComEA and the transmembrane DNA channel ComEC of the DNA uptake machinery. Deletion of comEC or comEA resulted in a dramatic drop in transformation frequency (Fig. 4B). The comEA mutant retained some residual activity (Fig. 4B), consistent with previous observations for other organisms, suggesting that another unidentified DNA binding protein contributes to the DNA uptake process. Microarray analysis and Northern blot analysis showed that expression of comEA is temperature dependent in the rnr mutant but not in the proQ and comR mutants. We then tested the competence of these three mutants at 30°C and 37°C, along with that of the Lp02 strain, which had previously been shown to be competent due the acquisition of an unidentified mutation (34) (Fig. 4C). While the comR and proQ mutants appeared to be constitutively competent regardless of the temperature, the rnr mutant was far less competent at 37°C, a situation that was predicted by the comEA Northern blot results and is similar to what had been reported for the Lp02 strain (34).
FIG. 4.
The rnr mutant is competent for genetic transformation. (A) Wild-type and rnr mutant strains of Legionella pneumophila were incubated for 2 days on nonselective plates with 1 μg of genomic DNA carrying an antibiotic resistance marker and then plated on selective and nonselective media. Transformation frequency represents the number of CFU on selective plates versus the number of CFU on nonselective plates. (B) Determination of transformability of the double rnr comEA and rnr comEC mutants at 30°C. (C) Effect of temperature on the transformability of Lp02 along with JR32 and its isogenic proQ, comR, and rnr mutants. When displayed, error bars represent standard deviations derived from at least three independent experiments. WT, wild type.
Regulation of comEA is posttranscriptional.
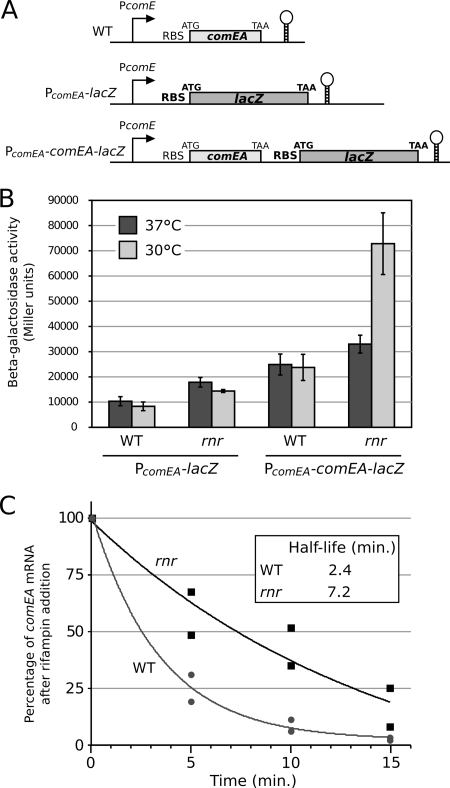
To further understand the induction of competence in the rnr mutant, we analyzed the source of the upregulation of comEA. We constructed chromosomal transcriptional lacZ fusions to the comEA promoter and to the whole comEA transcript in the wild-type strain and the rnr mutant (Fig. 5A). Determination of beta-galactosidase activity in the wild type and the rnr mutant showed only a minor increase in comEA promoter activity in the rnr mutant at both 37°C and 30°C (Fig. 5B). In contrast, a greater increase (>3-fold) in beta-galactosidase activity was detected in the rnr mutant at 30°C when the lacZ gene was placed at the end of the comEA transcript (Fig. 5B). This increase in comEA-lacZ expression was not observed for the rnr mutant at 37°C, which is a finding in agreement with the microarray results. Although we cannot rule out the possibility of the presence of a regulatory element within the comEA transcript, it seems that RNase R does not directly control the comEA promoter but rather controls comEA expression by affecting the steady-state level of the comEA mRNA. We analyzed the decay rate of the comEA mRNA in the wild type and the rnr mutant. The strains were grown at 30°C to exponential phase, transcription was stopped by the addition of rifampin, aliquots were taken at different time points, and amounts of the comEA mRNA were quantified by real-time reverse transcription-PCR (see Materials and Methods). We found that the decay rate for the rnr mutant is much lower than that for the wild type, indicating that RNase R plays a role in the turnover of the comEA mRNA (Fig. 5C). Taken together, these results show that RNase R does not directly control the promoter of comEA and that the increased expression of comEA in the rnr mutant is largely due to increased stability of its mRNA.
FIG. 5.
Expression of comEA in the rnr mutant. (A) Schematic representation of beta-galactosidase gene fusion to comEA in the chromosome of wild-type and rnr mutant strains of Legionella pneumophila. RBS, ribosome binding site. (B) Beta-galactosidase activities of the corresponding fusions from wild-type and rnr mutant strains of Legionella pneumophila grown on CYE plates at 30°C and 37°C. Error bars represent standard deviations derived from three independent experiments. (C) Decay of the comEA mRNA in the wild type and the rnr mutant at 30°C. Amounts of comEA mRNA were quantified by quantitative real-time reverse transcription-PCR at different times after rifampin addition. Data are expressed as percentages of the initial amount of mRNA. Half-lives of the mRNA were determined from two independent experiments. WT, wild type.
RNase R does not control the expression of the competence repressors ComR and ProQ.
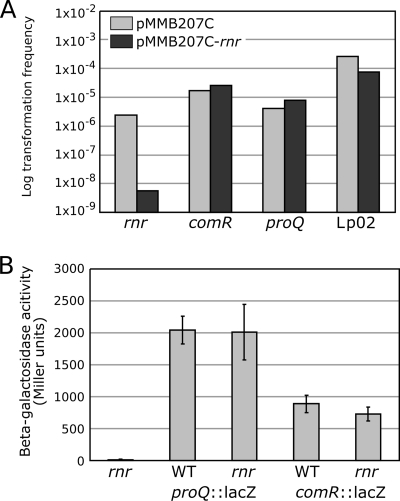
The identity of the genetic alteration that made the Lp02 strain competent has not been identified, but its dependence upon temperature raised the possibility that the rnr gene might be altered in Lp02. We tested this possibility by expressing RNase R in the Lp02 strain and testing its competence at 30°C. We also expressed RNase R in the comR and proQ mutants to find out if the comR and proQ mutations resulted in competence by downregulation of rnr. Expression of the rnr gene did not repress the competence of Lp02 or of the comR and proQ mutants (Fig. 6A). In addition, the rnr gene cloned from Lp02 could successfully complement the rnr mutation in JR32 (data not shown). The genetic alteration that makes Lp02 competent is therefore not associated with the rnr gene. However, the temperature dependency of the rnr mutant and the Lp02 strain suggests that the induction of competence in both strains is controlled by the same regulatory pathway. In contrast, the fact that the expression of the rnr gene in the comR and proQ mutants did not lower the level of competence suggests that the downstream pathway leading to competence in the comR and proQ mutants does not involve RNase R. Indeed, that pathway is not temperature dependent. Another possibility would be that RNase R somehow affects the expression of the competence repressors comR and proQ. To test this hypothesis, we took advantage of the translational lacZ fusions produced in the comR and proQ mutants by Tn903dIIlacZ insertions. The comR::Tn903dIIlacZ and proQ::Tn903dIIlacZ fusions were moved to the rnr background, and we measured the expression of the beta-galactosidase in the wild-type and rnr backgrounds. The inactivation of rnr did not result in significant changes in the expression of the ComR- and ProQ-LacZ fusions (Fig. 6B). In addition, the expressions of comR and proQ were determined by quantitative PCR in the wild-type and rnr backgrounds and were found to be identical (data not shown). Taken together, these data support the idea of a temperature-dependent pathway leading to the induction of competence in the rnr mutant that is different from the one involved in the comR and proQ mutants.
FIG. 6.
(A) Effect of RNase R overexpression on the transformability at 30°C of Lp02 and JR32 and of the isogenic proQ, comR, and rnr mutants of JR32. (B) Beta-galactosidase activities of wild-type (WT) and rnr mutant strains of Legionella pneumophila carrying translational fusions of ComR (comR::lacZ) and ProQ (proQ::lacZ) to beta-galactosidase. When displayed, error bars represent standard deviations derived from at least three independent experiments.
Enzymatically active RNase R is required to repress competence at 30°C.
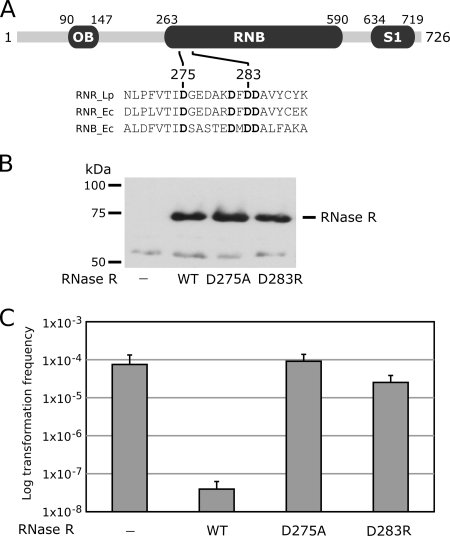
RNase R has two RNA binding domains (1, 4) in addition to the RNB catalytic domain (Fig. 7A). RNase R, RNase II, and PNPase (a phosphorolytic exoribonuclease) have an S1 RNA binding domain at the C termini. The enzymatic activity of PNPase is required for the optimal functioning of the Yersinia type three secretion system (31). Although PNPase is an exoribonuclease, its enzymatic activity was not required for the proper function of the type three secretion system, and the phenotype could be complemented by the expression of other S1 domains from unrelated RNase or RNA binding proteins. We tested the possibility that another RNase harboring an S1 RNA binding domain might be able to complement the L. pneumophila RNase R mutant for derepression of competence. Because Legionella does not have RNase II, PNPase is the only exoribonuclease harboring an S1 domain in Legionella. Even though overexpression of PNPase appeared to be slightly toxic, it could not repress competence (data not shown). We then hypothesized that the hydrolytic exoribonuclease activity of RNase R was necessary to repress competence. We generated point mutations of conserved aspartic residues D275A and D283R in the RNase R active site (Fig. 7A). These residues correspond to residues D201 and D209 of the Escherichia coli RNB domain of RNase II and coordinate magnesium ions. Mutation D201A significantly reduced exoribonuclease activity (4, 39), and mutation D209R abolished RNA activity while retaining RNA binding activity (1, 18). We then tested the mutant RNase R's for their ability to repress the competence of the rnr mutant. Even though the mutant enzymes seem to be stable and produced in amounts similar to those seen for the wild-type enzyme (Fig. 7B), they could not repress competence (Fig. 7C). This shows that a functional exoribonuclease domain is required for the repression of competence, indicating that the sole loss of exoribonuclease activity of RNase R can dramatically modify gene expression and result in competence induction.
FIG. 7.
Loss of exoribonuclease activity of RNase R induces competence. (A) Domain architecture of the L. pneumophila RNase R and alignment of the active site from members of the RNase R family showing conserved aspartic residues critical for exoribonuclease activity (RNR_Lp, RNase R of Legionella pneumophila; RNR_Ec, RNase R of Escherichia coli; RNB_Ec, RNase II of Escherichia coli). OB, oligosaccharide/oligonucleotide binding protein domain. (B) Western blot of wild-type and active-site mutants of RNase R expressed in the rnr mutant. (C) Effect of point mutations of the RNase R active site on genetic transformability at 30°C. Error bars represent standard deviations derived from three independent experiments. WT, wild type.
DISCUSSION
In E. coli, the highly conserved 3′-5′ exoribonuclease R (RNase R) has been shown to be involved in a variety of cell processes, including quality control of rRNA, processing of mRNA molecules with extensive secondary structure, degradation of mRNA on which ribosome stalling occurs, and maturation of the tmRNA. In spite of being involved in all of these important processes, an E. coli rnr mutant does not display any particular defect in cell growth at optimal growth temperature (37°C). This has been explained in part by the fact that the two other exoribonucleases, RNase II and PNPase, display enough functional overlap to perform the missing functions (14). In L. pneumophila, the rnr gene is not essential for the growth of the bacterium whether as a free-living organism or as an intracellular parasite inside macrophages or amoebas. However, the rnr mutant displays longer doubling time at 30°C, suggesting, as for E. coli, a role of RNase R in adaptation to low temperatures (5). We assumed that under these conditions the inactivation of RNase R would have an effect on the transcriptional profile of the bacteria. Interestingly, this experiment had never been performed with E. coli but proved to be highly informative for L. pneumophila. Regardless of growth temperature (30°C or 37°C), we observed changes in the steady-state level of mRNA in the rnr mutant. These changes affected a limited number of genes. Not surprisingly, growth at 30°C correlated with a moderate induction of a cold shock-like response, which was more pronounced in the rnr mutant. More surprisingly, the rnr mutation also resulted in the upregulation of competence-related genes.
Bacterial competence is a genetically programmed process which is triggered as a response to environmental stimuli. DNA uptake from the extracellular milieu by competence requires the expression of a complex machinery composed of a competence pseudopilus and a DNA translocase (8). In L. pneumophila, it has been reported that microaerophilic conditions (35) and genetic alteration of the competence repressors ComR and ProQ result in competence (34). Whereas growth under microaerophilic conditions results in a transformation frequency of about 10−6 with more than 100 μg of genomic DNA, the genetic alterations of proQ or comR result in a transformation frequency of 10−5 with only 1 μg of genomic DNA. This suggests that the environmental signal that triggers the expression of the competence regulon in L. pneumophila has yet to be found.
Many competence genes were found to be induced in the rnr mutant and represent a notable upregulated regulon. The DNA translocase-encoding genes comEA and comEC were both found to be upregulated in the rnr mutant. Although not observed from the microarray data, the comEC gene has been found to be upregulated by threefold by use of quantitative PCR (data not shown). Comparison of the expression profiles of the rnr mutant and the competent comR and proQ mutants showed that the induction of a competence regulon was temperature dependent. As predicted by the microarray analysis, the rnr mutant was found to be competent for genetic transformation only at 30°C but not at 37°C, a feature that is not shared by the constitutively competent comR and proQ mutants. Indeed, the induction of competence in the rnr mutant was found to be independent of the competence repressors ComR and ProQ. Because RNase R is involved in the tmRNA-dependent degradation of mRNA (5, 19, 26) and the degradation of repetitive extragenic palindrome (REP)-containing mRNA (10) and the ompA mRNA (2), the upregulation of the competence genes could have been attributed to the increase in the steady-state level of the mRNA targets of RNase R. In E. coli, the expression of RNase R can support the degradation of the cspA mRNA, which is stabilized by low temperature by the absence of the DEAD-box RNA helicase CsdA (3). The induction of competence in the rnr mutant could be the result of the stabilization of the mRNA of the competence regulon. Supporting this hypothesis, we found that the induction of comEA expression is not due to higher promoter activity. Instead, we found that the comEA mRNA has a longer half-life in the rnr mutant, indicating that it is a substrate of RNase R. Indeed, the sole inactivation of the exoribonuclease activity of RNase R at 30°C is sufficient to induce competence development. A recent genome-wide analysis of mRNA decay in an rnr mutant of Pseudomonas putida has revealed the important role of RNase R in mRNA turnover (17). Although RNase R is active on mRNAs containing REP sequences, the mRNAs affected by the lack of RNase R were not enriched in REP-containing sequences. We could not identify any REP sequence within the comEA mRNA and the mechanism underlying its increased stability is unclear. An alternative explanation would be that the comEA mRNA becomes a substrate of RNase R as a consequence of ribosome stalling. Ribosome stalling can be induced by rare codons or by a low availability of tRNA (30), and several tRNAs show reduced steady-state levels in the rnr mutant at 30°C (Table 3). The selective degradation by RNase R of mRNA on which the ribosome is stalled is dependent on the presence of the SmpB protein and tmRNA (29), and the absence of either SmpB or tmRNA results in an increased level of mRNA, inducing ribosome stalling. If the comEA mRNA is subject to RNase R because of ribosome stalling, a mutation of either smbB or tmRNA should also result in increased expression of comEA and eventually in competence development. Unfortunately, despite repeated attempts, we could not obtain ssrA or smpB mutants, suggesting that these genes are essential for viability in L. pneumophila.
TABLE 3.
List of strains, plasmids, and oligonucleotide probes
| Strain, plasmid, or probe | Description and/or genotype or sequence | Reference/source or characteristic/use |
|---|---|---|
| Strains | ||
| JR32 | Philadelphia-1; Smr; r− m+ | 32 |
| LELA559 | JR32 rnr::Tn903dIIlacZ, insertion at nt 513; Knr | This work |
| LELA559C | JR32 rnr::Tn903dIIlacZ::cat; Cmr | This work |
| LELA559G | JR32 rnr::Tn903dIIlacZ::aacC1; Gmr | This work |
| LELA559C ΔcomEA | LELA559C ΔcomEA::aptII; Knr | This work |
| LELA559C ΔcomEC | LELA559C ΔcomEC::aptII, Knr | This work |
| LELA3825 | JR32 proQ::Tn903dIIlacZ; Knr | This work |
| LELA3644 | JR32 comR::Tn903dIIlacZ; Knr | This work |
| LELA559G-3825 | LELA559G proQ::Tn903dIIlacZ; Knr | This work |
| LELA559G-3644 | LELA559G comR::Tn903dIIlacZ; Knr | This work |
| KS79 | JR32 ΔcomR | 13 |
| KS79 dotB | KS79 dotB::Tn903dIIlacZ; Knr | This work |
| KS79 dotB-hygro | KS79 dotB::Tn903dIIlacZ::hph; Hygr | This work |
| JR32 comEA-lacZ | JR32 PcomEA::lacZ; Knr | This work |
| JR32 comEA-lacZ2 | JR32 PcomEA-comEA::lacZ; Knr | This work |
| LELA559G comEA-lacZ | LELA559G PcomEA::lacZ; Knr | This work |
| LELA559G comEA-lacZ2 | LELA559G PcomEA-comEA::lacZ; Knr | This work |
| Plasmids | ||
| pMMB207C | Derivative of IncQ plasmid RSF1010 | Cmr; ΔmobA |
| pXDC36 | pMMB207C-rnr | rnr complementing plasmid |
| pXDC36-D275A | pMMB207C-rnr | Expresses RNaseR D275A |
| pXDC36-D283R | pMMB207C-rnr | Expresses RNaseR D283R |
| pXDC18 | pMMB207C::aacC1; Gmr | Gmr version of pMMB207C |
| pXDC18-comEA | pXDC18-comEA | comEA complementing plasmid |
| pXDC18-comEC | pXDC18-comEC | comEC complementing plasmid |
| pXDC44 | pGEM-Tn903dIIlacZ::cat; Ampr Cmr | Swapping cassette for Tn903dIIlacZ |
| pXDC46 | pGEM-Tn903dIIlacZ::aacC1; Ampr Gmr | Swapping cassette for Tn903dIIlacZ |
| pXDC57 | pGEM-Tn903dIIlacZ::hph; Ampr Hygr | Swapping cassette for Tn903dIIlacZ |
| Probes | ||
| comEA | CTACAAAACGCTGCCCTATACCCTTGACTTCCGCAAGC | |
| 16S-A | TGCTGGCACGGAGTTAGCCGGTGCTTCTTCTGTGGGTAACGTCCA | |
| 16S-B | ACTGAAAGTGCTTTACAACCCTCAGGCCTTCTTCACACACGCGGC |
Further work will be necessary to understand the mechanism of competence development in the rnr mutant and might help us understand the function of RNase R and the regulatory cascade leading to competence development in L. pneumophila. The induction of competence in the rnr mutant is a remarkable finding, since it is the first report of a gain of function due to the loss of RNase R. L. pneumophila may provide a convenient model system to study the function of RNase R, because the loss of RNase R activity has directly measurable transcriptional consequences.
Supplementary Material
Acknowledgments
We thank Murray Deutscher (Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine) for kindly providing polyclonal antibodies against RNase R. We thank the two anonymous reviewers for their constructive comments and detailed corrections. We also thank members of the Shuman lab for helpful discussion and comments.
This work was supported by NIH grants AI064481 and AI023549 awarded to H.A.S. X.C. was supported in part by a postdoctoral fellowship from the Fondation pour la Recherche Médicale (FRM) and by NIH funding to H.A.S. S.P.F. is supported by a postdoctoral fellowship from NSERC.
Footnotes
Published ahead of print on 10 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amblar, M., A. Barbas, P. Gomez-Puertas, and C. M. Arraiano. 2007. The role of the S1 domain in exoribonucleolytic activity: substrate specificity and multimerization. RNA 13317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, J. M., F. Cairrão, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60219-228. [DOI] [PubMed] [Google Scholar]
- 3.Awano, N., C. Xu, H. Ke, K. Inoue, M. Inouye, and S. Phadtare. 2007. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 1895808-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbas, A., R. G. Matos, M. Amblar, E. López-Viñas, P. Gomez-Puertas, and C. M. Arraiano. 2008. New insights into the mechanism of RNA degradation by ribonuclease II: identification of the residue responsible for setting the RNase II end product. J. Biol. Chem. 28313070-13076. [DOI] [PubMed] [Google Scholar]
- 5.Cairrão, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 501349-1360. [DOI] [PubMed] [Google Scholar]
- 6.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 6171-87. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and M. P. Deutscher. 2005. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 28034393-34396. [DOI] [PubMed] [Google Scholar]
- 8.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2241-249. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 3031358-1361. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Z., and M. P. Deutscher. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17313-318. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, Z., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 27721624-21629. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Z., and M. P. Deutscher. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. USA 1006388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Felipe, K. S., R. T. Glover, X. Charpentier, O. R. Anderson, M. Reyes, C. D. Pericone, and H. A. Shuman. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathogens 4e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutscher, M. P. 2006. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erova, T. E., V. G. Kosykh, A. A. Fadl, J. Sha, A. J. Horneman, and A. K. Chopra. 2008. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 1903467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonseca, P., R. Moreno, and F. Rojo. 2008. Genomic analysis of the role of RNase R in the turnover of Pseudomonas putida mRNAs. J. Bacteriol. 1906258-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazão, C., C. E. McVey, M. Amblar, A. Barbas, C. Vonrhein, C. M. Arraiano, and M. A. Carrondo. 2006. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443110-114. [DOI] [PubMed] [Google Scholar]
- 19.Hong, S., Q. Tran, and K. C. Keiler. 2005. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 57565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 2862165-2169. [DOI] [PubMed] [Google Scholar]
- 21.Karzai, A. W., and R. T. Sauer. 2001. Protein factors associated with the SsrA. SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA 983040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalonde, M. S., Y. Zuo, J. Zhang, X. Gong, S. Wu, A. Malhotra, and Z. Li. 2007. Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA 131957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mian, I. S. 1997. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 253187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikawa, J., and M. Kawabata. 1998. PCR- and ligation-mediated synthesis of marker cassettes with long flanking homology regions for gene disruption in Saccharomyces cerevisiae. Nucleic Acids Res. 26860-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purusharth, R. I., F. Klein, S. Sulthana, S. Jäger, M. V. Jagannadham, E. Evguenieva-Hackenberg, M. K. Ray, and G. Klug. 2005. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 28014572-14578. [DOI] [PubMed] [Google Scholar]
- 26.Purusharth, R. I., B. Madhuri, and M. K. Ray. 2007. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 28216267-16277. [DOI] [PubMed] [Google Scholar]
- 27.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381169-172. [DOI] [PubMed] [Google Scholar]
- 28.Qin, Y., N. Polacek, O. Vesper, E. Staub, E. Einfeldt, D. N. Wilson, and K. H. Nierhaus. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127721-733. [DOI] [PubMed] [Google Scholar]
- 29.Richards, J., P. Mehta, and A. W. Karzai. 2006. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 621700-1712. [DOI] [PubMed] [Google Scholar]
- 30.Roche, E. D., and R. T. Sauer. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 184579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenzweig, J. A., G. Weltman, G. V. Plano, and K. Schesser. 2005. Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280156-163. [DOI] [PubMed] [Google Scholar]
- 32.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 615361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 655057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sexton, J. A., and J. P. Vogel. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 1863814-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 1811395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsu, C. A., K. Kossen, and O. C. Uhlenbeck. 2001. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 7702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11641-653. [DOI] [PubMed] [Google Scholar]
- 38.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 291017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo, Y., H. A. Vincent, J. Zhang, Y. Wang, M. P. Deutscher, and A. Malhotra. 2006. Structural basis for processivity and single-strand specificity of RNase II. Mol. Cell 24149-156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.