Abstract
The genomes of two closely related thermophilic cyanobacterial isolates, designated Synechococcus isolate OS-A and Synechococcus isolate OS-B′, from the microbial mats of Octopus Spring (Yellowstone National Park) have been sequenced. An extensive suite of genes that are controlled by phosphate levels constitute the putative Pho regulon in these cyanobacteria. We examined physiological responses of an axenic OS-B′ isolate as well as transcript abundances of Pho regulon genes as the cells acclimated to phosphorus-limiting conditions. Upon imposition of phosphorus deprivation, OS-B′ stopped dividing after three to four doublings, and absorbance spectra measurements indicated that the cells had lost most of their phycobiliproteins and chlorophyll a. Alkaline phosphatase activity peaked and remained high after 48 h of phosphorus starvation, and there was an accumulation of transcripts from putative Pho regulon genes. Interestingly, the genome of Synechococcus isolate OS-B′ harbors a cluster of phn genes that are not present in OS-A isolates. The proteins encoded by the phn genes function in the transport and metabolism of phosphonates, which could serve as an alternative phosphorus source when exogenous phosphate is low. The phn genes were upregulated within a day of eliminating the source of phosphate from the medium. However, the ability of OS-B′ to utilize methylphosphonate as a sole phosphorus source occurred only after an extensive period of exposure to the substrate. Once acclimated, the cells grew rapidly in fresh medium with methylphosphonate as the only source of phosphorus. The possible implications of these results are discussed with respect to the ecophysiology of the microbial mats.
Significant advances have been made in characterizing the diversity and ecophysiology of microorganisms in the alkaline hot spring microbial mats of Yellowstone National Park (4, 53). However, relatively little is known about specific physiological adaptations and the acclimation responses of microbes in the mats with respect to nutrient limitation (1, 40, 42, 43). A unicellular cyanobacterium, Synechococcus sp., is the dominant, primary producer in these hot spring microbial mats at temperatures above 50°C and is found exclusively in the top green layer (comprising 1 to 2 mm of the ∼1.5-cm-thick mat) (53). Based on denaturing gradient gel electrophoresis analysis and the subsequent determination of the 16S rRNA/internal transcribed spacer sequence, it appears that Synechococcus sp. ecotypes are delineated along environmental gradients, including temperature and light (9, 10, 37). For instance, the Synechococcus OS-A isolate (hereafter OS-A) is abundant in the high-temperature region of the mat (58 to 65°C), while the OS-B′ isolate is abundant in the lower-temperature regions (50 to 60°C). OS indicates that the organisms were isolated from Octopus Spring (9).
We have complete genome sequences for Synechococcus OS-A and OS-B′ isolates, which have enabled examination of the genetic potential of these organisms to respond to fluctuating environmental conditions, such as nutrient levels, light intensities, and oxygen tension (3, 42, 43). Recently, we generated an axenic culture of OS-B′ and investigated acclimation responses to light intensities that they would experience in the microbial mat (19). It was found that OS-B′ cultures grew best at relatively low light intensities (25 to 200 μmol photon m−2 s−1), despite the high irradiation levels experienced by the cells in the natural environment (43). In fact, relatively high light elicited several acclimation responses, including chlorosis or cell bleaching, which reflects a reduction in the levels of chlorophyll a (chl a) and phycobiliprotein (PBP).
We are studying the ways in which axenic cultures of thermophilic cyanobacteria respond to a variety of defined environmentally relevant conditions in the laboratory. We have initiated such studies by defining the potential gene members of the Pho regulon and characterizing accumulation of transcripts from these genes in response to P deprivation. The levels of inorganic phosphate (Pi) in the effluent channel of OS have been determined to be as low as 0.37 μM (33), although much higher levels (17 μM) have also been reported (4). As a reference, P starvation responses of Escherichia coli are elicited when the extracellular Pi concentrations drops below 4 μM (49, 51). The dominant P source within the hot spring microbial mat is Pi, as determined by 31P-nuclear magnetic resonance analysis of the total P content (M. Adams and B. Cade-Menun, unpublished data); other P compounds detected include phosphonates and polyphosphate. Based on the 31P-nuclear magnetic resonance analysis, phosphonates could constitute up to 5% of the total P content in the microbial mat, and levels may fluctuate within the microbial mat over the diel cycle. However, the P composition of the aqueous environment surrounding the microbial mat (extracellular) was not investigated.
Pi metabolism has been studied extensively in E. coli (46, 49). The levels of over 80 identified proteins increase in response to P starvation, and many of these proteins are encoded by genes that are coregulated as part of the Pho regulon (47, 49). The multigene transcriptional induction associated with the Pho regulon is accomplished by a mechanism in which the transcriptional regulator PhoB binds to the Pho box sequence, which is an upstream activation sequence that precedes each operon of the Pho regulon (25, 49). Many of the transcriptional units that are part of the Pho regulon represent multigenic operons, such as the multigenic transport operon that encodes the four subunits (PstS, PstC, PstA, and PstB) of the Pst high-affinity Pi transport systems (25, 49). The PstSCAB transporter operon in bacteria often includes the gene encoding PhoU, which is thought to function as a negative regulator of the PhoR-PhoB two-component system (31). Genes of the Pho regulon also encode phosphatases (e.g., PhoA), which liberate Pi from organic sources in the environment, and the enzymes required for utilization of other P sources, including UGP-glycerol phosphate (41). The Pho regulon in Pseudomonas fluorescens Pf0-1 lacks a PhoA homolog but encodes a phosphatase, PhoX, which is predominantly responsible for extracellular phosphatase activity during P limitation (29). Secretion of PhoX by the twin-arginine transport pathway is a feature that distinguishes this family of phosphatases from the PhoA family (17).
The enzymes polyphosphate kinase 1 (PPK1) and exopolyphosphatase (PPX) have been shown to be members of the Pho regulon of E. coli K-12 (GenBank accession numbers NP_416996 and NP_416997 for PPK1 and PPX, respectively) (18) and are also present in the model cyanobacterium Synechocystis strain PCC 6803 (accession numbers NP_442590 [sll0290] and NP_442969 [sll1546] for PPK1 and PPX, respectively) (11). PPK1 is a highly conserved enzyme in prokaryotes, found in over 100 bacterial genomes to date, and is responsible for the reversible polymerization of ATP to make polyphosphate [poly(P)] (5). PPX and other enzymes with PPX activity, such as the periplasmic acid phosphatase SurE (36) or the guanosine pentaphosphate phosphohydrolase (18), degrade poly(P) in prokaryotes under conditions of P starvation.
Currently, the only well-studied cyanobacterial Pho regulon is that of Synechocystis strain PCC 6803, which features a sensor histidine kinase (SphS) and response regulator (SphR) that are orthologs of PhoR and PhoB, respectively, of E. coli (13). Three operons have been identified that are activated by SphR in Synechocystis strain PCC 6803, including two ABC-type high-affinity Pi uptake systems (pstS1-C1-A1-B1-B1 and pstS2-C2-A2-B2) and phoA-nucH, encoding an alkaline phosphatase and extracellular nuclease (45). Recently, in silico predictions of Pho box elements upstream of cyanobacterial genes have been performed for 19 different cyanobacteria that have fully sequenced genomes, including Synechococcus OS-A and OS-B′ isolates (44).
Polypeptides involved in the uptake and assimilation of P from molecules containing a phosphonate (Phn) bond are also part of the Pho regulon of E. coli (20, 27, 54). The proteins required for Phn utilization include transport components (PhnCDE) and a specific multisubunit C-P lyase (Phn-FGHIJKLMNOP) that is required for hydrolysis of the C-P bond (27). The C-P bond of Phns is recalcitrant to chemical hydrolysis, thermal denaturation, and photolysis, making these compounds markedly different from other P sources (32). Although Phns are found in organisms across phylogenetic groupings, only prokaryotic microorganisms have, so far, been shown to have the capability to cleave the Phn bond. Phns are ubiquitous and have been identified in eukaryotes, bacteria, and archaea as antibiotics and phosphonolipids, although the exact role of the latter within the lipid bilayer is still not clear (54). The most common naturally occurring Phn is aminoethylphosphonate (AEPhn), which is present in phosphonolipid head groups as well as in polysaccharide and glycoprotein side groups (54). In the marine environment, Phns can comprise as much as 25% of the high-molecular-weight dissolved organic P and serve as a P source for some bacteria (7).
Many Phns found in terrestrial and aquatic environments are anthropogenic in origin, as they are components of flame retardants, plasticizers, pharmaceuticals, and herbicides. Methylphosphonate is a component of several synthetic compounds, such as the flame extinguisher Pyrol 67, which is composed of vinylphosphonate and methylphosphonate. Glyphosate [N-(phosphonomethyl)glycine] is the active ingredient in the widely used agricultural herbicide Roundup (26). There are potential repercussions of the widespread use of Phns as herbicides, since some are potentially toxic and have long retention times in the environment (20).
In this study, we examined the physiological consequences of P deprivation on axenic isolates of the thermophilic cyanobacterium isolate Synechococcus OS-B′. We used a number of different methods, including assays for alkaline phosphatase activity, quantification of transcripts of the Pho regulon, and monitoring of cell doubling time, to assess the ability of OS-B′ to grow on Phn as the sole source of P. Under P deprivation conditions, the cells bleached and developed elevated levels of alkaline phosphatase (APase) activity. Transcripts from the putative Pho regulon genes (including phoA, phoD, phoX, and pstS) accumulated in cells soon after they were transferred to medium devoid of P. Furthermore, genes involved in the utilization of Phns were expressed in P-starved OS-B′ cells. These results are discussed in the context of cyanobacterial ecophysiology in hot spring microbial mats.
MATERIALS AND METHODS
Culture conditions.
The original Synechococcus isolate from Octopus Spring was designated JA-2-3B′a (2-13) (1). After generating an axenic culture of the strain, we designated the isolate Synechococcus sp. OS-B′ (CIW-10), which was grown and maintained at 55°C in liquid D medium (6) supplemented with 10 mM HEPES (pH 8.2 to 8.3) and Va vitamins (2). This growth medium, termed DH10, contains Pi as sodium phosphate at a concentration of 0.77 mM. For preparation of Pi-free medium, the sodium phosphate was replaced with an equimolar amount of sodium chloride. Cultures were bubbled with a mixture of 3% CO2 in air, in a 55°C incubator, and under continuous, relatively low light (∼75 μmol photon m−2 s−1). The irradiation was measured with a Li-Cor LI-189 quantum meter using a Li-Cor LI-193SA spherical sensor.
Growth measurements.
Growth rates and whole-cell absorbance spectra for OS-B′ were measured following growth in P-replete (+P) or P-free (−P) medium. Culture aliquots were collected at the same time every day; cell densities were estimated by counting cells using an Ultraplane Neubauer hemocytometer (Hausser Scientific, Pittsburgh, PA) and monitoring optical density of the culture at 750 nm (OD750) to generate a correlation between the OD750 and cell number. The OD750 showed a linear correlation with cell number over the first 200 h of growth in either +P or −P medium. Despite the characteristic loss of photosynthetic pigments during growth of OS-B′ under P starvation conditions, the relationship between cell density and OD750 remained relatively constant into stationary phase of the P-starved cells. OS-B′ cultures were started at an OD750 of 0.025 (∼106 cells/ml) and grown to late log phase (OD750 of 0.25 to 0.4, or ∼107cells/ml). Cells were washed twice in DH10-P medium to remove traces of Pi and then resuspended in fresh medium (either +P or −P) to an OD750 of 0.025 (∼106 cells/ml). The growth of OS-B′ was monitored into stationary phase, and the experiment was terminated at 192 h. All growth experiments were repeated at least three times.
Growth on MePhn.
The same methods were used to monitor growth of OS-B′ cells after they were transferred to +P or −P medium supplemented with methylphosphonate (MePhn), which was added to the medium by sterile filtration to a final concentration of 0.5 mM. This concentration of Phn was chosen because previous studies established that it supports optimal growth of E. coli, Pseudomonas stutzeri, and Rhizobium meliloti (34, 52). Other C-P compounds were also assessed as potential P sources, including ethylphosphonate (EthPhn), iN-(phosphonomethyl) glycine, AEPhn, and phosphonomycin. Of these, only EthPhn (like MePhn) could support the growth of OS-B′ as a sole source of P.
Cell viability.
Cell viability was monitored with the Live/Dead BacLight kit L-7012 (Molecular Probes, Inc., Eugene, OR), as previously described (24).
Alkaline phosphatase activity.
APase activity was measured at 24-h intervals over the course of an experiment by using a colorimetric assay modified from the methods of Ray et al. (39). Briefly, extracellular APase activity was assayed over the course of 15 min. A 500-μl aliquot of harvested cells at each respective time point was added to an equal amount of the colorimetric reagent p-nitrophenyl phosphate (pNPP) in Tris buffer, generating a reaction mix containing 3.6 mM pNPP with 200 mM Tris HCl, pH 7.0 to 9.0. Cells were removed by centrifugation, and the absorption of the supernatant was measured at 410 nm. APase activity was calculated as μg pNPP hydrolyzed per h per 1 × 106 cells.
Spectral data.
For each time point in the growth analyses, whole-cell absorbance spectra from 350 to 800 nm were measured to determine relative chl a and PBP contents (8). The spectra were normalized to the OD750 of the culture, which allowed comparisons of spectral features across different cell densities, since the OD750 could be used as a proxy for cell number under all growth conditions.
RNA extraction.
For analysis of transcript levels from putative Pho regulon genes over the time course of growth and under different P conditions (+P, −P, +P+MePhn, and −P+MePhn), cells were harvested at 24-h intervals throughout the logarithmic phase of growth (24, 48, 72, and 96 h). For experiments in which Pi was added to cultures following 72 h of growth in +P or −P medium, the Pi was filter sterilized and added to a concentration of 0.77 mM (if the concentration in the culture after the initial growth period was not considered). RNA was extracted from cells as previously described (42). Isolated RNA was subjected to DNase digestion (Turbo DNase; Ambion, Austin, TX), precipitated with ethanol, and tested by PCR for residual DNA contamination. Once the RNA was shown to be DNA free, it was either stored at −80°C or immediately used for reverse transcriptase quantitative PCR (RT-qPCR) (see below).
Reverse transcription and RT-qPCR.
Superscript III RT from Invitrogen (Carlsbad, CA) was used to reverse transcribe 1 μg of DNA-free RNA for each sample analyzed. The reaction mixture contained 1 μg of RNA, 200 ng random primer hexamers, and buffer supplied by Invitrogen in a final reaction volume of 20 μl. The RT reaction was performed in a PTC thermocycler (MJ Research Inc., St. Paul, MN). The RNA was denatured at 65°C for 5 min, the reaction mixture assembled, and the reaction allowed to proceed for 44 min at 55°C before being terminated by a 70°C incubation for 15 min (to inactivate the RT). The single-stranded cDNA product of the reaction was diluted 1:10 in nuclease-free water (final volume, 200 μl), and 2 μl was used in a 20-μl qPCR reaction, as previously described (19). The specificity of the qPCR was evaluated by performing a temperature melt curve analysis; a single sharp peak on the melt curve indicates the synthesis of a single specific product. The relative level of each specific transcript was quantified by comparing the cycle threshold (CT) values, determined for each PCR, with a standard curve of CT values generated using known amounts of DNA for the same target gene, over a 10-fold dilution series (42). Specific primers, shown in Table 1, were designed to amplify ∼200-bp segments of DNA from phoA, phoD, phoX, pstS-1, pstS-2, phnC-1, phnC-2, phnD-1, phnI, and phnJ. At least two biological and three technical replicates were performed for all RT-qPCRs. The results shown represent the three technical replicates (with the standard deviations) for one of the biological replicates. However, the trends and values for the qPCRs were very similar for the second biological replicate.
TABLE 1.
Primers used to quantify putative Pho regulon transcripts of OS-B′a
| Gene | Locus tag | Size
|
Forward primer | Reverse primer | |
|---|---|---|---|---|---|
| nt | aa | ||||
| phoA | CYB_1198 | 1,349 | 449 | TGGTGCAAACGGGATCCATCATTG | AATCTCCTCATAGTCGCTGCGCTT |
| phoD | CYB_0684 | 1,727 | 575 | GCCGCGGCGATATCGATTTCATTT | CAACAAAGGAAGTCCGGCCAAACA |
| phoX | CYB_1988 | 2,048 | 682 | ACGATGCCCGCTTTGAGTACATCT | CTTGGCCACATACAAGGTGCCATT |
| pstS-1 | CYB_1077 | 1,076 | 358 | CACAGGTGACTTTCCCGAAT | ATTCCACGTAGCCAATCGAG |
| pstS-2 | CYB_1915 | 1,061 | 353 | ATGCGAACCCTGCTTTCTGCTTTC | GAGATTTGCACGGTTTGGGCTTGA |
| phnC-1 | CYB_0159 | 791 | 263 | AAACAAGGTTGCCCTAAGGGAGGT | TGGCCTTCATGGAGAGGAAGAGAA |
| phnD-1 | CYB_0160 | 896 | 298 | GCTCCCATTGAAGCGTTCGTGAAA | TTGCCCTTGGCGTCTTCTAGAGTT |
| phnC-2 | CYB_1467 | 809 | 269 | TGGCTCAATGGAATCGACCTCACT | TAGCCCAACTGACCCGAAAGAACA |
| phnI | CYB_0164 | 696 | 231 | TGCTGGATTTGGAAATGGATCGCC | AGTGGCTCATCCAGTTGCTGAGAA |
| phnJ | CYB_0165 | 866 | 288 | GTAGCATATGAAACCCAAGCATAG | AACTCGAGAGGACCGCTCGTTT |
Included in the table are locus tags, the sizes of the gene in nucleotides (nt) and amino acids (aa), and both the forward and reverse primer sequences that were used.
Growth and physiology of MePhn-acclimated cultures.
Growth kinetics and P starvation responses of OS-B′ cells acclimated to MePhn were monitored and compared to responses of nonacclimated cells. The acclimated cells were defined as those grown on MePhn as the sole P source for >3 weeks, while nonacclimated cells were maintained on +P medium. Both MePhn-acclimated and nonacclimated cells were grown to late logarithmic phase, washed with P-free medium, and then used to start new cultures at a low cell density (106 cells/ml, or an OD750 of 0.020 to 0.025); the cells were subcultured into +P, −P, −P+MePhn, and +P+MePhn media (biological triplicates were used). APase activities and whole-cell absorbance spectra were routinely monitored during cell growth. The loss of acclimation to MePhn was evaluated at various times following the replacement of MePhn with Pi in cell cultures that had been growing for at least 1 month on MePhn as sole P source.
RESULTS
Putative Pho regulon genes.
The two completely sequenced genomes of the Synechococcus sp. isolated from the hot spring microbial mats of Yellowstone National Park were examined for putative pho genes. Based on the annotation available through GenBank (analysis updated June 2008) and prior knowledge of well-studied Pho regulon genes, we identified 42 genes in the OS-B′ genome (23 of which have homologs in OS-A) that are putative members of the Pho regulon (Table 2). Recently, an in silico analysis of Pho regulons in cyanobacteria was performed by Su et al., who used a scanning algorithm designed to search for putative Pho box sequences (sequences that bind PhoB) based on those that had been characterized experimentally in various bacteria and cyanobacteria (44). These computational predictions of PhoB binding sites were also used for pho gene identifications in the OS-A and OS-B′ genomes. According to this analysis, the predicted cyanobacterial Pho box sequences are comprised of three tandem direct repeats of 8 bp with the consensus sequence CTTAACCT and which are separated by A/T-rich 3-bp linker sequences. This differs from the well-defined Pho box of E. coli, which consists of at least two tandem direct repeats (Pho boxes) of 8 bp with the consensus sequence CTGTACTA separated by an A/T-rich 3-bp linker that is located 10 bp upstream of the −10 consensus σ70 TAN3T/A binding site (49).
TABLE 2.
Putative Pho regulon genes of the OS-B′ genomea
| Name of gene(s) | Locus tag | Putative encoded protein | Closest homolog | % AAID |
|---|---|---|---|---|
| phoR | CYB_0858 | Sensor histidine kinase | Nostoc strain PCC 7120 | 41 |
| phoB | CYB_2856 | Response regulator | Gloeobacter violaceus PCC 7421 | 75 |
| phoA | CYB_1198 | Alkaline phosphatase | Chlorobium chlorochromatii CaD3 | 45 |
| phoX | CYB_1988 | Alkaline phosphatase | Hahella chejuensis KCTC 2396 | 54 |
| phoD | CYB_0684 | Phosphodiesterase | Bacillus subtilis | 28 |
| surE-1 | CYB_0884 | Acid phosphatase | Thermosynechococcus elongatus BP-1 | 60 |
| surE-2 | CYB_1427 | Acid phosphatase | Lyngbya strain PCC 8106 | 51 |
| phoH | CYB_2320 | PhoH family protein | Thermosynechococcus elongatus BP-1 | 61 |
| npp | CYB_0274 | 5′-Nucleotidase phosphatase | Cyanothece strain PCC 7424 | 53 |
| nucH | CYB_2765 | Putative secreted nuclease | Roseiflexus castenholzii DSM 13941 | 46 |
| ppx | CYB_1493 | Exopolyphosphatase | Anabaena variabilis ATCC 29413 | 58 |
| ppk | CYB_2082 | Polyphosphate kinase | Cyanothece strain PCC 8801 | 62 |
| pstS-1, pstC-1, pstA-1, pstB-1 | CYB_1077-74 | High-affinity ABC-type Pi transporter | Cyanothece strain CCY 0110b | 50, 51, 53, 70 |
| pstS-2, pstC-2, pstA-2, pstB-2 | CYB_1915-12 | High-affinity ABC-type Pi transporter | Cyanothece strain ATCC 51142b | 45, 50, 70, 70 |
| phoU | CYB_2526 | Regulatory protein | Nostoc strain PCC 7120 | 61 |
| phnC-1, phnD-1, phnE-1 | CYB_0159-61 | Phn ABC-type transporter proteins | Cyanothece strain PCC 8801b | 44, 31, 45 |
| phnG-phnM | CYB_0162-68 | C-P lyase | Roseiflexus strain RS-1b | 48, 32, 38, 59, 45, 41, 39 |
| phnD,cphnD-2, phnD-3, phnC-2, phnE-2, phnE-3 | CYB_1464-69 | Phn ABC-type transporter proteins | Dinoroseobacter shibae DFL 12b | 69, 51, 53, 72, 54, 56 |
| phnE-4, phnD-4, phnC-3 | CYB_0012-11, 09 | Phn ABC-type transporter proteins | Cyanothece strain PCC 7424b | 36, 27, 69 |
| ugpB, ugpA | CYB_2477-78 | Glycerol-3-phosphate transporter | Deinococcus geothermalis DMS 11300b | 39, 46 |
Gene names, locus tags, predicted products, and percent AAID to the closest bacterial homolog in all available sequenced organisms (excluding OS-A) are given. Genes associated with putative operons are grouped in the list. Genes shown in bold are preceded by a high-ranking predicted Pho box (44).
For gene clusters, we show the percent AAID relative to the respective homologs from a single organism.
PhnD had the highest AAID to the Sinorhizobium meliloti 1021 homolog.
The 42 putative pho genes identified on the OS-B′ genome were examined for the presence of a putative PhoB binding site, based on the consensus sequence (44) (Fig. 1A, inset). Six genes have highly ranked Pho box sequences upstream of their coding region, and the genes downstream of them are likely to be part of a Pho-regulated operon (Table 2). Based on the in silico Pho box predictions, it is likely that there are several other genes, not investigated in this study, that may be part of the Pho regulon (44). The OS-A and OS-B′ orthologs associated with the Pho regulon, based on a mutual best BLAST hit analysis (updated June 2008), share high identity at the nucleotide (NAID) (83 to 98%) and amino acid (AAID) (86 to 98%) levels (see Table S1 in the supplemental material). There is also a relatively high conservation of predicted Pho box sequences between orthologs of OS-A and OS-B′ (40; Z. Su, personal communication). This is not surprising, since these two isolates are closely related (3). On the other hand, we found that there is relatively low identity between the OS-A and OS-B′ Pho regulon genes and their closest putative orthologs in other cyanobacteria or bacteria (from 27 to 75% AID), as shown in Table 2.
FIG. 1.
Genomic organization of phn genes and comparison of the regions flanking the phn gene clusters in OS-B′ and OS-A. (A) The top row of genes shows the main phn gene cluster of OS-B′, with the ABC phosphonate (Phn) transporter (phn C-1, phn D-1, and phn E-1) and the C-P lyase (phnG-phnM) genes. The bottom row shows the second (phnD, phn D-2, phn D-3 phn C-2, and phn E-3) and third (phnE-4, phnD-4, and phnC-3) phn clusters. See Table 2 for accession numbers. The Phn transporter genes include the ATPase component (phnC; red), the substrate binding protein (phnD; yellow), and the membrane permease component (phnE; green). A putative Pho box is located upstream of phnC-1, phnD, and phnC. Asterisks indicate genes that overlap with the next contiguous gene. (Inset) Logo representation of the profile of the top 10 ranked Pho boxes of OS-B′, as predicted by phylogenetic footprinting (40). The logo was generated by using the Weblogo server (http://weblogo.berkeley.edu/logo.cgi). (B) Organization of flanking regions around the main phn gene cluster (gray arrows) in OS-B′ compared to the analogous region in OS-A. (C) Organization of flanking regions around the second phn cluster (phnD, phnD-2, phnD-3, phnC-2, phnE-2, and phnE-3) in OS-B′ compared to the analogous region in OS-A. (D) Organization of flanking regions around the third phn cluster (phnE-4, phnD-4, and phnC-3) in OS-B′ compared to the analogous region in OS-A. Gray arrows represent the phn genes, and solid colored arrows (but not black arrows) indicate syntenic sequences in OS-A relative to OS-B′, while the broken arrows indicate genes where there is a break in synteny between the two genomes. Black arrows in the OS-B′ genome represent genes not present in OS-A. The scale is indicated in panel B.
Two-component regulatory system.
The OS-A and OS-B′ genomes encode the sensor histidine kinase PhoR (CYA_2352 and CYB_0858) and the response regulator PhoB (CYA_1033 and CYB_2856), which constitutes the two-component system that controls the activity of the Pi starvation-responsive Pho regulon. There is relatively low identity of the putative PhoB/PhoR polypeptides of OS-B′ to the analogous proteins of E. coli; the highest identity is to PhoB of Gloeobacter violaceus PCC 7421 (75% AAID) and PhoR of Nostoc strain PCC 7120 (41% AAID) (Table 2).
High-affinity ABC-type Pi transport systems.
The OS-A and OS-B′ genomes both contain two copies of a putative pstSCAB operon that are located at different genomic locations: pstS-1, pstC-1, pstA-1, and pstB-1 (CYA_1558-CYA_1555 and CYB_1077 to CYB_1074) and pstS-2, pstC-2, pstA-2, and pstB-2 (CYA_1735 to CYA_1732 and CYB_1915 to CYB_1912) (Table 2). Moreover, the putative PhoB binding site upstream of the second pstS-2 C-2 A-2 B-2 gene cluster in OS-B′ (CYB_1915 to CYB_1912) was the highest-ranked Pho box sequence predicted by in silico analysis (44). The regulatory gene phoU (CYA_0182 and CYB_2562) is not associated with either of the pstSCAB operons (in either OS-B′ or OS-A).
Phosphatases.
There are at least seven putative phosphatases (phoA, phoX, phoD, surE-1, surE-2, nucH, and npp) encoded by the OS-A and OS-B′ genomes (56). High-scoring Pho box sequences were predicted upstream of the phoA, phoX, phoD, and nucH genes in OS-B′, suggesting that they are likely to be regulated by PhoB binding (44). The phoA gene (CYA_0781 and CYB_1198), present in both OS-A and OS-B′, encodes an APase with a predicted molecular mass of 48 kDa. The highest identity/similarity of the OS-B′ PhoA is to the APase of the symbiotic green sulfur bacterium Chlorobium chlorochromatii CaD3 (45% AAID, 65% amino acid similarity). Both the OS-A and OS-B′ genomes contain a putative nucH homolog (CYA_0117 and CYB_2765), although it is not contiguous with phoA, and the OS-B′ NucH is most similar to the putative secreted nuclease of the thermophilic, filamentous, anoxygenic phototroph Roseiflexus castenholzii DSM 13941 (46% AAID), which is also present in the OS microbial mat.
The OS-A and OS-B′ genomes also encode a putative extracellular alkaline phosphatase, PhoX (CYA_1696 and CYB_1988), which is most similar to the PhoX of the marine, gammaproteobacterium Hahella chejuensis KCTC 2396 at 54% AAID. The phoX gene is not present in all completely sequenced cyanobacterial genomes; it has been identified on the genomes of Gloeobacter violaceus, Nostoc strain PCC 7120, and Synechococcus strain WH 8102. Genes encoding the acid phosphatases SurE-1 (CYA_0967 and CYB_0884) and SurE-2 (CYA_0017 and CYB_1427) are also present on the OS-A and OS-B′ genomes, and the orthologs exhibit 95% and 92% AAID to each other, respectively (see Table S1 in the supplemental material). These phosphatases are thought to function in liberating Pi from phosphate esters when cells enter stationary phase (38).
Poly(P) synthesis and utilization.
The OS-A and OS-B′ genomes contain the ppk gene (CYA_2477 and CYB_2082), which encodes polyphosphate kinase 1, and the ppx gene (CYA_2432 and CYB_1493), which encodes an exopolyphosphatase (Table 2). The presence of the ppk and ppx genes suggests that these hot spring cyanobacteria can acclimate to environmental P fluctuations by modulating the formation and metabolism of cellular poly(P) reserves. The putative PPK and PPX that are encoded in the OS-A and OS-B′ genomes are 94% and 91% identical to each other, respectively, but the OS-B′ homolog has only 62% AAID to the putative PPK of Cyanothece strain PCC 8801 and 58% AAID to the PPX of the filamentous, heterocyst-forming cyanobacterium Anabaena variabilis ATCC 29413.
The phn gene clusters.
The OS-B′ genome contains a cluster of contiguous genes associated with Phn uptake and metabolism. This cluster, designated the main phn cluster, represents a putative operon consisting of 10 genes (phnC-1, phnD-1, phnE-1, phnG, phnH, phnI, phnJ, phnK, phnL, and phnM; CYB_0159 to CYB_0168) and spans ∼8 kb. The phn genes of OS-B′ are organized similarly to that of the well-characterized 14-gene phn operon of E. coli, with genes encoding the transport proteins at the 5′ end of the putative operon followed by genes encoding the C-P lyase (Fig. 1A, top gene cluster) (27). However, these polypeptides associated with Phn utilization in OS-B′ show relatively low AAID (between 31 and 59%) to polypeptides encoded by other phn genes and are not present in the OS-A genome (Table 2). Neither the OS-B′ nor the OS-A genome contains putative homologs of other genes that would allow for Pho-independent phosphonate catabolism, including phnA, phnB, phnW, phnX, and phnY.
Interestingly, 8 of the 10 genes in the OS-B′ phn operon overlap by 7 to 31 bp, with overlapping sequences found between phnD-1 and phnE-1 (31 bp), phnH and phnI (15 bp), phnI and phnJ (16 bp), phnJ and phnK (7 bp), and phnL and phnM (19 bp). Putative Pho box elements have been predicted upstream of the phn gene cluster; the sequence of the PhoB binding site upstream of this cluster was ranked the second highest relative to all such sites on the OS-B′ genome (44).
In addition to the main phn gene cluster, there are two additional clusters (“secondary” clusters) with genes encoding putative Phn transporter components (Fig. 1A, gene clusters in the second row). The second phn cluster contains six transport-related genes (phnD, phnD-2, phnD-3, phnC-2, phnE-2, and phnE-3; CYB_1464 to CYB_1469), while the third phn cluster contains three genes (phnE-4, phnD-4, and phnC-3; CYB_0012 and -11 and CYB_0009). There is no C-P lyase gene associated with either of these clusters. None of these phn genes is present in the OS-A genome, and the OS-B′ Phn polypeptides show relatively low AAID to polypeptides encoded by phn genes in other organisms (Table 2). Putative Pho box sequences were also predicted upstream of these phn clusters, albeit with much lower scores, based on a scanning algorithm (Z. Su, personal communication), than that obtained for the Pho box sequence of the main phn gene cluster. The putative Phn transporter proteins encoded by the secondary clusters exhibit low AAID to potential paralogs of the main cluster (see Table S2 in supplemental material). However, the polypeptides encoded by the secondary clusters have higher AAIDs to Phn transport polypeptides than to any of the other transport polypeptides, including those associated with Pi transport in various bacteria (data not shown). These results suggest that the secondary cluster polypeptides are involved in Phn uptake, although they may be tailored for the uptake of specific Phn substrates.
While comparison of the two sequenced genomes of the Synechococcus isolates from OS indicates that the OS-A genome lacks any gene or gene fragment with significant similarity to phn genes, the genomic region that flanks the 8-kb main phn gene cluster of OS-B′ is present in OS-A; the genes that immediately border the phn genes are contiguous in the OS-A genome (Fig. 1B). Similarly, the regions flanking the second (Fig. 1C) and third (Fig. 1D) phn clusters in OS-B′ are also contiguous in the OS-A genome. The proteins encoded by these flanking genes are relatively highly conserved between the genomes (ranging from 78 to 96% AAID), which may indicate mobility of phn gene clusters within the mat microbial population and explain the presence of these genes in one isolate and their absence in the other. However, it is difficult to determine how these clusters may have been transferred; a single transposase-like gene (CYB_0018) is found on the genome contiguous to one of the putative Phn transporter clusters (CYB_0009, CYB_0012-11).
P deprivation alters cell growth and physiology.
The abilities of axenic OS-B′ cultures to cope with P deprivation and to utilize MePhn as a sole P source were analyzed. OS-B′ grown in +P medium had a doubling time of ∼24 h up to the point at which the cells reached stationary phase, going from ∼1 × 106 to nearly 1 × 108 cell/ml (between six and seven doublings) in 192 h (Fig. 2A). Cells in +P medium showed low-level, constitutive APase activity (<0.02 μg pNPP hydrolyzed/h/1 × 106 cells), which did not change significantly throughout the growth period (Fig. 2B). When the +P cells were transferred to −P conditions, growth stopped after ∼72 h (i.e., ∼three to four doublings) at a density of ∼1 × 107 cells/ml, as shown in Fig. 2A, while the APase activity reached maximal levels (∼0.3 μg pNPP hydrolyzed/h/1 × 106 cells) at ∼48 h following the transfer to −P conditions; this level of activity was sustained throughout the stationary phase of growth (Fig. 2B). Furthermore, P starvation of OS-B′ caused changes in whole-cell absorption spectra that have been associated with macronutrient deprivation in other cyanobacteria (8); there was a decrease in both chl a (maximal absorbance at 430 and ∼675 nm) and PBP pigments (maximal absorbance at 620 nm) on a per cell basis, measured after 96 h in −P medium (Fig. 2C). This loss of photosynthetic pigment is most likely due to dilution of PBP and chl after termination of synthesis, assuming that Pi starvation affects the photosynthetic apparatus of OS-B′ in a manner similar to that of Synechococcus strain PCC 7942 (8). Further quantifications of pigments and their synthesis and turnover would be required to clearly address this point.
FIG. 2.
Growth response, APase activity, and absorbance spectra of OS-B′ under various P conditions. (A) Late-logarithmic-phase cells grown in +P medium were transferred growth under four different conditions: (i) +P, (ii) −P, (iii) +P medium with 0.5 mM MePhn (+P+MePhn), and (iv) −P medium with 0.5 mM MePhn (−P+MePhn). Growth of all cultures was monitored for ∼192 h. Note the log scale on the y axis for cells per ml data. (B) APase activity and growth measurements were quantified in cell cultures once every 24 h. The APase activity was measured as μg of pNPP hydrolyzed per h per 1 × 106 cells. (C) Whole-cell absorption spectra of each culture between the wavelengths of 400 and 800 nm, normalized to the OD750, at 96 h after cell transfer. Results for growth and APase activity show the means and standard deviations (error bars) from measurements taken from biological triplicates.
When cells were transferred to −P medium supplemented with MePhn as the sole P source, they ceased growth within 24 h; this is in contrast to growth of OS-B′ in medium lacking P, in which cells doubled three to four times (Fig. 2A). These cultures also exhibited markedly lower APase activity relative to the −P cultures (Fig. 2B). When both MePhn and Pi were included in the medium, cell division continued for at least 192 h. The growth rate for the +P+MePhn cells was comparable to that of the +P cells for the first 24 h and then slowed relative to cells grown in +P medium between 24 and 120 h. Approximately the same final cell densities for the +P+MePhn and the +P cells were attained when the experiment was terminated at 192 h. Both the −P and the −P+MePhn cultures stopped growing sooner than either the +P or +P+MePhn cultures and never reached as high a cell density. The lowest cell density was observed for the −P+MePhn-grown cells, which appeared to enter stationary phase at a density of 2.5 × 106 to 3.2 × 106 cells/ml. Like the −P cultures, the −P+MePhn cultures exhibited a decrease in chl a and PBP content, based on whole-cell absorbance spectra. However, only the −P+MePhn cultures showed a notable change in absorbance at 480 to 550 nm, suggesting an increase in carotenoid accumulation after 96 h of starvation (Fig. 2C). We conclude that MePhn is inhibitory to cell growth over the time period of 200 h and exacerbates the physiological responses of cells to the starvation conditions.
Pho regulon transcripts.
We used qPCR with specific primers (Table 1) to quantify the level of transcripts for several genes of OS-B′ of the putative Pho regulon. The cultures were initially grown under nutrient-replete conditions and subsequently transferred to four different media, as done for the experiment presented in Fig. 2: +P (nutrient replete medium), −P (no P added to medium), +P+MePhn (nutrient replete supplemented with MePhn), and −P+MePhn (MePhn as the sole P source). The density of the cells transferred to the new growth conditions was very low (<106 cells/ml) to ensure that the cells were in the logarithmic phase of growth and that they would have the capacity to maintain logarithmic growth over the course of the experiment.
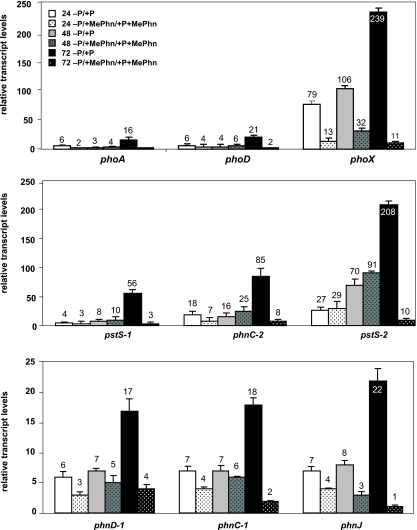
The ratios of individual transcripts in cells grown under the different P conditions tested are shown in Fig. 3 and 4. A similar trend in transcript accumulation was obtained for all putative Pho regulon transcripts that were quantified. This trend was also verified by a biological replicate (data not shown), and all technical replicates had standard deviations from the mean of 5% or less. An increase in transcript levels was observed after 24 h under −P conditions (expressed as the ratio of −P/+P or −P+MePhn/+P+MePhn transcript levels) and remained high at 72 h of P deprivation. The increase in transcript abundance observed in cells transferred to −P+MePhn relative to +P+MePhn was generally not as great as that observed for cells transferred to −P relative to +P, particularly at the 72-h time point. As shown in Fig. 3 (top panel), transcripts from phoA (CYB_1198) and phoD (CYB_0684) were both sixfold higher after 24 h of P starvation. The highest transcript levels were observed 72 h after the transfer of cells to −P conditions, with 16-fold and 21-fold changes for phoA and phoD transcripts, respectively. Transcripts for the phoX alkaline phosphatase (CYB_1988) increased within 24 h and continued to accumulate over the time course in −P relative to +P conditions; the increase in relative transcript level was >200-fold after 72 h.
FIG. 3.
Quantification of relative Pho regulon transcript accumulation following Pi starvation. Specific Pho regulon transcripts, including those of the phn gene cluster, were measured over 24-h intervals under various P conditions. The top panel represents transcripts for genes encoding phosphatases, the middle panel is transcripts for genes associated with transport systems, and the bottom panel is transcripts for genes involved in Phn transport and metabolism. For each gene, relative transcript levels, presented as a ratio under −P and +P conditions, were determined at 24, 48, and 72 h following the transfer of cells to the new growth medium. In the presence of MePhn, relative transcript levels representing −P+MePhn compared to +P+MePhn are shown at 24 h, 48 h, and 72 h. The qPCR results show the means and standard deviations (error bars) for data from three technical replicates.
FIG. 4.
Quantification of Pho regulon transcript accumulation after Pi addition to P-starved cultures. Pi was added back to the growth medium after 72 h of starvation, and transcript levels were measured (as described in Fig. 3) at 96 h (i.e., 24 h after adding Pi back). Relative transcript levels are shown that compare −P to +P after Pi addition and the control, i.e., −P compared to +P prior to Pi addition but with no Pi added back. The qPCR results show the means and standard deviations (error bars) for data from three technical replicates.
Similar trends in transcript abundance were observed for the Pho regulon genes encoding the periplasmic Pi binding proteins associated with the high-affinity transport systems pstS-1 (CYB_1074) and pstS-2 (CYB_1911) (Fig. 3, middle panel), although the increase for pstS-2 was significantly more than for pstS-1. Transcripts for phnC-2 (CYB_1467), encoding the ATP binding component of the Phn transporter, also exhibited a >80-fold increase by 72 h of P deprivation.
In a separate experiment, cells were grown in −P medium for 72 h and then were either supplemented with Pi (an addition to 0.77 mM Pi) or maintained in −P medium; growth was continued for an additional 24 h for each of the cultures before transcript analysis. The addition of Pi to the cultures after 72 h of P starvation caused a rapid decrease in putative Pho regulon transcript levels within 24 h; decreases were observed for the phosphatase (phoA, phoD, and phoX) and ABC-type transporter periplasmic substrate-binding protein transcripts (pstS-1, phnC-1, and phnC-2) (Fig. 4) in comparison to the respective transcript levels at 72 h (Fig. 4). As expected, the transcript levels remained high for cells maintained under −P conditions for the additional 24 h (Fig. 4), although in some cases there was some decrease. This decrease could reflect a reduction in the growth and metabolism at the later time following the imposition of P deprivation.
The phn gene cluster is part of the Pho regulon.
We also monitored changes in several transcripts from the phn gene clusters during growth in +P and −P growth medium, as well as in the presence and absence of MePhn (Fig. 3, middle and bottom panels). After 72 h in −P medium, there was an increase in transcript levels of 18-fold for phnC-1 and 17-fold for phnD-1 (Fig. 3, bottom panel); these genes are likely part of the same operon (Fig. 1A, first row). Transcripts of the second Phn transport cluster exhibited a greater change in relative transcript levels than those of the main phn cluster; there was a >80-fold increase of phnC-2 by 72 h (Fig. 3, middle panel). An increase in transcripts from genes encoding the C-P lyase was also observed when the cells experienced P deprivation. Transcripts for phnJ increased 22-fold after 72 h of growth in −P compared to +P medium. Similarly, an increase of transcript levels was observed for cells grown in −P+MePhn relative to cells grown in +P+MePhn, although the ratio was generally lower than that observed for cultures that were not supplemented with MePhn, and the 72-h time point generally exhibited the lowest ratio (Fig. 3, middle and bottom panels). Finally, the addition of Pi to the P-starved cultures caused a rapid decrease in phn gene transcript accumulation within 24 h, as quantified at the 96-h time point for phnC-1 and phnC-2 (Fig. 4); the transcripts remained high in cultures to which no Pi was added (Fig. 4).
MePhn addition alters Pho regulon transcript levels.
To examine how MePhn affects levels of transcripts after cells are transferred to −P medium, we compared levels of transcripts for the putative Pho regulon genes following 72 h of growth in −P medium relative to −P+MePhn medium; the ratios of the transcript abundances measured under these two conditions are presented in Table 3. For all investigated genes that were affected by P starvation, the transcript levels following 72 h of growth on −P medium were between 4- and 18-fold higher than in cells grown for 72 h in −P medium supplemented with MePhn. These results demonstrate that MePhn suppresses accumulation of Pho regulon transcripts during P deprivation, although as demonstrated in Fig. 3, there is still an increase in transcript levels when cultures supplemented with MePhn are placed into medium lacking P.
TABLE 3.
Effect of MePhn on transcript accumulationa
| Gene | Relative transcript level (at 72 h) |
|---|---|
| phoX | 18 |
| phnJ | 14 |
| pstS-2 | 11 |
| phnC-1 | 8 |
| phoA | 8 |
| pstS-1 | 7 |
| phnC-2 | 5 |
| phnD-1 | 5 |
| phoD | 4 |
Acclimation to MePhn and its utilization.
The capacity of OS-B′ to metabolize MePhn as a sole P source was tested. We observed that cells transferred to −P+MePhn showed a slight initial growth during the first 24 h and then stopped growing, with no additional growth for the first 16 to 20 days (Fig. 2A). The cells also showed significant bleaching (Fig. 2C), indicating a loss of photosynthetic pigments on a per cell basis. However, after ∼20 days in −P+MePhn medium, the OS-B′ cells slowly appeared to regain pigmentation and grow. During this 3- to 4-week “acclimation” period, the cells attained doubling times comparable to those of cells kept in nutrient-replete medium. After the −P cells supplemented with MePhn reached stationary phase, we considered them acclimated to growth on MePhn and they were subcultured into fresh −P medium supplemented with MePhn.
To further examine this phenomenon, we initiated growth experiments in which OS-B′ cultures were either acclimated to phosphonate (maintained in −P+MePhn medium) or maintained under nonacclimated conditions (in +P medium with no exposure to MePhn). Both the nonacclimated and acclimated cells were subcultured during logarithmic growth into +P, −P, or −P+MePhn medium, to investigate the effects of MePhn on the growth of both nonacclimated and acclimated cells.
Placing nonacclimated OS-B′ cells into −P+MePhn medium strongly inhibited cell division compared to cells placed in −P medium (Fig. 2A and 5). Nonacclimated cells stopped growing at 24 to 48 h after their transfer to −P+MePhn medium. In contrast, cultures transferred to −P medium (not supplemented with MePhn) kept growing for ∼72 h and reached much higher cell densities (Fig. 2A and 5). Growth characteristics of the Phn-acclimated cells were also monitored in +P, −P, and −P+MePhn media (Fig. 5). The Phn-acclimated OS-B′ cultures exhibited a doubling time of ∼24 h in the presence of +P or −P+MePhn; the doubling time of the acclimated cells in −P+MePhn was only slightly lower than that of acclimated cells grown in +P medium. Phn-acclimated cells continued to double with Pi or MePhn supplied as a P source and reached stationary phase at ∼120 h. The growth kinetics of the Phn-acclimated cells in +P or −P+MePhn was similar to that of nonacclimated cells grown in +P medium. Furthermore, the Phn-acclimated cells grown in −P+MePhn medium continued to divide over the entire 192-h growth period and reached a final cell density comparable to that of either nonacclimated or acclimated cells grown in +P medium. These results suggest that the acclimated cells can use Phn as a sole P source and, furthermore, that utilization of this P source results in rates of growth and final cell densities that are comparable to those observed when the cells are grown in medium containing Pi.
FIG. 5.

Growth of cultures in MePhn. Growth responses of MePhn-acclimated (triangles) and nonacclimated (squares) OS-B′ cells to +P, −P, and −P+MePhn conditions. The graph shows the means and standard deviations (error bars) from measurements taken from biological triplicates. Starting cultures were initially grown to logarithmic phase in either +P or −P+MePhn medium for 20 days. Note the log scale on the y axis for cells per ml data.
DISCUSSION
Studies of the Pho regulon and P starvation responses in model bacterial systems, including E. coli (46, 49) and Synechocystis sp. strain PCC 6803 (13, 45), have established a wealth of information that has guided us in investigations of P starvation responses in the thermophilic cyanobacterium Synechococcus OS-B′ isolates. Complete genome sequence information coupled with physiological studies and transcript analyses of OS-B′ have yielded insights as to how thermophilic cyanobacteria cope with P limitation and may utilize alternative P sources within the microbial mat consortium. This investigation demonstrated that genes of the putative Pho regulon of OS-B′, a key photosynthetic organism in the microbial mats of Mushroom Spring and Octopus Spring, are responsive to P starvation. The genome also contains phn gene clusters, which appear to be part of the Pho regulon and are likely involved in the ability of the cells to grow on MePhn (or other Phns in the environment) when no other P source is available. Indeed, one of the major findings of this study is that OS-B′ is capable of acclimating to P starvation conditions by developing the ability to utilize MePhn as a sole P source.
Initially, we addressed the question of whether OS-B′ is physiologically adapted to cope with P limitation. Axenic cultures of OS-B′ continued to progress through three to four cell divisions after Pi was removed from the growth medium; this contrasts with the results obtained for other microorganisms, such as E. coli, which rapidly enter stationary phase upon removal of P from the medium (35). The continued growth of OS-B′ after removal of P from the medium suggests that the storage and metabolism of intracellular poly(P) may be one mechanism that enables the organism to cope with low exogenous Pi, as has been demonstrated for other cyanobacteria (12, 21, 22). Consistent with this possibility, we have measured poly(P) levels and found that there are large poly(P) pools present in OS-B′, but these levels are much lower in cells maintained on MePhn (M. R. Gomez-Garcia, M. Adams, A. Grossman, and D. Bhaya, unpublished data). A similar result has also recently been reported for the chemolithoautotroph Acidithiobacillus ferrooxidans, which is also able to use MePhn as an alternative P source; during growth on Phn there is a drastic reduction in poly(P) levels relative to the level in cells grown in the presence of Pi (48).
Pi limitation of OS-B′ was found to elicit the accumulation of extracellular APase activity and increased levels of transcripts encoding putative phosphatases. The phosphatase genes that appeared to be activated at the transcriptional level included phoA, phoD, and phoX, which encode enzymes that can cleave Pi from phosphate monoesters and diesters (the transcriptional activities for nucH, surE-1, surE-2, and npp were not examined in this study). The PhoX of OS-B′ may be responsible for most of the extracellular phosphatase activity assayed during P deprivation, similar to the PhoX of Pseudomonas fluorescens Pf0-1, as transcript levels of phoX exhibited a much greater change than those of phoA or phoD following P starvation of OS-B′. The role of PhoX as the primary phosphatase is also in line with the highly ranked Pho box predicted to precede the phoX gene (CYB_1198), based on in silico analysis.
Transcripts encoding the high-affinity ABC-type Pst transport systems also accumulated in response to P depletion. Upregulation of both sets of pstS transcripts suggests that OS-B′ has two Pi transport systems that become more abundant when environmental Pi concentrations decline. In comparison, E. coli has a single Pst transport system plus a Pit transport system; the latter is active when Pi is in excess (35). Examination of 19 complete cyanobacterial genome sequences showed that all contained at least one set of genes encoding the Pst transporter. The genomes of the hot spring Synechococcus isolates OS-B′ and OS-A, as well as Anabaena variabilis, Synechocystis strain PCC 6803, Nostoc strain PCC 7120, and Gloeobacter violaceus PCC 7421, contain two clusters of these genes (44), although it is not clear if both of the Pst transporters encoded on the cyanobacterial genomes have a high affinity for their substrate. None of the genomes of any marine Prochlorococcus spp. or Trichodesmium erythraeum IMS101 contain two full pst transporter gene clusters, although Prochlorococcus does contain some duplicate gene homologs. Perhaps selection for a single high-affinity Pst system has occurred in the very stable, low-Pi marine environments, in comparison to other environments where bioavailable Pi may fluctuate more over time and it may be advantageous to have transporters with different kinetic and regulatory characteristics.
Another set of genes involved in P assimilation, which are also responsive to P depletion, are those encoding proteins involved in the transport and assimilation of Phns. Clusters of phn genes are present on the genome of OS-B′ and, similar to the phn operon of E. coli, Pi starvation elicits accumulation of phn transcripts in OS-B′. Most microorganisms investigated thus far exhibit phn gene expression when the cells are starved for P, so there is no apparent requirement for induction of the system by Phn molecules (20). This is also the case for OS-B′, for which transcripts of all investigated phn genes increase when P is eliminated from the growth medium (no Phn supplementation). However, when MePhn was added to cells deprived of Pi, it caused a depression in the levels of phn transcripts, relative to cells exposed to −P conditions without MePhn supplementation. Interestingly, Phn transporters in some bacteria have been shown to have relaxed substrate specificity, with an affinity not only for Phns but also for Pi (54) and other reduced P substrates (55), which raises the possibility that the phn genes of OS-B′ may encode proteins with the ability to acquire a broader range of substrates than just Phn. This is likely the case for OS-B′, which has three separate phn clusters on the genome encoding putative Phn transporter components that exhibit low homologies both among themselves and with Phn proteins in other organisms.
While all putative Pho regulon genes of OS-B′ are responsive to Pi levels, the polypeptides encoded by these genes in many cases have low AAIDs with the analogous proteins from other bacteria, such as E. coli, Thermosynechococcus elongatus, and Roseiflexus strain RS-1. For example, PhoA of OS-B′ is highly divergent from the PhoA in all other bacteria. This contrasts with the acid phosphatases (SurE-1 and SurE-2) of OS-B′, which share 60% AAID with the SurE-1 of Thermosynechococcus elongatus BP-1 and 51% AAID to the SurE-2 of Lyngbya strain PCC 8106. Despite the lack of strong sequence conservation among many bacterial Pho regulon proteins with those of OS-B′ and OS-A, there is a relatively high NAID and AAID between the pho orthologs of OS-A and OS-B′. Furthermore, many Pho regulon genes that are present in the hot spring cyanobacteria, including phoX and the phn gene cluster, are only present on a few other cyanobacterial genomes, which suggests environmental or niche-specific adaptation of P metabolism. Databases such as NCBI and IMG now contain complete genome sequences of cyanobacteria with diverse physiological potentials from a range of habitats, e.g., terrestrial and marine, unicellular and filamentous, and several nitrogen-fixing organisms. At this point, a full phn gene cluster (encoding both the transporter and C-P lyase) has only been identified in the genomes of the filamentous, heterocyst-forming Nostoc strain PCC 7120 and the filamentous, nitrogen-fixing, nonheterocystous Trichodesmium erythraeum IMS101 (15), as well as the unicellular, diazotrophic OS-B′ Synechococcus isolate. All sequenced unicellular marine cyanobacterial genomes contain genes encoding putative Phn transporter components (PhnC, PhnD, and PhnE), but surprisingly, they appear to lack known genes encoding putative C-P lyase subunits (30).
The genes of the phn cluster of OS-B′ are tightly packed on the genome, and there is significant overlap between adjacent coding sequences. This observation suggests that the environment imposes selective pressure to maintain the operon in a relatively condensed state. It has been suggested that the occurrence of overlapping genes within an operon may be one characteristic of sequences prone to lateral gene transfer (LGT) events (23). The potential for LGT of the phn cluster is also consistent with the finding that it is scattered across prokaryotic phylogenies and, moreover, that phn genes are not universally present within the cyanobacterial lineage (15). The potential for LGT of phn genes is also corroborated by the finding that while phn genes are not present on the OS-A genome, the genes flanking the phn cluster in OS-B′ are syntenic on the OS-A genome; this observation suggests a relatively recent loss or gain of the phn cluster. Furthermore, preliminary observations (M. Davison and D. Bhaya, unpublished data) suggest that there is an uncharacterized subpopulation of OS-A that contains the phn genes. Additional sequencing of environmental samples from Octopus Spring and Mushroom Spring will likely provide new information concerning the distribution and diversity of phn genes in the microbial mat community of the hot springs.
Unlike the phn operon of E. coli, the OS-B′ gene cluster does not contain phnF, phnN, phnO, or phnP. PhnF is thought to be a regulatory factor that controls phn gene transcription, while PhnN functions in the formation of ribose-phosphate, perhaps an intermediate in some aspect of Phn metabolism (14). The function of PhnO and PhnP are less clear. None of these “missing” genes is required for C-P bond cleavage; they most likely regulate the uptake and/or metabolism of Phns (28). Despite low AAIDs to other Phn polypeptides and the absence of some of the E. coli phn genes on the OS-B′ genome, the OS-B′ phn gene cluster likely functions in Phn assimilation, since the transcripts accumulate in response to P deprivation and the cells can grow with MePhn as the sole P source, albeit after a long acclimation period. The lack of the putative phn regulatory factors in OS-B′ raises questions about mechanisms associated with controlling Phn uptake and assimilation in cyanobacteria of the microbial mats. We observed that the transcripts from the second phn gene cluster are strongly upregulated under −P conditions. However, it is not absolutely clear if these transporters are responsible for the uptake of MePhn or other substrates. It has been suggested that the long and variable acclimation phase during which cells grow slowly when Phn is supplied as a sole source of P may be the consequence of steps that are limiting in Phn degradation, particularly transport systems that may take varied amounts of time to ready for the transport of different Phn compounds (20). Since OS-B′ has the potential to synthesize multiple putative Phn transporters, it is necessary to determine the substrate specificities and efficiencies of the various transporters and the extent to which they accumulate in the cytoplasmic membrane.
Even though phn transcripts of OS-B′ rapidly accumulate in response to P starvation, OS-B′ cannot effectively use Phn as a sole P source until the cells have acclimated for approximately 3 weeks. The initial addition of MePhn into the medium suppresses activation of Pho regulon genes that are typically transcribed when P is absent from the medium and also blocks continued cell division. Furthermore, cells that are exposed to MePhn appear to have an exacerbated starvation response, as indicated by reduced cell growth and their increased tendency to bleach and accumulate carotenoids, as monitored by whole-cell absorbance spectra. The negative effects of the MePhn on OS-B′ did not result in significant cell death (as visualized via the Live/Dead BacLight system) (data not shown). In cells for which MePhn was the sole source of P, cellular growth arrest and chlorosis was followed by a second phase in which there was steady and slow growth. After about 400 h (∼20 days), the cells began to grow more rapidly. This result was potentially a consequence of abiotic degradation of MePhn (although this compound is extremely stable). This explanation was eliminated by demonstrating that once the cells had acclimated, upon transfer into fresh medium containing MePhn as a sole source of P they initiated growth immediately and attained a doubling time similar to that of cells using Pi as their sole P source. We attempted to grow Phn-acclimated OS-B′ cells on other Phn substrates, including AEPhn and glyphosphate, but consistent growth was only observed with MePhn. However, recent preliminary experiments have demonstrated that cells can also grow on EthPhn and do not appear to require an extended acclimation period, as observed with MePhn. A requirement for an extended acclimation period has been observed for E. coli; the metabolism of MePhn and cell growth is much delayed relative to the rapid accumulation of the phn transcripts (50). The E. coli K-12 strain is cryptic for MePhn utilization, and variants that can grow with MePhn as the sole P source are selected for after exposure to the Phn substrate followed by a relatively long lag phase, similar to the situation for OS-B′. The basis for this extended lag period of OS-B′ is not known, although in E. coli, the lag phase represents the time that it takes to generate a slip strand event in the phnE gene; this lesion deletes one of three direct repeat sequences, thereby restoring the function of a membrane component of the Phn transporter (16). The OS-B′ MePhn acclimation phenomenon could also represent a genetically based mechanism, as recent growth results indicate that acclimation requires the MePhn substrate presence in addition to Pi starvation and that acclimation to the substrate is not lost over a prolonged period following growth of the cells on Pi in the absence of MePhn (data not shown). In the case of OS-B′, it is not clear whether the entire cell population acclimates or whether a subpopulation becomes responsive during the lag phase and ultimately outgrows the cells that were unable to acclimate and utilize Phn. Interestingly, the physiological responses of OS-B′ to MePhn suggest that Phn actually inhibits the accumulation of transcripts from genes that comprise the Pho regulon. The reduced ability to respond to Pi starvation when MePhn is present in the medium indicates that Phns may block transcription of Pho regulon genes (and perhaps other genes as well), either directly or indirectly. For example, it is possible that MePhn interacts with the Pi transport system and thereby suppresses the signaling that is elicited by transport components that work in conjunction with the PhoB/PhoR regulatory elements.
In conclusion, we have investigated how OS-B′, a thermophilic cyanobacterium recently isolated from hot springs in Yellowstone National Park, responds to Pi limitation, and we have demonstrated that MePhn can serve as a sole source of P for these organisms. This work also describes the extended period of acclimation required for growth of the cells on MePhn. The phn gene cluster has been found in divergent microorganisms isolated from a variety of ecosystems, including some in which Phns constitute a substantial fraction of dissolved organic P of the total P pool (7). The capacity to utilize Phns when other sources of P are limiting would confer an adaptive advantage to the OS-B′ ecotype in an environment where Pi may be scarce. Indeed, Pi levels in the Octopus Spring effluent channel drop below concentrations required for activation of the Pho regulon as characterized in E. coli, although it not known if the available Pi fluctuates on a temporal or spatial scale. Starvation on a daily or seasonal time frame may allow OS-B′ to acclimate to low-P conditions, which includes an increased capability for utilizing phosphonates to satisfy the P demand. If fluctuations in the Pi concentration are frequent, the cells may remain in the acclimated state even when availability is elevated over a short time interval. Further investigations are needed to understand the biogeochemical role of Phns in hot spring environments, the distribution and expression of the phn genes among the different bacterial ecotypes, and the exact molecular mechanisms associated with the extended acclimation period that precedes efficient utilization of Phns by OS-B′. We have recently developed in situ approaches to identify gene expression patterns directly in microbial mat samples (42, 43). Now that we have a set of genes that can be used as markers for the P status of these thermophilic cyanobacteria, we will be able to monitor the expression of these genes to determine whether transcripts involved in P acquisition and utilization stay constant or fluctuate over the diel cycle, which will shed light on the different environmental cues that might account for any observed fluctuations.
Supplementary Material
Acknowledgments
We thank Fariba Fazeli for her technical expertise in generating the axenic OS-B′ cultures and also Dave Ward and Mary Bateson for providing the initial OS-B′ enrichment cultures. We thank Anne-Soisig Steunou for her expertise with molecular techniques and for helpful discussions. We thank Oliver Kilian, Jeffrey Moseley, and Michelle Davison (Carnegie Institution for Science), Barbara Cade-Menun (Agriculture and Agri-Food Canada), and George Somero (Stanford University) for helpful discussions. Matthew Blain-Hartung is acknowledged for his assistance in growth experiments.
This research was supported by the Frontiers in Integrative Biology Program of the National Science Foundation (grant EF-0328698). M.M.A. was supported by a National Science Foundation graduate research fellowship.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allewalt, J. P., M. M. Bateson, N. P. Revsbech, K. Slack, and D. M. Ward. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the octopus spring microbial mat community of Yellowstone National Park. Appl. Environ. Microbiol. 72544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 2005. Handbook of media for environmental microbiology. Chapman & Hall/CRC Press/Kluwer, New York, NY.
- 3.Bhaya, D., A. R. Grossman, A. S. Steunou, N. Khuri, F. Cohan, N. Hamamura, M. C. Melendrez, M. M. Bateson, D. M. Ward, and J. F. Heidelberg. 2007. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 1703-713. [DOI] [PubMed] [Google Scholar]
- 4.Brock, T. D. 1978. Thermophilic microorganisms and life at high temperatures. Springer-Verlag, New York, NY.
- 5.Brown, M. R. W., and A. Kornberg. 2004. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 10116085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castenholz, R. W. 1981. Isolation and cultivation of thermophilic cyanobacteria, p. 243. In M. P. Starr, H. Stolp, H. G. Turner, A. Balows and H. G. Schlegel (ed.), The prokaryotes: a handbook on habitats, isolation and identification of bacteria. Springer-Verlag, Berlin, Germany.
- 7.Clark, L. L., E. D. Ingall, and R. Benner. 1998. Marine phosphorus is selectively remineralized. Nature 393426. [Google Scholar]
- 8.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 1744718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 631375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferris, M. J., M. Kühl, A. Wieland, and D. M. Ward. 2003. Cyanobacterial ecotypes in differing optical microenvironments of a 68°C hot spring mat community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 692893-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-García, M. R., M. Losada, and A. Serrano. Concurrent transcriptional activation of ppa and ppx genes by phosphate deprivation in the cyanobacterium Synechocystis sp. strain PCC 6803. Biochem. Biophys. Res. Commun. 302601-609. [DOI] [PubMed]
- 12.Grillo, J. F., and J. Gibson. 1979. Regulation of phosphate accumulation in the unicellular cyanobacterium Synechococcus. J. Bacteriol. 140508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirani, T. A., S. Suzuki, N. Murata, H. Hayashi, and J. J. Eaton-Rye. 2001. Characterization of a two-component signal transduction system involved in the induction of alkaline phosphatase under phosphate-limiting conditions in Synechocystis sp. PCC 6803. Plant Mol. Biol. 45133-144. [DOI] [PubMed] [Google Scholar]
- 14.Hove-Jensen, B., T. J. Rosenkrantz, A. Haldimann, and B. L. Wanner. 2003. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. J. Bacteriol. 1852793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, J., S. Zhengchang, and Y. Xu. 2005. The evolution of microbial phosphonate degradative pathways. J. Mol. Evol. 61682-690. [DOI] [PubMed] [Google Scholar]
- 16.Iqbal, S., G. Parker, H. Davidson, E. Moslehi-Rahmani, and R. L. Robson. 2004. Reversible phase variation in the phnE gene, which is required for phosphonate metabolism in Escherichia coli K-12. J. Bacteriol. 1866118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin-Ru, W., S. Jui-Hung, K. S. Happy, H. Chung-Chi, G. Shuen-Rong, C. Ling-Yun, and C. Poa-Chun. 2007. Cloning of the gene and characterization of the enzymatic properties of the monomeric alkaline phosphatase (PhoX) from Pasteurella multocida strain X-73. FEMS Microbiol. Lett. 267113-120. [DOI] [PubMed] [Google Scholar]
- 18.Keasling, J. D., L. Bertsch, and A. Kornberg. 1993. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. USA 907029-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilian, O., A.-S. Steunou, F. Fazeli, S. Bailey, D. Bhaya, and A. R. Grossman. 2007. Responses of a thermophilic Synechococcus isolate from the microbial mat of Octopus Spring to light. Appl. Environ. Microbiol. 734268-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kononova, S. V., and M. A. Nesmeyanova. 2002. Phosphonates and their degradation by microorganisms. Biochemistry (Moscow) 67220-233. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 6889-125. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, B. A., C. Suarez, A. DePina, E. Click, N. H. Kolodny, and M. M. Allen. 1998. Two internal pools of soluble polyphosphate in the cyanobacterium Synechocystis sp. strain PCC 6308: an in vivo 31P NMR spectroscopic study. Arch. Microbiol. 169195-200. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5355-359. [DOI] [PubMed] [Google Scholar]
- 24.Leuko, S., A. Legat, S. Fendrihan, and H. S. Lotter. 2004. Evaluation of the LIVE/DEAD BacLight kit for detection of extremophilic archaea and visualization of microorganisms in environmental hypersaline samples. Appl. Environ. Microbiol. 706884-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, K., H. Shinagawa, M. Amemura, S. Kimua, N. Atsuo, and A. Ishihama. 1988. Regulation of the phosphate regulon of Eschericia coli: activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 20385-95. [DOI] [PubMed] [Google Scholar]
- 26.Malik, J., G. Barry, and G. Kishore. 1989. The herbicide glyphosate. Biofactors 217-25. [PubMed] [Google Scholar]
- 27.Metcalf, M. W., and B. L. Wanner. 1993. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 12927-32. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf, M. W., and B. L. Wanner. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J. Bacteriol. 1753430-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monds, R. D., P. D. Newell, J. A. Schwartzman, and G. A. O'Toole. 2006. Conservation of the Pho regulon in Pseudomonas fluorescens Pf01. Appl. Environ. Microbiol. 721910-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, L. R., M. Ostrowski, D. J. Scanlan, K. Feren, and T. Sweetsir. 2005. Ecotypic variation in phosphorus-acquisition mechanisms within marine picocyanobacteria. Aquat. Microb. Ecol. 39257-269. [Google Scholar]
- 31.Muda, M., N. N. Rao, and A. Torriani. 1992. Role of PhoU in phosphate transport and alkaline phosphatase regulation. J. Bacteriol. 1748057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowack, B. 2003. Environmental chemistry of phosphonates. Water Res. 372533-2546. [DOI] [PubMed] [Google Scholar]
- 33.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5650-659. [DOI] [PubMed] [Google Scholar]
- 34.Parker, G. F., T. P. Higgins, T. Hawkes, and R. L. Robson. 1999. Rhizobium (Sinorhizobium) meliloti phn genes: characterization and identification of their protein products. J. Bacteriol. 181389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, C. N., M. J. Mandel, and T. J. Silhavy. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 1877549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot, M., E. Kuznetsova, G. Brown, N. N. Rao, M. Kitagawa, H. Mor, A. Savchenko, and A. F. Yakunin. 2004. General enzymatic screens identify three new nucleotidases in Escherichia coli. J. Biol. Chem. 27954687-54694. [DOI] [PubMed] [Google Scholar]
- 37.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical stratification of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 661038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 1781394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray, J. M., D. Bhaya, M. A. Block, and A. R. Grossman. 1991. Isolation, transcription, and inactivation of the gene for an atypical alkaline phosphatase. J. Bacteriol. 1734297-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revsbech, N. P., and D. M. Ward. 1984. Microelectrode studies of intersitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, H., and W. Boos. 1985. Regulation of ugp, the sn-glycerol-3-phosphate transport system of Escherichia coli K-12 that is part of the pho regulon. J. Bacteriol. 163392-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steunou, A.-S., D. Bhaya, M. M. Bateson, M. C. Melendrez, D. M. Ward, E. Brecht, J. W. Peters, M. Kuhl, and A. R. Grossman. 2006. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 1302398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steunou, A.-S., S. I. Jensen, E. Brecht, E. Becraft, M. M. Bateson, O. Kilian, D. Bhaya, D. M. Ward, J. W. Peters, A. R. Grossman, and M. Kühl. 2008. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J. 2364-378. [DOI] [PubMed] [Google Scholar]
- 44.Su, Z., V. Olman, and Y. Xu. 2007. Computational prediction of Pho regulons in cyanobacteria. BMC Genomics 81-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, S., A. Ferjani, I. Suzuki, and N. Murata. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 27913234-13240. [DOI] [PubMed] [Google Scholar]
- 46.Torriani-Gorini, A., E. Yagil, and S. Silver. 1994. Phosphate in microorganisms: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 47.VanBogelen, R. A., P. Sankar, R. L. Clark, J. A. Bogan, and F. C. Neidhardt. 1996. Gene-protein database of Escherichica coli K-12, p. 2067-2117. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichica coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 48.Vera, M., F. Pagliai, N. Guiliani, and C. A. Jerez. 2008. The chemolithoautotroph Acidithiobacillus ferrooxidans can survive under phosphate-limiting conditions by expressing a C-P lyase operon that allows it to grow on phosphonates. Appl. Environ. Microbiol. 741829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 50.Wanner, B. L. 1994. Molecular genetics of carbon-phosphorous bond cleavage in bacteria. Biodegradation 5175-184. [DOI] [PubMed] [Google Scholar]
- 51.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 5147-54. [DOI] [PubMed] [Google Scholar]
- 52.Wanner, B. L., and W. W. Metcalf. 1992. Molecular genetic studies of a 10.9-kb operon in Escherichia coli for phosphonate uptake and biodegradation. FEMS Microbiol. Lett. 100133-140. [DOI] [PubMed] [Google Scholar]
- 53.Ward, D. M., T. Papke, U. Nubel, and M. C. McKitrick. 2002. Natural history of microorganisms inhabiting hot spring microbial mat communities: clues to the origin of microbial diversity and implications for microbiology and macrobiology, p. 25-49. In R. Mitchell (ed.), Biodiversity of microbial life. Wiley-Liss, New York, NY.
- 54.White, A. K., and M. W. Metcalf. 2007. Microbial metabolism of reduced phosphorus compounds. Annu. Rev. Microbiol. 61379-400. [DOI] [PubMed] [Google Scholar]
- 55.White, A. K., and W. W. Metcalf. 2004. Two C-P lyase operons in Pseudomonas stutzeri and their roles in oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 1864730-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zalatan, J. G., T. D. Fenn, A. T. Brunger, and D. Herschlag. 2006. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution. Biochemistry 459788-9803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.