Abstract
Streptomyces clavuligerus produces at least five different clavam metabolites, including clavulanic acid and the methionine antimetabolite, alanylclavam. In vitro transposon mutagenesis was used to analyze a 13-kb region upstream of the known paralogue gene cluster. The paralogue cluster includes one group of clavulanic acid biosynthetic genes in S. clavuligerus. Twelve open reading frames (ORFs) were found in this area, and mutants were generated in each using either in vitro transposon or PCR-targeted mutagenesis. Mutants with defects in any of the genes orfA, orfB, orfC, or orfD were unable to produce alanylclavam but could produce all of the other clavams, including clavulanic acid. orfA encodes a predicted hydroxymethyltransferase, orfB encodes a YjgF/YER057c/UK114-family regulatory protein, orfC encodes an aminotransferase, and orfD encodes a dehydratase. All of these types of proteins are normally involved in amino acid metabolism. Mutants in orfC or orfD also accumulated a novel clavam metabolite instead of alanylclavam, and a complemented orfC mutant was able to produce trace amounts of alanylclavam while still producing the novel clavam. Mass spectrometric analyses, together with consideration of the enzymes involved in its production, led to tentative identification of the novel clavam as 8-OH-alanylclavam, an intermediate in the proposed alanylclavam biosynthetic pathway.
The biochemistry and genetics behind the production of β-lactam antibiotics by microbes have long been an area of active research due to the clinical importance of these compounds. Streptomyces clavuligerus is a bacterium capable of producing a number of β-lactam metabolites, including, most notably, the β-lactamase inhibitor clavulanic acid (9). Although clavulanic acid has only weak antimicrobial activity, it inhibits serine β-lactamase enzymes, which are responsible for much of the penicillin resistance that is seen in clinical settings. When used in combination with conventional β-lactam antibiotics, clavulanic acid provides a powerful tool to combat infections by antibiotic-resistant pathogens. Understanding the complete biosynthetic pathway of clavulanic acid could lead to the design of new strains of S. clavuligerus able to produce higher levels or more potent variants of clavulanic acid.
In addition to clavulanic acid, S. clavuligerus also produces the following other β-lactam metabolites that are structurally similar to clavulanic acid: clavam-2-carboxylate, 2-formyloxymethylclavam, 2-hydroxymethylclavam, and alanylclavam (10, 42) (Fig. 1). All of these β-lactam compounds, including clavulanic acid, are called clavams by virtue of containing a β-lactam ring fused to a five-membered oxazolidine ring. While this bicyclic nucleus in clavulanic acid has 5R stereochemistry, the other clavams mentioned above have a 5S configuration and are thus called the 5S clavams. The β-lactamase inhibitory activity displayed by clavulanic acid is associated with its 5R stereochemistry (3), and therefore the 5S clavams exhibit antifungal or antibacterial activities but do not have β-lactamase inhibitory properties (10, 42).
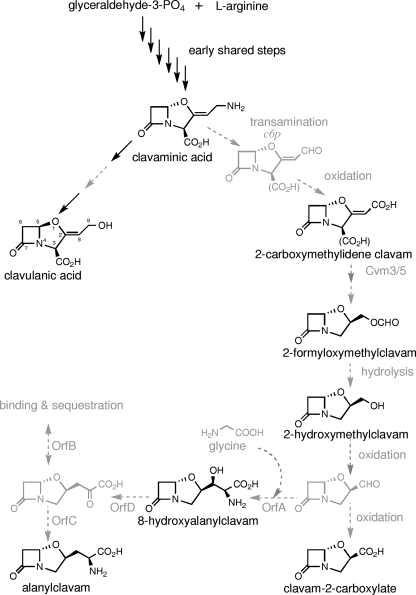
FIG. 1.
Proposed biosynthetic pathway leading to clavulanic acid and the 5S clavams in S. clavuligerus.
Clavulanic acid and the 5S clavams share the same early steps in their biosynthesis. However, once clavaminic acid is synthesized, the pathway splits into two branches, one specific for clavulanic acid production and the other for 5S clavam production (13) (Fig. 1). Two reactions from the late stages of the pathway leading to clavulanic acid have been characterized (2, 21, 35), while only one intermediate leading specifically to the 5S clavams, 2-carboxymethylideneclavam, has been identified (51).
Three unlinked gene clusters containing genes involved in clavam biosynthesis have been identified in S. clavuligerus (52). These are the clavulanic acid, the clavam, and the paralogue gene clusters. The clavulanic acid gene cluster contains genes involved in the early steps required for the biosynthesis of clavaminic acid, as well as genes specific to the late steps of clavulanic acid production (17, 21, 23, 29, 31, 40). The clavam gene cluster carries primarily genes required for the late steps of 5S clavam biosynthesis but also includes cas1 encoding clavaminate synthase, an early biosynthetic enzyme. cas1 is a paralogue of the cas2 gene from the clavulanic acid gene cluster (34, 53). Except for cas1, paralogues for all of the other early biosynthetic genes of the clavulanic acid gene cluster are found in the paralogue gene cluster (22, 53). The paralogue gene cluster also contains genes involved specifically in the late steps of 5S clavam biosynthesis (22, 51, 53).
Five genes required specifically for the late steps of 5S clavam biosynthesis have been described. In the clavam gene cluster, cvm1, cvm2, and cvm5 are necessary for the production of all 5S clavams (34, 51). cvm1 encodes a putative aldo-keto reductase, cvm2 shows similarity to isomerases, and cvm5 encodes a predicted monooxygenase. The roles of these three genes in 5S clavam biosynthesis are uncertain, but Cvm5 may catalyze a Baeyer-Villiger-type oxidation leading to 2-formyloxymethylclavam production. Two other genes, c6p and c7p, are found in the paralogue gene cluster and are also required for the production of all the 5S clavams (51). c6p encodes an aminotransferase that is predicted to catalyze the first step from clavaminic acid toward the 5S clavams, while c7p encodes a probable transcriptional regulator of 5S clavam biosynthesis. While the activities of these gene products allow an outline of a pathway to the 5S clavams to be envisioned, it is apparent that additional activities are required to account for the diversity of the 5S clavam products formed.
In an effort to identify additional genes involved in 5S clavam biosynthesis, a region upstream of the known paralogue gene cluster was sequenced, and genes found in this area were subjected to mutagenesis. In vitro transposon mutagenesis was used to facilitate both DNA sequence analysis and mutagenesis, thus establishing the utility of this technique in S. clavuligerus. Analysis of the mutant phenotypes revealed four genes required specifically for alanylclavam production and suggested a putative biosynthetic pathway for alanylclavam.
MATERIALS AND METHODS
Bacterial strains, plasmids, cosmids, and culture conditions.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were grown in Luria broth (LB) or on LB agar plates at 37°C (45). Strains carrying resistance determinants were grown in medium supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), apramycin (Apr; 50 μg/ml), or chloramphenicol (25 μg/ml), as appropriate.
Strains of S. clavuligerus were grown on maltose-yeast extract-malt extract agar (50) or ISP4 (Difco) agar plates at 28°C. Liquid cultures used for determining antibiotic production levels were grown as previously described (21). Briefly, seed cultures were prepared by inoculating 108 spores from glycerol stocks into 25-ml amounts of Trypticase soy broth supplemented with 1% (wt/vol) soluble starch. These cultures were incubated for 40 h and then inoculated at 2% (vol/vol) into soy flour medium (21). Culture supernatants were collected after 72 and 96 h for assessment of metabolite levels. All S. clavuligerus liquid cultures were grown at 28°C and shaken at 250 rpm. Mutant strains were grown at least three times to determine metabolite-producing capabilities. Antibiotic-resistant strains of S. clavuligerus were grown with Apr (25 μg/ml) or thiostrepton (2.5 to 10 μg/ml).
DNA manipulations.
Plasmid and cosmid DNA isolation from E. coli, restriction endonuclease digestions, PCRs, Southern analyses, and transformations of E. coli were carried out using standard techniques (45).
Isolation of genomic DNA from S. clavuligerus was performed using a standard technique (25). Plasmid and cosmid DNA was introduced into S. clavuligerus through intergeneric conjugation from E. coli ET12567(pUZ8002) as previously described (25). Exconjugants were isolated on AS-1 plus 10 mM MgCl2 medium (4) agar using nalidixic acid (25 μg/ml) to counterselect the donor E. coli strain, and an appropriate antibiotic was used to select for exconjugants.
Both Southern analysis and mycelial PCR were used to confirm mutations in S. clavuligerus. PCRs were typically carried out using mycelia from 40-h seed cultures that had been washed twice with 10.3% sucrose, suspended to their original volume in dimethyl sulfoxide, and then added directly to PCR mixtures to give a final dimethyl sulfoxide concentration of 5% (vol/vol). Oligonucleotide primers used in this study are listed in Table S2 in the supplemental material.
In vitro transposon mutagenesis.
In vitro transposition reactions were carried out according to the manufacturer's specifications (Epicentre, Madison, WI). Briefly, Tn5062 (carrying Aprr, egfp, and oriT) was liberated from pQM5062 (8) by restriction digestion and combined with cosmid 6G9, transposase, and reaction buffer. The reaction was incubated at 37°C for 2 h and then terminated by the addition of a stop solution and heating at 70°C for 10 min. Reaction mixtures were introduced into E. coli Transformax EC100, and transformants were selected on Apr-containing agar.
DNA sequence analysis.
Cosmid 6G9 carries the paralogue gene cluster and its flanking regions. Within 6G9, an 11-kb EcoRI fragment comprises much of the region upstream of the paralogue cluster, and this was targeted for sequencing. Mutant forms of 6G9 carrying individual transposon insertions in the 11-kb EcoRI fragment were identified via restriction analysis. Thirty such mutant cosmids were used as templates, together with primers that bind to the upstream and downstream edges of Tn5062, to sequence outward from each insertion. This provided overlapping sequence information for most of the region upstream of the paralogue gene cluster. When required, sequence-specific primers were used to fill in gaps and to sequence the ends of the 11-kb EcoRI fragment.
A portion of the region immediately upstream of ceaS1 was not contained on the 11-kb EcoRI fragment. Plasmid p2.8-18 carries a 2.8-kb EcoRI fragment comprising the 5′ end of ceaS1 and upstream DNA, and it abuts the 11-kb EcoRI fragment. It was used as a sequencing template to bridge the gap between the paralogue gene cluster and the 11-kb EcoRI fragment.
Sequence analysis was performed using the DYEnamic ET Terminator cycle sequencing system (Roche, Basel, Switzerland) by the Molecular Biology Service Unit, University of Alberta. The double-stranded sequence was obtained for all novel genomic DNA sequences. Open reading frames (ORFs) were predicted using the online program FramePlot 3.0beta (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl) (19), and similarity searches were performed using the BLAST programs made available by the National Institute for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (1). GeneTools version 2.0 (BioTools, Inc., Edmonton, Alberta, Canada) was used to produce DNA sequence assemblies, perform sequence alignments, and identify restriction endonuclease sites. Protein domains were located using the InterProScan Sequence Search (57) provided by the European Molecular Biology Laboratory-European Bioinformatics Institute (http://www.ebi.ac.uk/InterProScan/).
Generation of mutants.
Mutant versions of cosmid 6G9 carrying transposon insertions in all of the ORFs in the upstream region of interest, except for orfB, were obtained by in vitro transposon mutagenesis. The mutant cosmids were conjugated into S. clavuligerus, and Aprr kanamycin-sensitive exconjugants (indicating a double crossover had taken place) were isolated to give mutant strains of S. clavuligerus with insertions in each of the genes orfA, orfC, orfD, orfI, orfJ, orfK, and orfL. However, mutant strains with transposon insertions in orfE, orfF, orfG, and orfH were never successfully generated by this process, despite repeated efforts, and so mutants in these ORFs and in orfB were instead generated by employing a PCR-targeting protocol (16). A second orfC mutant (in addition to the one generated by transposon mutagenesis) was also generated by PCR targeting. For mutants generated by PCR targeting, cosmid 14E10 rather than 6G9 was used as the template, because the genes targeted for mutagenesis are more centrally located in the insert of this cosmid than they are in cosmid 6G9. All mutants were confirmed using Southern analysis or mycelial PCR.
Construction of pSET-AT-orfC.
The tsr gene from pDA504 was inserted as a 1.1-kb BglII/BamHI fragment into the BamHI site of pSET152 to give pSET-AT. A 1.9-kb AgeI/EcoRI fragment containing orfC and its upstream region was isolated from cosmid 6G9 and inserted into pSET-AT after passage through pSL1180 to pick up compatible restriction sites. This construct, pSET-AT-orfC, was then conjugated into an S. clavuligerus orfC::Tn5062 mutant.
Bioassays.
Clavulanic acid production was detected using a disc-diffusion bioassay with Klebsiella pneumoniae ATCC 29665 as the indicator organism (34). Alanylclavam and 2-hydroxymethylclavam were detected using a disc-diffusion bioassay with Bacillus sp. ATCC 27860 as the indicator organism (42).
HPLC and LC-MS analyses.
Culture filtrates were analyzed for clavulanic acid and 5S clavams by high-performance liquid chromatography (HPLC) after imidazole derivatization using a Bondclone C18 column (100 by 8.0 mm, 10 μm; Phenomenex, Torrance, CA) under previously described conditions (34). Culture filtrates containing novel metabolites were further analyzed using LC-mass spectrometry (MS) as previously described (21).
Analysis of the novel clavam.
Five-milliliter amounts of culture supernatants from both wild-type and orfD mutants of S. clavuligerus grown in soy flour medium for 96 h were acidified by the addition of 20 μl of 5 N HCl and then centrifuged for 5 min at 13,000 × g to remove precipitated matter. Supernatants were passed through SepPak Classic C18 cartridges (Waters Scientific, Milford, MA), and the eluant (unretained fraction) containing the clavam metabolites was immediately neutralized with NaOH. Samples (0.5 ml) of each neutralized eluant were reduced to dryness in a SpeedVac, dissolved in 100 μl of water, and derivatized by the addition of 25 μl of imidazole reagent (7). After 15 min at 21°C, 5-μl samples of each were analyzed by HPLC on an XTerra column (0.21 by 10 cm; Waters Scientific) operating under isocratic conditions in 0.1% acetic acid adjusted to pH 3.7 with NH4OH (96%), methanol (4%) at 0.2 ml/min with detection at 311 nm and on-line analysis of effluents by electrospray MS using a ZMD-2 single quadrupole spectrometer (Waters Scientific).
Nucleotide sequence accession number.
The nucleotide sequence of this region of the S. clavuligerus chromosome has been appended to the existing file in GenBank for the paralogue gene cluster under accession number AY426768.
RESULTS
In vitro transposon mutagenesis of S. clavuligerus.
The paralogue gene cluster of S. clavuligerus includes genes involved in the early stages of biosynthesis of all clavam metabolites as well as genes specific for the production of 5S clavam metabolites. In order to analyze the region of the S. clavuligerus chromosome located upstream of the paralogue cluster, in vitro transposon mutagenesis of cosmid 6G9 was undertaken. The resulting mutant cosmids were introduced into E. coli, and thousands of clones carrying Tn5062 in 6G9 were obtained. Among these, 156 mutant clones were subjected to restriction analysis to determine the Tn5062 insertion point.
6G9 can be digested into the following six EcoRI fragments: an 8.2-kb fragment (pWE15) and 20-kb, 11-kb, 5.7-kb, 2.8-kb, and ∼100-bp fragments derived from the genomic DNA insert. The relative positions of the 5.7- and 2.8-kb fragments were already known from prior sequencing, and Southern analysis revealed that the 11-kb fragment (Fig. 2) carries the region of interest lying upstream of the paralogue gene cluster (data not shown). Thirty of the 156 mutant 6G9 cosmids carried Tn5062 in the 11-kb upstream fragment, as determined by restriction analysis. The number of Tn5062 insertions in the different EcoRI fragments is indicated in Table 1. Only 134 cosmids are represented in the table because 22 of the mutant cosmids were not readily interpretable by analysis of EcoRI restriction digests. For the 134 mutant cosmids, frequencies of insertion correlate well with the size of each fragment.
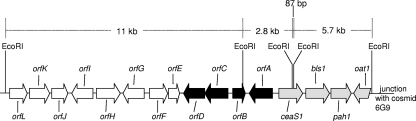
FIG. 2.
The paralogue gene cluster. Gray arrows represent ORFs of the previously described paralogue gene cluster. Black arrows represent newly discovered ORFs specifically required for alanylclavam biosynthesis. White arrows indicate newly discovered ORFs not required for alanylclavam biosynthesis.
TABLE 1.
Frequencies of transposon insertion into the different EcoRI fragments of cosmid 6G9
| EcoRI fragments of cosmid 6G9b | No. of cosmids with Tn5 insertion in a specific EcoRI fragmenta | Frequency of Tn5 insertion (%) | Fragment size as a proportion of total cosmid size (%) |
|---|---|---|---|
| ∼20 | 68 | 50.7 | 41.9 |
| 11 | 30 | 22.4 | 23.1 |
| 8.2 | 16 | 12 | 17.2 |
| 5.7 | 14 | 10.4 | 11.9 |
| 2.8 | 6 | 4.5 | 5.9 |
| 87 | 0 | 0 | 0 |
Results are based on the 134 transposon-containing cosmids examined.
All values are kilobases except the last entry, which is shown as base pairs.
Sequencing and analysis of the region upstream of the paralogue gene cluster.
The 30 6G9 cosmids carrying Tn5062 in the 11-kb EcoRI fragment were used as templates to sequence bidirectionally outward from each transposon insertion point. Gaps in the resulting assembled sequence were filled by sequencing from custom primers. The end of the 2.8-kb EcoRI segment that abuts the 11-kb EcoRI fragment was also sequenced to bridge the gap between the known paralogue gene cluster and the newly sequenced region. Taken together, this provided 13,003 bp of new sequence upstream of the paralogue gene cluster.
Examination of the sequence revealed 12 new ORFs (Fig. 2; Table 2). Of these, four encode products with similarities to proteins involved in amino acid metabolism. orfA encodes a putative serine hydroxymethyltransferase, an enzyme typically involved in the reversible conversion of serine to glycine (46). orfA is also notable in that it contains a TTA codon as the 126th codon of the 391-codon ORF. The product encoded by orfC strongly resembles aminotransferases, which catalyze the transfer of amino groups from amino acids to oxoacids, such that the ketone oxygen is replaced by an amino group (30). orfD encodes a protein similar to threonine dehydratases. Threonine dehydratases (also called threonine deaminases) catalyze the dehydration and deamination of threonine or serine to yield 2-ketobutyrate or pyruvate, respectively (15). Finally, the protein encoded by orfJ shows similarity to dihydrofolate reductases, which carry out the reduction of dihydrofolate to tetrahydrofolate (48). This reaction is an essential step in the production of glycine, purines, and deoxythymidine phosphate.
TABLE 2.
Newly sequenced ORFs from the region upstream of the paralogue cluster of S. clavuligerus
| ORF | Size (aa)a | Most similar protein, accession no., similarity/identity (%)b | Proposed function |
|---|---|---|---|
| orfA | 390 | Serine hydroxymethyltransferase from Methanocaldococcus jannaschii, AAB99615, 32/47 over 380 aa | Hydroxymethyltransferase |
| orfB | 126 | YjgF/YER057c/UK114 family protein from Saccharopolyspora erythraea, YP_001107461, 55/70 over 126 aa | Amino acid biosynthesis regulator |
| orfC | 393 | Aminotransferase from “Solibacter usitatus,” YP_824873, 37/53 over 344 aa | Aspartic/tyrosine/aromatic aminotransferase |
| orfD | 351 | Threonine dehydratase from an Acidobacteria bacterium, YP_591159, 39/56 over 314 aa | Threonine dehydratase |
| orfE | 102 | Hypothetical protein from S. erythraea, YP_001104250, 54/69 over 102 aa | AzlD-type amino acid permease |
| orfF | 253 | Branched-chain amino acid permease from Rhodococcus sp., 53/66 over 204 aa | AzlD-type amino acid permease |
| orfG | 364 | Hypothetical protein from Streptomyces avermitilis, NP_821993, 51/65 over 353 aa | Serine/threonine-type protein kinase |
| orfH | 416 | Transporter from Streptomyces ambofaciens, CAK50890, 45/60 over 397 aa | AraJ-type arabinose efflux permease |
| orfI | 356 | LysR-type transcriptional regulator from S. erythraea, YP_001107820, 48/62 over 294 aa | Transcriptional regulator |
| orfJ | 201 | Dihydrofolate reductase-type protein from Frankia alni, YP_714368, 84/91 over 189 aa | Bifunctional deaminase-reductase |
| orfK | 277 | Hypothetical protein from Frankia sp., YP-481895, 36/49 over 240 aa | Uncharacterized conserved protein |
| orfL | 229 | ArsR-type transcriptional regulator from Rhodococcus sp., YP_707049, 66/82 over 195 aa | Transcriptional regulator |
aa, amino acids.
Percent similarities/identities were determined using the online BLASTp program.
Two of the new ORFs are similar to amino acid transport proteins. Proteins encoded by orfE and orfF have similarity to AzlD and AzlC, respectively. In Bacillus subtilis, AzlC and AzlD cause 4-azaleucine resistance when overproduced and are believed to be involved in branched-chain amino acid transport (5).
orfB, orfI, and orfL code for proteins which show similarities to regulatory proteins. OrfB, based on its amino acid sequence, belongs to the YjgF/YER057c/UK114 family of proteins. In eukaryotes, proteins in this family have been implicated in RNase activity, mitochondrial genome maintenance, and isoleucine biosynthesis (33, 26, 37). In prokaryotes, this family of proteins has been found to be involved in the repression of purine and isoleucine biosynthesis (14, 43). OrfI appears to be a LysR-type transcriptional regulator. LysR-type transcriptional regulators are found in many prokaryotic systems. In fact, ClaR, a protein that positively regulates clavulanic acid biosynthesis in S. clavuligerus, is itself a LysR-type protein (38, 41). OrfL shows similarity to the ArsR family of transcriptional repressors, proteins that regulate expression of arsenical resistance operons (12).
The three other ORFs in the region upstream of the paralogue gene cluster do not have any commonalities with the other ORFs or with each other. The protein encoded by orfG shows very limited similarity to protein kinases, the orfH protein product is similar to multidrug resistance transporters, and orfK codes for a protein with limited similarity to activators of Hsp90 ATPase.
Generation and analysis of mutants.
To determine whether any of the newly discovered ORFs play a role in the biosynthesis of 5S clavams or other metabolites produced by S. clavuligerus, mutants with defects in each of the ORFs were obtained. Mutants in orfA, orfC, orfD, orfI, orfJ, orfK, and orfL were generated by conjugating cosmid 6G9 derivatives carrying Tn5062 in the ORF of interest into S. clavuligerus. Recombination between the mutant cosmids and the chromosome allowed gene conversion to occur, thus transferring the mutations to the chromosomal copy of the gene. However, even after repeated attempts, no orfE, orfF, orfG, or orfH mutations could be obtained via this route. Although this initially suggested that these ORFs may be essential for cellular functions, PCR-targeting mutagenesis was subsequently performed successfully to generate mutants (16). Since there were no mutant cosmids carrying a transposon insertion in orfB, the mutation of orfB was also carried out via PCR targeting. Thus, transposon-bearing mutants were generated in orfA, orfC, orfD, orfI, orfJ, orfK, and orfL, and mutants carrying an antibiotic resistance gene cassette replacing the ORF of interest were prepared in orfB, orfE, orfF, orfG, and orfH. All mutations were confirmed through either Southern analyses or mycelial PCR.
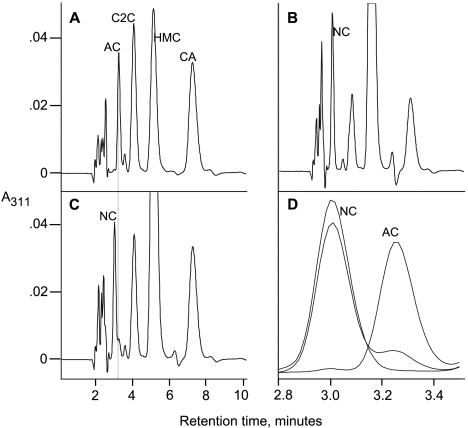
None of the mutants showed any morphological effects or dramatic changes in either cephamycin C or clavulanic acid production (data not shown). Most of the mutants also showed no changes in production of 5S clavams. However, mutants with defects in orfA, orfB, orfC, and orfD were specifically defective in the production of alanylclavam (Fig. 3). Interestingly, analysis of culture filtrates of orfC and orfD mutants showed the presence of an additional clavam-type peak eluting just before the alanylclavam retention time on HPLC chromatograms, indicating that these mutants were producing a novel metabolite (Fig. 3).
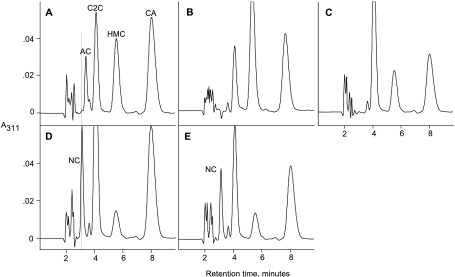
FIG. 3.
HPLC analysis of clavam metabolites produced by wild-type and mutant strains of S. clavuligerus. Wild type (A), orfA mutant (B), orfB mutant (C), orfC mutant (D), and orfD mutant (E) are shown. AC, alanylclavam; C2C, clavam-2-caboxylate; HMC, 2-hydroxymethylclavam; CA, clavulanic acid; NC, novel clavam. The faint dotted line shows the expected location of the novel clavam in the HPLC profile from the wild-type strain.
Mass determination of the novel clavam.
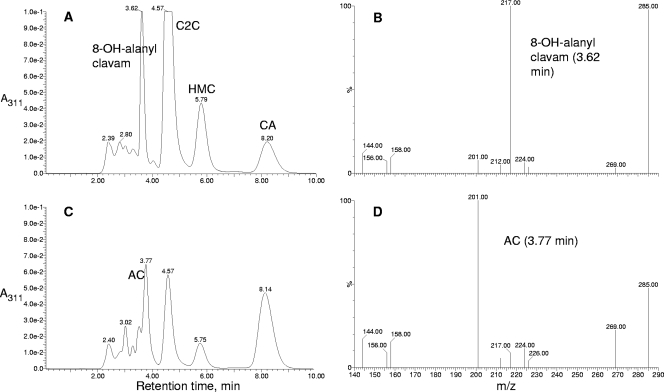
Underivatized clavam metabolites have poor retention on reverse-phase HPLC columns, and so to investigate the structure of this new metabolite, imidazole-derivatized clavams from wild-type and orfD culture supernatants were subjected to HPLC with on-line electrospray MS analysis (Fig. 4). Analysis of wild-type culture supernatants showed an alanylclavam peak that gave the expected prominent mass ions of 269 (intact imidazole derivative) and 201 (fragmented derivative), whereas the novel clavam from orfD mutant culture supernatants gave a peak with mass ions of 285 (intact imidazole derivative) and 217 (fragmented derivative). These masses are consistent with the novel clavam having a structure of 8-OH-anylclavam. The HPLC profile of the wild-type culture supernatant shows a shoulder on the leading edge of the alanylclavam peak corresponding in retention time to that of 8-OH-alanylclavam. That shoulder peak also shows low levels of mass ions at 285 and 217, indicating that 8-OH-alanylclavam may normally be present in wild-type culture supernatants but at low levels compared to those of alanylclavam. In contrast, the 8-OH-alanylclavam peak from the orfD mutant culture supernatant showed no appreciable levels of alanylclavam (Fig. 4A).
FIG. 4.
MS analysis of the novel clavam accumulated by orfC and orfD mutants of S. clavuligerus. HPLC profile of orfD mutant of S. clavuligerus (A), mass spectrum of the novel clavam peak (retention time, 3.62 min) seen in panel A (B), HPLC profile of wild-type S. clavuligerus (C), and mass spectrum of the alanylclavam peak (retention time, 3.77 min) seen in panel C (D) are shown. Abbreviations for metabolite peaks are given in legend to Fig. 3.
Complementation of the orfC mutant.
Mutants with defects in either orfC or orfD accumulate the same novel clavam, suggesting either that the OrfC and OrfD proteins are both needed for a single reaction in alanylclavam synthesis or that the genes are transcribed together in an operon and that transposon insertion in the upstream orfC has polar effects on the expression of orfD. This latter possibility seemed most likely, since the orfC stop codon overlaps the orfD start codon, and this in turn suggested that the loss of OrfD alone could be responsible for accumulation of the novel metabolite.
To test whether these two genes are transcribed in an operon, the orfC mutant was complemented. orfC was inserted into pSET-AT, which confers thiostrepton resistance and integrates into the chromosome at the ΦC31 phage attachment site (6). orfC mutant strains carrying pSET-AT-orfC were fermented, and culture filtrates were analyzed (Fig. 5). Chromatograms revealed that the complemented mutants still produced primarily the novel clavam (retention time, 3.0 min), but a small peak (retention time, 3.3 min) consistent with the presence of alanylclavam was also seen (Fig. 5C). This shoulder peak is not found in the uncomplemented orfC mutant (Fig. 5B).
FIG. 5.
HPLC analysis of clavam metabolites produced by the complemented orfC mutant of S. clavuligerus. Wild type (A), orfC mutant (B), complemented orfC mutant (C), and overlay and expansion of panels A to C in the region of retention times from 2.8 to 3.5 min (D) are shown. The faint dotted line indicates the presence of low levels of alanylclavam in the complemented orfC mutant. Abbreviations for metabolite peaks are given in legend to Fig. 3.
Complementation of the orfC mutant with pSET-AT-orfC was able to restore only trace levels of alanylclavam production, supporting the hypothesis that the orfC mutation was polar on orfD. To obtain additional evidence, we attempted to produce an orfC in-frame deletion mutant, which would not be polar on orfD. As a first step, a ΔorfC::apra mutant was created by PCR-targeted mutagenesis. HPLC analysis of culture filtrates from this mutant showed the same chromatographic profile as was seen in the orfC transposon mutant, with accumulation of the novel clavam (data not shown). However, when the ΔorfC::apra mutant was subjected to flp-mediated recombination to remove the Aprr cassette and replace it with an 81-bp in-frame scar (16), no in-frame mutants were recovered, despite two independent attempts.
DISCUSSION
The known sequence of the paralogue gene cluster has been increased to 23,328 bp through the use of in vitro transposon mutagenesis, the first time this technology has been applied to S. clavuligerus. Within the 13 kb of new DNA sequence, 12 new ORFs were located. Using a combination of in vitro transposon mutagenesis and PCR-targeted mutagenesis, mutations were generated in each of the newfound ORFs.
For unknown reasons, Tn5062 could not be introduced into all of the genes on the chromosome. Mutant cosmids with Tn5062 inserted into orfE, orfF, orfG, and orfH were obtained, but transfer of these mutations into the S. clavuligerus chromosome was unsuccessful, despite repeated attempts. This suggested that the ORFs might be essential for S. clavuligerus survival, but PCR-targeted mutagenesis subsequently proved that not to be the case. The Tn5062 system is currently being used to mutate the entire Streptomyces coelicolor genome, and no indication of difficulties has been reported in that species (8, 18). Although the exact cause is not known, in vitro transposon mutagenesis using Tn5062 may not be ideal for saturation mutagenesis of S. clavuligerus, but it is still useful for DNA sequencing.
Of the 12 newly described ORFs, four were specifically required for alanylclavam biosynthesis: orfA, orfB, orfC, and orfD. Each of these ORFs encodes a protein with similarities to those involved in amino acid regulation or biosynthesis. OrfA shows similarity to serine hydroxymethyltransferases, enzymes that catalyze the reversible interconversion of glycine and 5,10-methylenetetrahydrofolate to serine and tetrahydrofolate. While OrfA contains three of the four amino acids involved in glycine binding, it lacks five of the six amino acids involved in 5,10-methylenetetrahydrofolate binding (46). This suggests that OrfA may catalyze a reaction between glycine and a substrate other than 5,10-methylenetetrahydrofolate.
OrfB belongs to the YjgF/YER057c/UK114 family of proteins (14, 26, 33, 37, 43). While these proteins have been implicated in many cellular functions in both eukaryotes and prokaryotes, involvement in regulation of isoleucine biosynthesis has been observed in a number of organisms. YjgF from Salmonella enterica serovar Typhimurium is necessary for isoleucine biosynthesis in that it is required for normal IlvB (an aminotransferase directly involved in isoleucine biosynthesis) activity (14, 47). As well, members of the YjgF/YER057c/UK114 family in Haemophilus influenzae and E. coli are capable of binding to α-keto acids, such as 2-ketobutyrate, the first intermediate of isoleucine biosynthesis (11, 39). This led to the formulation of a model in which YjgF binds to an α-keto acid, possibly an intermediate in the isoleucine biosynthetic pathway such as 2-ketobutyrate, that would, in its unbound form, repress IlvB activity (47). By binding to the α-keto acid, YjgF sequesters the metabolite and prevents it from inhibiting the aminotransferase, thus allowing isoleucine production to continue (47).
Examination of the OrfB amino acid sequence reveals that it contains all of the eight residues implicated in ligand binding (Y18, G32, Q33, F86, N90, P102, R104, and E118) as determined from the crystal structures of E. coli and human YjgF/YER057c/UK114 family proteins (32, 55). Although C107 in TdcF (an E. coli family member) has also been identified as an important residue for 2-ketobutyrate binding, it is not as conserved as the other eight active site residues and is not found in either the human YjgF/YER057c/UK114 family protein nor OrfB (11, 55). Therefore, OrfB is likely to be capable of binding to α-keto acids.
Analysis of the OrfC amino acid sequence shows that OrfC belongs to family I of the aminotransferases (20, 30). Family I contains aspartate, alanine, and aromatic amino acid aminotransferases (30), but OrfC contains only two of the six amino acids involved in substrate binding found in similar family I aminotransferases (24, 36, 44), suggesting that OrfC may catalyze aminotransferase reactions but use atypical substrates.
OrfD is similar to threonine and serine dehydratases. It contains all of the residues likely involved in substrate binding found in the E. coli biosynthetic threonine dehydratase (15), the S. enterica serovar Typhimurium threonine deaminase TdcB (49), and the rat liver serine dehydratase (56). For this reason, it is probable that OrfD catalyzes a reaction similar to that carried out by threonine and serine dehydratases, involving a substrate structurally analogous to threonine or serine.
From the apparent activities of OrfA, OrfB, OrfC, and OrfD, it is possible to envision a putative alanylclavam biosynthetic pathway. OrfA may use glycine as a substrate and fuse it to an as-yet-unobserved aldehyde-containing clavam intermediate in a reaction analogous to that catalyzed by serine hydroxymethyltransferase (Fig. 1). This would produce 8-OH-alanylclavam, which has the same mass as the novel clavam accumulated by orfC and orfD mutants. OrfD, by analogy to threonine deaminase, would then carry out the dehydration/deamination of 8-OH-alanylclavam to produce a clavam intermediate with a pyruvyl side chain attached at C2 (Fig. 1). Alanylclavam would finally be generated by transamination of the keto group on the pyruvyl side chain through the action of OrfC (Fig. 1). In this putative pathway, the role of OrfB is regulatory in nature. If the pyruvylclavam intermediate (an α-keto acid) inhibits OrfC activity, analogous to the way α-keto acids inhibit aminotransferase activity in isoleucine biosynthesis, OrfB may function to sequester this intermediate, thereby allowing OrfC to remain active (Fig. 1). Although more detailed analyses will be required to confirm the identity of 8-OH-alanylclavam, the proposed structure is in agreement with the observed mass of the compound and is consistent with the proposed biosynthetic pathway.
The proposed pathway can also help to explain the observed phenotypes of the orfC and orfD mutants and of the complemented orfC mutant. In the orfD mutant, 8-OH-alanylclavam would be expected to accumulate, since its further conversion requires OrfD. Because orfC and orfD form an apparent operon, the polar effects of orfC disruption on orfD expression would also result in a buildup of 8-OH-alanylclavam in the orfC mutant. However, despite the operon arrangement, orfD may be expressed at low levels in the orfC mutants due to some level of transcription originating within the transposon or to the presence of a weak promoter located within orfC but after the point of Tn5062 insertion. In an orfC mutant, this low level of OrfD would generate correspondingly small amounts of the pyruvylclavam intermediate (not observed), but the main product would be 8-OH-alanylclavam. In the complemented orfC mutant, OrfD would remain at the same low level, and so even full restoration of OrfC would still result in 8-OH-alanylclavam as the main product, although low levels of alanylclavam would now also be produced. The fermentation profiles observed for these mutants and complemented strains are consistent with this proposed pathway and the transcriptional arrangement of the orfC and orfD genes. Interestingly, our unsuccessful attempts to generate an in-frame orfC mutant, which should have decoupled expression of orfD from that of orfC, suggests that a buildup of the predicted α-keto acid product of OrfD might be toxic to the cell.
Regulation of alanylclavam production is also of interest in view of the observation that orfA contains a TTA codon. This TTA codon represents only the second such codon found in any of the genes associated with production of β-lactam metabolites in S. clavuligerus. The only other TTA codon is found in ccaR, a gene that encodes a pathway-specific transcriptional regulator controlling cephamycin and clavulanic acid production. bldA is a rare leucyl tRNA for the translation of UUA codons in Streptomyces spp. (27). Genes containing TTA codons are typically strongly dependent on bldA for expression and are usually genes that are expressed during the later stages of growth, such as those involved in secondary metabolite production (28).
Examination of S. clavuligerus bldA mutants has revealed that they can produce all of the clavams except alanylclavam (unpublished results). orfA would make a logical target for regulation of alanylclavam biosynthesis by bldA, since it encodes the first dedicated enzyme of the proposed alanylclavam branch of the 5S clavam biosynthetic pathway, but additional studies will be required to confirm its involvement, since ccaR expression was found unexpectedly to proceed in a bldA mutant of S. clavuligerus, despite the presence of a TTA codon (54).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). N.J.Z. was supported by a studentship from NSERC.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arulanantham, H., N. J. Kershaw, K. S. Hewitson, C. E. Hughes, J. E. Thirkettle, and C. J. Schofield. 2006. ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. J. Biol. Chem. 281279-287. [DOI] [PubMed] [Google Scholar]
- 3.Baggaley, K. H., A. G. Brown, and C. J. Schofield. 1997. Chemistry and biosynthesis of clavulanic acid and other clavams. Nat. Prod. Rep. 14309-333. [DOI] [PubMed] [Google Scholar]
- 4.Baltz, R. H. 1980. Genetic recombination by protoplast fusion in Streptomyces. Dev. Ind. Microbiol. 2143-54. [Google Scholar]
- 5.Belitsky, B. R., M. C. Gustafsson, A. L. Sonenshein, and C. Von Wachenfeldt. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 1795448-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierman, M., R. Logan, E. T. O'Brien, E. T. Seno, N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. E., J. M. Bellis, and B. C. Gasson. 1982. Spectrophotometric assay of clavulanic acid by reaction with imidazole. Analyst 1071241-1245. [Google Scholar]
- 8.Bishop, A., S. Fielding, P. Dyson, and P. Herron. 2004. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, A. G., D. Butterworth, M. Cole, G. Hanscombe, J. D. Hood, C. Reading, and G. N. Robinson. 1976. Naturally occurring beta-lactamase inhibitors with antibacterial activity. J. Antibiot. 29668-669. [DOI] [PubMed] [Google Scholar]
- 10.Brown, D., J. R. Evans, and R. A. Fletton. 1979. Structures of three novel β-lactams isolated from Streptomyces clavuligerus. J. Chem. Soc. Chem. Commun. 1979282-283. [Google Scholar]
- 11.Burman, J. D., C. E. Stevenson, R. G. Sawers, and D. M. Lawson. 2007. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct. Biol. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27131-143. [DOI] [PubMed] [Google Scholar]
- 13.Egan, L. A., R. W. Busby, D. IwataReuyl, and C. A. Townsend. 1997. Probable role of clavaminic acid as the terminal intermediate in the common pathway to clavulanic acid and the antipodal clavam metabolites. J. Am. Chem. Soc. 1192348-2355. [Google Scholar]
- 14.Enos-Berlage, J. L., M. J. Langendorf, and D. M. Downs. 1998. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 1806519-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher, D. T., G. L. Gilliland, G. Xiao, J. Zondlo, K. E. Fisher, D. Chinchilla, and E. Eisenstein. 1998. Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6465-475. [DOI] [PubMed] [Google Scholar]
- 16.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 1001541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgson, J. E., A. P. Fosberry, N. S. Rawlinson, H. N. M. Ross, R. J. Neal, J. C. Arnell, A. J. Earl, and E. J. Lawlor. 1995. Clavulanic acid biosynthesis in Streptomyces clavuligerus: gene cloning and characterization. Gene 16649-55. [DOI] [PubMed] [Google Scholar]
- 18.Hoskisson, P. A., S. Rigali, K. Fowler, K. C. Findlay, and M. J. Buttner. 2006. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J. Bacteriol. 1885014-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174251-253. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, R. A., and W. Gu. 1996. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 1782161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, S. E., A. S. Paradkar, R. H. Mosher, C. Anders, P. H. Beatty, M. J. Brumlik, A. Griffin, and B. Barton. 2004. Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 48192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, S. E., A. Wong, A. Griffin, and B. Barton. 2004. Streptomyces clavuligerus has a second copy of the proclavaminate amidinohydrolase gene. Antimicrob. Agents Chemother. 48514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, S. E., K. J. Elder, K. A. Aidoo, and A. S. Paradkar. 2000. Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 44720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamitori, S., A. Okamoto, K. Hirotsu, T. Higuchi, S. Kuramitsu, H. Kagamiyama, Y. Matsuura, and Y. Katsube. 1990. Three-dimensional structures of aspartate aminotransferase from Escherichia coli and its mutant enzyme at 2.5 Å resolution. J. Biochem. 108175-184. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 26.Kim, J. M., H. Yoshikawa, and K. Shirahige. 2001. A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells 6507-517. [DOI] [PubMed] [Google Scholar]
- 27.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 11305-1310. [DOI] [PubMed] [Google Scholar]
- 28.Leskiw, B. K., E. J. Lawlor, J. M. Fernandez-Abalos, and K. F. Chater. 1991. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA 882461-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, R., N. Khaleeli, and C. A. Townsend. 2000. Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J. Bacteriol. 1824087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214549-561. [DOI] [PubMed] [Google Scholar]
- 31.Mellado, E., L. M. Lorenzana, M. Rodriguez-Saiz, B. Diez, P. Liras, and J. L. Barredo. 2002. The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 1491427-1438. [DOI] [PubMed] [Google Scholar]
- 32.Mistiniene, E., N. Pozdniakovaite, V. Popendikyte, and V. Naktinis. 2005. Structure-based ligand binding sites of protein p14.5, a member of protein family YER057c/YIL051c/YjgF. Int. J. Biol. Macromol. 3761-68. [DOI] [PubMed] [Google Scholar]
- 33.Morishita, R., A. Kawagoshi, T. Sawasaki, K. Madin, T. Ogasawara, T. Oka, and Y. Endo. 1999. Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J. Biol. Chem. 27420688-20692. [DOI] [PubMed] [Google Scholar]
- 34.Mosher, R. H., A. S. Paradkar, C. Anders, B. Barton, and S. E. Jensen. 1999. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 431215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson, N. H., K. H. Baggaley, R. Cassels, M. Davison, S. W. Elson, M. Fulston, J. W. Tyler, and S. R. Woroniecki. 1994. Evidence that the immediate biosynthetic precursor of clavulanic acid is its N-aldehyde analog. J. Chem. Soc. Chem. Commun. 19941281-1282. [Google Scholar]
- 36.Okamoto, A., Y. Nakai, H. Hayashi, K. Hirotsu, and H. Kagamiyama. 1998. Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network. J. Mol. Biol. 280443-461. [DOI] [PubMed] [Google Scholar]
- 37.Oxelmark, E., A. Marchini, I. Malanchi, F. Magherini, L. Jaquet, M. A. Hajibagheri, K. J. Blight, J. C. Jauniaux, and M. Tommasino. 2000. Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol. Cell. Biol. 207784-7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paradkar, A. S., K. A. Aidoo, and S. E. Jensen. 1998. A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol. Microbiol. 27831-843. [DOI] [PubMed] [Google Scholar]
- 39.Parsons, L., N. Bonander, E. Eisenstein, M. Gilson, V. Kairys, and J. Orban. 2003. Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family. Biochemistry 4280-89. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Redondo, R., A. Rodriguez-Garcia, J. F. Martin, and P. Liras. 1999. Deletion of the pyc gene blocks clavulanic acid biosynthesis except in glycerol-containing medium: evidence for two different genes in formation of the C3 unit. J. Bacteriol. 1816922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Redondo, R., A. Rodriguez-Garcia, J. F. Martin, and P. Liras. 1998. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211311-321. [DOI] [PubMed] [Google Scholar]
- 42.Pruess, D. L., and M. Kellett. 1983. Ro-22-5417, a new clavam antibiotic from Streptomyces clavuligerus. I. Discovery and biological activity. J. Antibiot. 36208-212. [DOI] [PubMed] [Google Scholar]
- 43.Rappu, P., B. S. Shin, H. Zalkin, and P. Mantsala. 1999. A role for a highly conserved protein of unknown function in regulation of Bacillus subtilis purA by the purine repressor. J. Bacteriol. 1813810-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee, S., M. M. Silva, C. C. Hyde, P. H. Rogers, C. M. Metzler, D. E. Metzler, and A. Arnone. 1997. Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate. J. Biol. Chem. 27217293-17302. [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Scarsdale, J. N., S. Radaev, G. Kazanina, V. Schirch, and H. T. Wright. 2000. Crystal structure at 2.4 Å resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate. J. Mol. Biol. 296155-168. [DOI] [PubMed] [Google Scholar]
- 47.Schmitz, G., and D. M. Downs. 2004. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnell, J. R., H. J. Dyson, and P. E. Wright. 2004. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 33119-140. [DOI] [PubMed] [Google Scholar]
- 49.Simanshu, D. K., H. S. Savithri, and M. R. Murthy. 2006. Crystal structures of Salmonella typhimurium biodegradative threonine deaminase and its complex with CMP provide structural insights into ligand-induced oligomerization and enzyme activation. J. Biol. Chem. 28139630-39641. [DOI] [PubMed] [Google Scholar]
- 50.Stuttard, C. 1982. Temperate phages of Streptomyces venezuelae: lysogeny and host specificity shown by phages SV1 and SV2. J. Gen. Microbiol. 128115-121. [Google Scholar]
- 51.Tahlan, K., C. Anders, A. Wong, R. H. Mosher, P. H. Beatty, M. J. Brumlik, A. Griffin, C. Hughes, J. Griffin, B. Barton, and S. E. Jensen. 2007. 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chem. Biol. 14131-142. [DOI] [PubMed] [Google Scholar]
- 52.Tahlan, K., H. U. Park, and S. E. Jensen. 2004. Three unlinked gene clusters are involved in clavam metabolite biosynthesis in Streptomyces clavuligerus. Can. J. Microbiol. 803-810. [DOI] [PubMed]
- 53.Tahlan, K., H. U. Park, A. Wong, P. H. Beatty, and S. E. Jensen. 2004. Two sets of paralogous genes encode the enzymes involved in the early stages of clavulanic acid and clavam metabolite biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 48930-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trepanier, N. K., S. E. Jensen, D. C. Alexander, and B. K. Leskiw. 2002. The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology 148643-656. [DOI] [PubMed] [Google Scholar]
- 55.Volz, K. 1999. A test case for structure-based functional assignment: the 1.2 Å crystal structure of the yjgF gene product from Escherichia coli. Protein Sci. 82428-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada, T., J. Komoto, Y. Takata, H. Ogawa, H. C. Pitot, and F. Takusagawa. 2003. Crystal structure of serine dehydratase from rat liver. Biochemistry 4212854-12865. [DOI] [PubMed] [Google Scholar]
- 57.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17847-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.