Abstract
The two optical forms of aldohexose galactose differing at the C-1 position, α-d-galactose and β-d-galactose, are widespread in nature. The two anomers also occur in di- and polysaccharides, as well as in glycoconjugates. The anomeric form of d-galactose, when present in complex carbohydrates, e.g., cell wall, glycoproteins, and glycolipids, is specific. Their interconversion occurs as monomers and is effected by the enzyme mutarotase (aldose-1-epimerase). Mutarotase and other d-galactose-metabolizing enzymes are coded by genes that constitute an operon in Escherichia coli. The operon is repressed by the repressor GalR and induced by d-galactose. Since, depending on the carbon source during growth, the cell can make only one of the two anomers of d-galactose, the cell must also convert one anomer to the other for use in specific biosynthetic pathways. Thus, it is imperative that induction of the gal operon, specifically the mutarotase, be achievable by either anomer of d-galactose. Here we report in vivo and in vitro experiments showing that both α-d-galactose and β-d-galactose are capable of inducing transcription of the gal operon with equal efficiency and kinetics. Whereas all substitutions at the C-1 position in the α configuration inactivate the induction capacity of the sugar, the effect of substitutions in the β configuration varies depending upon the nature of the substitution; methyl and phenyl derivatives induce weakly, but the glucosyl derivative does not.
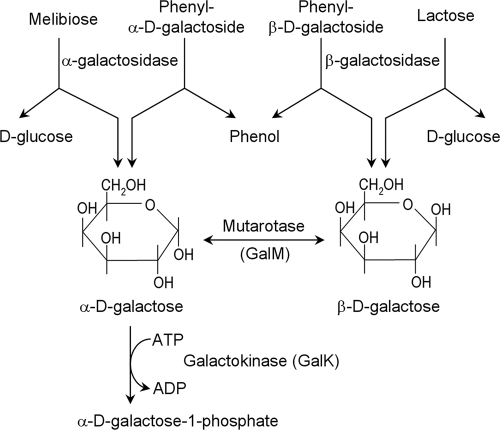
d-Galactose, also known as cerebrose, is a natural aldohexose which is ubiquitous in bacteria, plants, and animals, including human brains (28). It occurs also as part of more complex carbohydrates, e.g., oligo- and polysaccharides, glycoproteins, and glycolipids, and as components of bacterial cell walls. d-Galactose has two anomeric forms, α-d-galactopyranose and β-d-galactopyranose (Fig. 1) (36). Although the anomers spontaneously interconvert slowly in aqueous solution, an enzyme (mutarotase or aldose-1-epimerase) is responsible for interconverting the two optical varieties in monosaccharide forms in vivo (6, 21). Mutarotases are present throughout the prokaryotic and eukaryotic phyla (6, 31, 34, 37). Mutarotase syntheses are also induced in response to specific signals. For example, galactose mutarotase synthesis is induced by retinoic acid in human myeloid cells (30). In the bacterium Escherichia coli, enzymes of d-galactose metabolism (15, 20), including the mutarotase (6), are encoded in an operon which is induced by d-galactose. It has been suggested previously that only β-d-galactose is the inducer of the gal operon (7). Since d-galactose moieties in complex carbohydrates are anomerically specific, α or β, we hypothesize that the mutarotase enzyme, which interconverts the α and β anomers, must be induced for the interconversion when either one of the two anomers is present in the cell, say, by hydrolysis of d-lactose, which generates β-d-galactose, or of d-melibiose, which generates α-d-galactose. In this study, we showed by both in vivo and in vitro experiments that both of the anomers of d-galactose, as proposed above, can induce gal transcription by inactivating the gene regulatory repressor GalR.
FIG. 1.
Generation of α and β anomers of d-galactose in cellular metabolism.
MATERIALS AND METHODS
α-d-Galactose, β-d-galactose, d-galactosides, and other sugars.
α and β anomers of d-galactopyranose were purchased from Omicron Biochemicals, Inc. (South Bend, IN). Methyl-α-d-galactopyranoside (catalog no. 66916), methyl-β-d-galactopyranoside (catalog no. M0285), α-d-galactose-1-phosphate (catalog no. G0380), UDP-α-d-galactose (catalog no. U4500), UDP-α-d-glucose (catalog no. 94335), α-d-melibiose (catalog no. M269-1), d-fucose (catalog no. 47880), 2-deoxy-d-galactose (catalog no. D-4407), and 4-nitrophenyl-β-d-glucuronide (catalog no. N1627) were from Sigma-Aldrich Corp. (St. Louis, MO). Lactose (catalog no. L-107) and d-galactose (catalog no. G-106) were purchased from Pfanstiehl Laboratory, Inc. (Waukegan, IL). Phenyl-β-d-galactopyranoside (catalog no. MP023920801) and phenyl-α-d-galactopyranoside (catalog no. MP032090801) were purchased from Carbosynth, Ltd. (Berkshire, United Kingdom).
Bacteria, phage, and plasmids.
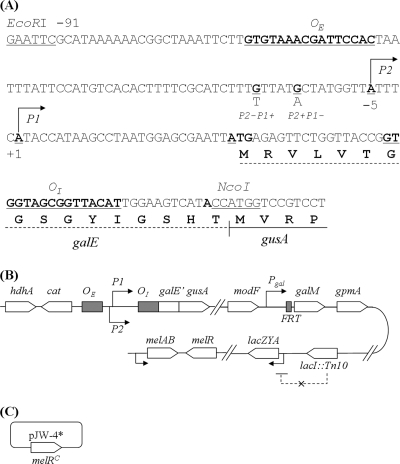
The E. coli strains, phage, and plasmids used in this study are listed in Table 1. E. coli strains with individual galE, galK, and galM open reading frame deletions were obtained from the Keio Collection (2). The promoter regions of the galE gene (−115 to +48 from the start codon) were amplified by PCR from pI24, pSA809, and pSA810 with an EcoRI-containing primer at the 5′ end and an NcoI-containing primer at the 3′ end (Fig. 2A). The fragments were cloned into EcoRI and NcoI sites in the backbone of pSA809 to make a translational fusion of the galE segment containing OI and the entire gusA gene (Fig. 2A), generating pSL305, pSL306, and pSL307, respectively. In addition, the upstream region (−1540 to −939) of gusA and a chloramphenicol marker (+699 to −147 region of the cat gene from plasmid pACYC184 [New England BioLabs, Inc., Beverly, MA]) were placed in the upstream region of the gal promoters of pSL305, pSL306, and pSL307 to make pSL310 (P2+P1+ gusA), pSL311 (P2+P1− gusA), and pSL312 (P2−P1+ gusA), respectively (Fig. 2).
TABLE 1.
Bacterial strains, phage, and plasmids used in this study
| Strain, phage, or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| MG1655 | F−ilvG rfb-50 rph-1 | NIH stock |
| NM18 | lacI::Tn10 | 26 |
| BW25113 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ−rph-1 Δ(rhaD-rhaB)568 hsdR514 | 13 |
| JW0742 | BW25113 galE::FRT-Kmr-FRT | 2 |
| JW0740 | BW25113 galK::FRT-Kmr-FRT | 2 |
| JW0739 | BW25113 galM::FRT-Kmr-FRT | 2 |
| SL300 | BW25113 galETK::FRT-Kmr-FRT galM+ | This study |
| SL301 | BW25113 galETKM::FRT-Kmr-FRT | This study |
| SL310 | BW25113 gal P2+ P1+gusA (Cmr) | This study |
| SL311 | BW25113 gal P2+ P1−gusA (Cmr) | This study |
| SL312 | BW25113 gal P2− P1+gusA (Cmr) | This study |
| SL330 | MG1655 lacI::Tn10 gal P2+ P1+gusA (Cmr) | This study |
| SL331 | MG1655 lacI::Tn10 gal P2+ P1−gusA (Cmr) | This study |
| SL332 | MG1655 lacI::Tn10 gal P2− P1+gusA (Cmr) | This study |
| SL3304 | MG1655 lacI::Tn10 gal P2+ P1+gusA (Cmr) galETK galM+/pJW-4* | This study |
| SL3305 | MG1655 lacI::Tn10 gal P2+ P1−gusA (Cmr) galETK galM+/pJW-4* | This study |
| SL3306 | MG1655 lacI::Tn10 gal P2− P1+gusA (Cmr) galETK galM+/pJW-4* | This study |
| SL3307 | MG1655 lacI::Tn10 gal P2+ P1+gusA (Cmr) galETKM/pJW-4* | This study |
| SL3308 | MG1655 lacI::Tn10 gal P2+ P1−gusA (Cmr) galETKM/pJW-4* | This study |
| SL3309 | MG1655 lacI::Tn10 gal P2− P1+gusA (Cmr) galETKM/pJW-4* | This study |
| Phage P1 vir | vir mutations | NIH stock |
| Plasmids | ||
| pJW-4* | pJW15 derivative carrying melR(Con) mutations (Y25D, F53Y, N183I, F191S) that is active in the absence of melibiose; Apr | 18 |
| pKD46 | Temperature-sensitive plasmid containing lambda recombinase inducible by l-arabinose; Apr | 13 |
| pCP20 | Temperature-sensitive plasmid having FLP recombinase capable of recognizing FRT sequence; Apr | 9 |
| pSA934 | gal P2− P1+ | D. Lewis |
| pDL1004 | gal P2+ P1− | D. Lewis |
| pI24 | pBR322 Δ(EcoRI-PvuII) gal+ | S. Adhya |
| pSA809 | pI24 gal P2+ P1−gusA+ | 22 |
| pSA810 | pI24 gal P2− P1+gusA+ | 22 |
| pSL310 | gal P2+ P1+gusA Cmr | This study |
| PSL311 | gal P2+ P1−gusA Cmr | This study |
| pSL312 | gal P2− P1+gusA Cmr | This study |
FIG. 2.
(A) Sequence of the translational fusion of gal promoters (P2+ P1+, P2+ P1−, and P2− P1+) to the gusA reporter gene. (B) Genotype of E. coli SL3304 showing the relevant chromosomal markers, gal promoters fused to gusA at the gus locus, gal promoters directly fused to the galM gene but with the galETK region deleted, lacI::Tn10, and the mel locus. (C) Plasmid pJW-4* having the melR(Con) mutation.
Chromosomal integration.
PCR was performed to amplify the galE-gusA cassettes containing chloramphenicol marker and homologous regions by using pSL310, pSL311, and pSL312 as templates. The DNA products were electroporated into E. coli BW25113 containing plasmid pKD46 in which the lambda recombinase was highly expressed by l-arabinose induction (13). Colonies, which were selected on agar plates containing chloramphenicol (10 μg/ml), were confirmed by chromosomal DNA sequencing and named E. coli SL310, SL311, and SL312, respectively (Table 1).
To make a precise deletion of the galETK segment, kanamycin resistance cassettes and the left-flanking 500 bp of galE and the right-flanking 500 bp of galK were amplified by PCR with chromosomal DNAs of strains JW0742 and JW0740, respectively. The two PCR products were mixed, and PCRs were performed to generate overlapping PCR products which have a kanamycin resistance marker and homologous regions upstream of galE and downstream of galK. The overlapping PCR products were electroporated into strain BW25113, where lambda recombinase was induced by l-arabinose, to make E. coli SL300. The same strategy was employed to make a deletion of the entire galETKM operon. PCR products obtained by using chromosomes of JW0742 and JW0739 as templates were mixed, and overlapping PCR was performed. The final PCR products were electroporated into lambda recombinase-overexpressing strain BW25113 to make strain SL301 (Table 1). All constructions were confirmed by DNA sequencing.
To achieve constitutive expression of β-galactosidase, the lacI gene, which encodes the LacI repressor, was disrupted by a Tn10 insertion in strain MG1655 by P1 transduction of the marker lacI::Tn10. P1 lysates of strains SL310, SL311, and SL312 were next used to transduce the gal P1+P2+, P2+, and P1+ promoters fused to the gusA reporter gene into MG1655 (lacI::Tn10) to make strains SL330, SL331, and SL332, respectively. P1 lysates of strains SL300 and SL301 were used to transduce galETK and galETKM mutant markers into SL330, SL331, and SL332, respectively, to make six strains which were transformed with plasmid pCP20 to eliminate the FRT-flanked kanamycin resistance marker, which enables galM+ to be under the control of the gal promoters (2). Plasmid pCP20 were cured at 42°C. Finally, plasmid pJW-4* having the melR(Con) gene was introduced for constitutive expression of α-d-galactosidase in the six strains to obtain strains SL3304, SL3305, SL3306, SL3307, SL3308, and SL3309.
Enzyme assays.
β-Glucuronidase activities were measured as described previously (22). Cells were grown to log phase in 15-ml tubes containing M63 medium (5 ml) supplemented with 0.3% (wt/vol) disodium succinate, 0.1% (wt/vol) Casamino Acids, and 0.0001% (wt/vol) vitamin B1. At an optical density (OD) at 600 nm of 0.25, as measured with an Ultrospec 3100 (Amersham Biosciences, Piscataway, NJ), d-galactose, lactose, melibiose, phenyl-α-d-galactopyranoside, or phenyl-β-d-galactopyranoside (final concentration, 2 mM) was added. Two hours after inducer addition, ODs were recorded. The activity of β-glucuronidase was measured with the SpetraMAX plus microplate spectrophotometer system (Molecular Devices, Sunnyvale, CA). The rate of 4-nitrophenyl-β-d-glucuronide hydrolysis was determined at 405 nm at 37°C. The rates were divided by cell ODs to obtain specific activities.
Protein purification.
His6-tagged GalR was expressed in BL21(DE3) cells at 18°C with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction at an OD of 0.5 for 16 h. The resulting pellets were lysed with sonication in a buffer containing 20 mM NaPO4 (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 5 mM imidazole. The clarified extract was passed over a 22-ml column of chelating Sepharose resin charged with nickel ion. GalR was eluted over a 5 to 500 mM imidazole gradient. GalR was eluted in the range of 200 to 400 mM as pooled fractions, which were dialyzed against storage buffer (0.6 M KCl, 25 mM Tris HCl [pH 7.5], 1 mM dithiothreitol, 1 mM EDTA, 50% glycerol). Purified GalR was stored at −80°C in 100-μl aliquots.
Nuclear magnetic resonance (NMR) analysis.
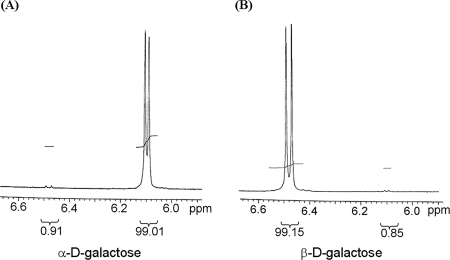
Anomeric composition was quantified by 300-MHz 1H NMR spectrophotometry referenced to dimethyl sulfoxide (DMSO)-d6 at room temperature. The samples of α- and β-d-galactose were dissolved in DMSO. Anomeric hydrogen peak regions (6.0 to 6.6 ppm) in NMR spectra were integrated for anomeric composition.
In vitro transcription assay.
Transcription reactions were carried out as described previously (23). Supercoiled pSA934 (P2− P1+) template DNA (2 nM) was preincubated at 37°C for 5 min in transcription buffer (50 μl) containing 20 mM Tris acetate (pH 7.8), 10 mM magnesium acetate, 200 mM potassium glutamate, 1 mM dithiothreitol, 1 mM ATP, 0.8 U μl−1 rRNasin (Promega), and 20 nM RNA polymerase (USB; specific activity, 2.4 × 103 U mg−1, 1 U μl−1). Inducers (final concentration, 10 mM) were added prior to GalR (200 nM). To initiate the reactions, nucleotides were added to final concentrations of 0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP, and 5 μCi [α-32P]UTP (MP Biomedicals; specific activity, 3,000 μCi mmol−1, 10 μCi μl−1). The reaction mixtures were incubation at 37°C for an additional 10 min, and then the reactions were terminated by the addition of 50 μl of loading dye (90% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). After incubation at 90°C for 2 to 3 min, samples were chilled on ice. Sample aliquots of 10 μl were loaded onto an 8% sequencing gel and electrophoresed at 60 W. The 106- and 108-nucleotide RNAI transcripts were used as internal controls to quantify the relative amount of gal transcripts (38). To compare the kinetics of inducibility of α- and β-d-galactose, supercoiled plasmid pDL1004 (P2+ P1−) (24) was preincubated at 37°C for 5 min in transcription buffer containing GalR (200 nM). Ten minutes after nucleotides were added, inducers (final concentration, 10 mM) dissolved in DMSO were added. Aliquots were terminated at 0 to 300 s by the addition of loading dye.
RESULTS
The gal operon in E. coli can be expressed from either the P1 or the P2 promoter (29). The promoters are repressed by GalR by two different mechanisms. Either (i) GalR represses P1 by inhibiting open complex formation at the promoter-bound RNA polymerase (33) or (ii) P2 is repressed by a DNA loop that makes the promoter inadequate for transcription initiation (10, 11). The presence of d-galactose inactivates GalR to release the repression in each case (25). In the following experiments, we attempted to identify the anomeric configuration (α or β) of d-galactose active as an inducer in vivo and in vitro.
In vivo induction of the gal operon. We constructed gal promoter fusions to a gusA reporter gene to assay the expression of the operon by measuring the level of β-d-glucuronidase activity in the cell (22). We used the wild-type P1+ P2+ promoters, as well as two mutants in which one of the two promoters was mutated (P1+P2− and P1−P2+) (4). In the three fusions used, P1+P2+ gusA, P1+P2− gusA, and P1−P2+ gusA, the gusA open reading frame was fused to the 16th codon of galE, the first cistron of the gal operon. In essence, the promoter of the gusA gene (at 36.4 min) in the E. coli chromosome was replaced with the above three gal promoter variants by recombineering (13) (see Materials and Methods). We constructed two sets of such reporter fusions, one in which the wild-type gal operon at its normal chromosomal location (17 min) was deleted and another in which galETK, but not galM, was deleted. Expression of the lac operon was rendered constitutive by disruption of the lacI gene, which encodes the Lac repressor (12). The strains were also transformed with a plasmid carrying a mutant allele of the melR gene [melR(Con)] that allows constitutive expression of the mel operon (18). The strains generate β-d-galactose and α-d-galactose intracellularly in the presence of lactose and melibiose, respectively. The ability of the intracellularly generated α-d-galactose and β-d-galactose individually to induce the gal operon was studied in vivo by assaying the β-d-glucuronidase level. The absence of the mutarotase gene was expected to prevent the interconversion of the d-galactose anomers intracellularly. Because the activity of water in cytosol is much less than 1.0, the spontaneous mutarotation in the cytosol is much slower than in an aqueous solution (6). The genes that encode enzymes of d-galactose metabolism, GalK, GalT, and GalE, were deleted to prevent the intracellular generation of any d-galactose by the basal level of galactose enzymes (27). The results are shown in Table 2. When the cells were grown in minimal medium containing succinate as a carbon source, the basal levels of gusA activity from the P1P2, P1, or P2 promoter in exponentially growing cells were very low, as expected. The addition of commercial d-galactose, which is a mixture of 28% α-d-galactose and 72% β-d-galactose, resulted in the synthesis of about 6 and 20 U of β-d-glucuronidase activity from the P2 and P1 promoters, respectively. The reason for lower P2 activity compared to P1 activity is most likely that cells grown in succinate medium accumulate a high level of cyclic AMP (cAMP) (5). The cAMP receptor protein-cAMP complex is known to lower P2 promoter activity (1, 8, 29). When both the P1 and P2 promoters were present, approximately 13 U of enzyme activity was made. These results suggest that, under the conditions used, the gal operon is expressed roughly 50% from each promoter when both are active.
TABLE 2.
In vivo induction of the gal operon with a gusA reporter gene
| Inducer |
galETK galM+
|
galETKM
|
||||
|---|---|---|---|---|---|---|
| P1P2 (SL3304)a | P2 (SL3305) | P1 (SL3306) | P1P2 (SL3307) | P2 (SL3308) | P1 (SL3309) | |
| None | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 |
| d-Galactose | 13.2 ± 0.2 (1.00)b | 5.7 ± 0.5 (1.00) | 21.0 ± 1.0 (1.00) | 14.7 ± 0.3 (1.00) | 5.7 ± 0.2 (1.00) | 15.7 ± 0.6 (1.00) |
| d-Lactose | 4.5 ± 0.2 (0.34) | 5.8 ± 0.9 (1.01) | 5.7 ± 0.5 (0.27) | 5.0 ± 0.2 (0.34) | 5.4 ± 0.7 (0.93) | 4.3 ± 1.3 (0.27) |
| d-Melibiose | 7.1 ± 0.7 (0.54) | 6.3 ± 1.6 (1.09) | 11.3 ± 0.9 (0.54) | 8.2 ± 0.2 (0.56) | 5.1 ± 0.4 (0.88) | 9.4 ± 0.8 (0.60) |
| β-PGc | 16.6 ± 0.7 (1.26) | 7.6 ± 0.7 (1.32) | 22.3 ± 0.7 (1.06) | 17.8 ± 2.0 (1.21) | 7.0 ± 0.3 (1.21) | 15.6 ± 1.0 (0.99) |
| α-PGd | 14.1 ± 1.6 (1.07) | 3.7 ± 1.3 (0.65) | 18.3 ± 0.6 (0.87) | 11.4 ± 1.4 (0.77) | 4.1 ± 0.6 (0.71) | 13.9 ± 0.5 (0.88) |
Relevant gal promoters. Shown in parentheses is strain used for GusA assays.
The values shown are means ± standard deviations of GusA specific activity. Shown in parentheses is the relative GusA specific activity with the specific activity of each promoter induced by d-galactose set as 1.
β-PG, phenyl-β-d-galactoside.
α-PG, phenyl-α-d-galactoside.
Because commercial d-galactose, as mentioned, is a mixture of the α anomer and the β anomer and identical results were obtained in both galM+ and galM mutant strains (Table 2), no light is shed on the question of whether one of the two anomers of d-galactose or both are active as inducers. To investigate the issue further, we assayed β-d-glucuronidase levels in strains grown in the presence of d-lactose or d-melibiose. The strains made β-galactosidase (from the lac operon) and α-galactosidase (from the mel operon) constitutively because of the presence of the lacI and melR(Con) mutations, respectively. The cells generated β-d-galactose intracellularly from the hydrolysis of d-lactose by β-galactosidase or α-d-galactose from hydrolysis of d-melibiose by α-galactosidase. The mutarotation of the intracellular d-galactose anomer was prevented by a galM mutation. The results (Table 2) show that both anomers of d-galactose are capable of inducing the gal promoters, although the induction of the P1 promoter was slightly less efficient, particularly when the β anomer was the inducer. Since the results were the same both in the presence and in the absence of the mutarotase enzyme, we conclude that both anomers of d-galactose act as inducers of the gal operon. The hydrolysis of lactose and melibiose also generates glucose. Therefore, the presence of glucose inside the cell may have affected gal induction in an undefined way. Hence, we tested gal inducibility by growing cells in a d-lactose analogue (phenyl-β-galactoside) or a d-melibiose analogue (phenyl-α-d-galactoside) which does not produce glucose after its hydrolysis. The two substrates, after hydrolysis, generate phenol and the corresponding anomer of d-galactose. Phenol is excreted into the culture medium and does not accumulate in the cell. When cells were grown in the presence of one of the two analogues, the results of induction were the same as those obtained with commercial d-galactose in the absence of mutarotase.
These results confirm that both the α- and β-d-galactose anomers can induce the gal promoters in vivo. Incidentally, the level of P1 induction in the presence of lactose or melibiose was lower than in the presence of the corresponding phenyl-d-galactoside derivatives, suggesting that the inhibitory effect on P1 is likely due to the generation of intracellular glucose.
Induction in vitro.
We tested the capabilities of various d-galactose analogues to induce the gal operon in an in vitro transcription system. Our assay system consisted of supercoiled plasmid DNA carrying the gal P1 promoter which, when transcribed, produced gal RNA products of a discrete size (125 bp) because of the presence of a transcription termination signal located downstream of the promoter (23). The results are shown in Fig. 3. In the presence of GalR, about 90% of the transcription from P1 was repressed (lanes 1 and 2). The level of P1 transcription was restored by the presence of d-galactose, which lifted P1 repression completely (lane 3). The inducing abilities of other derivatives of d-galactose on P1 transcription are shown in lanes 4 to 14. Derivatives at the C-1 position of α-d-galactose, methyl-α-d-galactoside (lane 4), phenyl-α-d-galactoside (lane 6), and glucosyl-α-d-galactoside (melibiose) (lane 8), did not show any induction in vitro. Derivatives at the C-1 position of β-d-galactose showed mixed results; the methyl (lane 5) and phenyl (lane 7) derivatives showed partial induction, whereas the glucosyl derivative (lactose) (lane 9) did not induce P1. The two d-galactose metabolic intermediates α-d-galactose-1-phosphate (lane 10) and UDP-α-d-galactose (lane 11) did not show any induction, most likely because these derivatives of the α anomers at the C-1 position inactivate the inducing property of d-galactose, as was observed for the α derivatives above. UDP-α-d-glucose did not affect promoter repression (lane 12). Whereas 6-deoxy-d-galactose (d-fucose) induced transcription very well (lane 13), 2-deoxy-d-galactose induced transcription poorly (lane 14), suggesting that the C-2 position affects the induction process. The C-1 carbon in the latter two sugars is in equilibrium, as in d-galactose, between the α and β positions.
FIG. 3.
In vitro transcription assays of gal (P2− P1+) template DNA in the presence of GalR and various forms of d-galactose. α-MG, methyl-α-d-galactopyranoside; β-MG, methyl-β-d-galactopyranoside; α-PG, phenyl-α-d-galactopyranoside; β-PG, phenyl-β-d-galactopyranoside; 2DG, 2-deoxy-d-galactose.
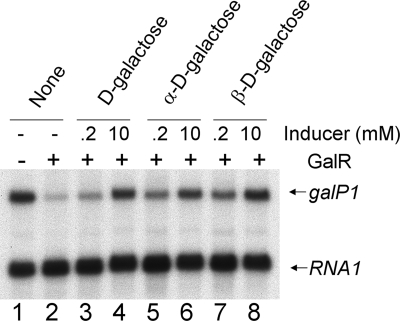
Since the d-galactose used above is a mixture of the α and β anomers, we also tested the inducibility of gal repression in vitro by using the purified α and β anomers of d-galactose. When the pure anomers are dissolved in water, spontaneous mutarotation starts immediately, with a rate of 0.028 min−1 (21). This was prevented by dissolving the optically pure sugars in DMSO instead of water and immediately using them in an in vitro transcription reaction (3). It has been reported that the half-maximal concentration of d-galactose as an inducer is at least 2 mM in in vitro transcription assays (35). Two different concentrations of anomers of d-galactose (0.2 and 10 mM) were tested, and the reactions were carried out for 10 min. We reasoned that the slow spontaneous mutarotation of the pure anomers of d-galactose in the transcription buffer would not influence the induction process because it has been shown that d-galactose mutarotation takes at least several hours to reach equilibrium under aqueous conditions at room temperature (19, 21, 32). The results showed that both anomeric varieties of d-galactose can lift repression well, at least at the higher concentration (Fig. 4).
FIG. 4.
In vitro derepression of the gal P1 promoter by α- and β-d-galactose.
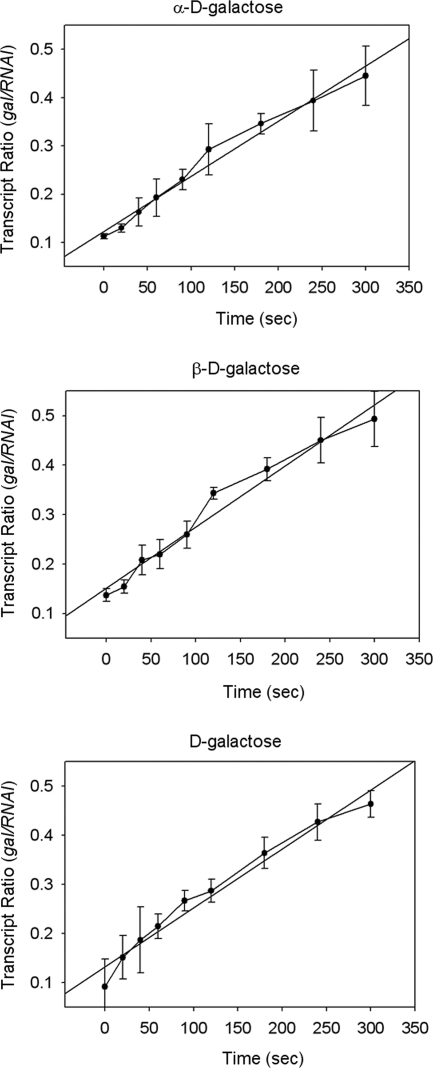
We followed the kinetics of dissociation of GalR from a gal operator in the presence of optically pure anomers of d-galactose. Binding of GalR to OI alone causes a roadblock to transcription elongation from the gal promoter (24). We tested the effect of the galactose anomers on the transcription elongation block by GalR from the P2 promoter kinetically (Fig. 5). In all cases, the elongation block disappeared with equal kinetics. The rates at which gal RNA was made relative to a control RNAI were 0.069 ± 0.013 min−1 for α-d-galactose, 0.074 ± 0.008 min−1 for β-d-galactose, and 0.072 ± 0.014 min−1 for commercial d-galactose. GalR was inactivated by both anomers even within 1 min (Fig. 5). α- and β-d-galactose had 99.01 and 99.15% anomeric purities, respectively (Fig. 6). The rate of spontaneous d-galactose mutarotation is 0.028 min−1. The results demonstrate that both anomers of d-galactose are equally efficient at dissociating GalR from DNA.
FIG. 5.
Kinetics of dissociation of GalR from OI by α- and β-d-galactose.
FIG. 6.
1H NMR spectra of α- and β-d-galactose.
DISCUSSION
Since the sugar d-galactose can be generated intracellularly in either its α-anomeric or its β-anomeric form, the synthesis of the enzymes of d-galactose metabolism, including the mutarotase that interconverts them, should be available for specific incorporation of each anomer into complex carbohydrates. The expression of the gal operon, which encodes the d-galactose-metabolizing enzymes, is repressed by binding of GalR to its multipartite DNA binding sites in the operon (14, 16, 17). d-Galactose induces transcription of the operon by binding and inactivating the GalR protein (25). By studying the binding to GalR of d-galactose derivatives (methylated at the C-1 position) by fluorescence spectroscopy, it was shown previously that only methyl-β-d-galactose, and not the corresponding α derivative, binds to GalR (7). From these studies, the authors suggested that the β form of d-galactose is the actual inducer (7). Since α- or β-d-galactose can be generated exclusively within the cell by hydrolysis of a disaccharide, we propose that each anomer should be capable of inactivating GalR and inducing the operon to make enzymes of mutarotation and d-galactose metabolism. We attempted to identify whether one or both anomers of d-galactose can derepress gal transcription in vivo and in vitro.
Our in vivo results showed that both α- and β-d-galactose can induce gal transcription in vivo. The results were identical in galM+ and galM mutant cells. This conclusion was further confirmed by assaying the gal induction level in the above-mentioned cells grown in phenyl-α-d-galactoside or phenyl-β-galactoside, which generates α- and β-d-galactose and phenol, respectively. The two galactoside derivatives induced the gal operon with equal efficiency. Both promoters of the operon were inducible. As expected from in vivo results, both α- and β-d-galactose were able to inhibit GalR-mediated repression of P1. Both anomers inactivated the repressor efficiently and with equal kinetics. Interestingly, substitution at C-1 in the α configuration completely inactivated the ability of the sugar to remove repression whereas substitution in the β configuration showed about 80% reduction of the inducing capacity of the sugar. These results agree with the findings of Brown et al. (7), who showed that GalR binds to methyl-β-d-galactoside but not to methyl-α-galactoside. However, the affinity of the β anomer to GalR relative to free d-galactose is not known. We assume that, compared to free d-galactose, the affinity of the methyl-β-d-galactoside is much less and the ability of a galactose-derivative to derepress transcription reflects its affinity for GalR. Substitution at the C-1 position in the α configuration eliminates its binding affinity, while the effect of substitution in the β configuration depends on the nature of the substitution; the methyl or phenyl derivative retains some binding ability, while the glucosyl derivative completely eliminates it. Incidentally, d-galactose metabolic intermediates, e.g., α-d-galactose-1-phosphate, UDP-α-d-galactose, and UDP-α-d-glucose fail to act as inducers. These results are expected, since both galactose-1-phosphate and UDP-galactose have substitutions at the C-1 position in the α configuration.
In nature, E. coli encounters α- or β-galactosides and generates only one of the two optical anomers of d-galactose. Since both anomers are needed for various biosynthetic reactions, intracellularly generated α- or β-d-galactose must be able to induce the gal regulon members, including the gal operon, to make mutarotase to convert one to the others. Consistently, our results show that this is indeed the case.
Acknowledgments
We thank S. Busby for providing plasmid pJW-4* and B. Wanner for providing pKD46 and pCP20. We are also grateful to the Keio Collection for mutant strains. We thank M. Yarmolinsky for critical reading of the manuscript. We thank our colleagues in the laboratory for discussions and technical help.
This research was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, the Center for Cancer Research.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Adhya, S., and W. Miller. 1979. Modulation of the two promoters of the galactose operon of Escherichia coli. Nature 279492-494. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol. Syst. Biol. 22006.2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballash, N. M., and E. B. Robertson. 1973. The mutarotation of glucose in dimethylsulfoxide and water mixtures. Can. J. Chem. 51556-564. [Google Scholar]
- 4.Bingham, A. H., S. Ponnambalam, B. Chan, and S. Busby. 1986. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene 4167-74. [DOI] [PubMed] [Google Scholar]
- 5.Botsford, J. L., and M. Drexler. 1978. The cyclic 3′,5′-adenosine monophosphate receptor protein and regulation of cyclic 3′,5′-adenosine monophosphate synthesis in Escherichia coli. Mol. Gen. Genet. 16547-56. [DOI] [PubMed] [Google Scholar]
- 6.Bouffard, G. G., K. E. Rudd, and S. L. Adhya. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244269-278. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. P., N. Shaikh, M. Brenowitz, and L. Brand. 1994. The allosteric interaction between d-galactose and the Escherichia coli galactose repressor protein. J. Biol. Chem. 26912600-12605. [PubMed] [Google Scholar]
- 8.Busby, S., H. Aiba, and B. de Crombrugghe. 1982. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J. Mol. Biol. 154211-227. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 10.Choy, H. E., R. R. Hanger, T. Aki, M. Mahoney, K. Murakami, A. Ishihama, and S. Adhya. 1997. Repression and activation of promoter-bound RNA polymerase activity by Gal repressor. J. Mol. Biol. 272293-300. [DOI] [PubMed] [Google Scholar]
- 11.Choy, H. E., S. W. Park, P. Parrack, and S. Adhya. 1995. Transcription regulation by inflexibility of promoter DNA in a looped complex. Proc. Natl. Acad. Sci. USA 927327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulondre, C., and J. H. Miller. 1977. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 117525-567. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz, H. J., H. Bicknase, B. Gleumes, C. Heibach, S. Rosahl, and R. Ehring. 1983. Characterization of two mutations in the Escherichia coli galE gene inactivating the second galactose operator and comparative studies of repressor binding. EMBO J. 22129-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden, H. M., I. Rayment, and J. B. Thoden. 2003. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 27843885-43888. [DOI] [PubMed] [Google Scholar]
- 16.Irani, M., L. Orosz, S. Busby, T. Taniguchi, and S. Adhya. 1983. Cyclic AMP-dependent constitutive expression of gal operon: use of repressor titration to isolate operator mutations. Proc. Natl. Acad. Sci. USA 804775-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irani, M. H., L. Orosz, and S. Adhya. 1983. A control element within a structural gene: the gal operon of Escherichia coli. Cell 32783-788. [DOI] [PubMed] [Google Scholar]
- 18.Kahramanoglou, C., C. L. Webster, M. S. El-Robh, T. A. Belyaeva, and S. J. Busby. 2006. Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon. J. Bacteriol. 1883199-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaffa, L., E. Fekete, C. Gamauf, A. Szentirmai, C. P. Kubicek, and B. Seiboth. 2006. d-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 1521507-1514. [DOI] [PubMed] [Google Scholar]
- 20.Leloir, L. F. 1951. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. 33186-190. [DOI] [PubMed] [Google Scholar]
- 21.Levy, G. B., and E. S. Cook. 1954. A rotographic study of mutarotase. Biochem. J. 5750-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, D. E., M. Geanacopoulos, and S. Adhya. 1999. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 31451-461. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, D. E. A. 2003. Identification of promoters of Escherichia coli and phage in the transcription section plasmid pSA850. Methods Enzymol. 370618-645. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, D. E. A., N. Komissarova, P. Le, M. Kashlev, and S. Adhya. 2008. DNA sequences in gal operon override transcription elongation blocks. J. Mol. Biol. 382843-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar, A., S. Rudikoff, and S. Adhya. 1987. Purification and properties of Gal repressor:pL-galR fusion in pKC31 plasmid vector. J. Biol. Chem. 2622326-2331. [PubMed] [Google Scholar]
- 26.Mandal, N., W. Su, R. Haber, S. Adhya, and H. Echols. 1990. DNA looping in cellular repression of transcription of the galactose operon. Genes Dev. 4410-418. [DOI] [PubMed] [Google Scholar]
- 27.Miller, Z., H. E. Varmus, J. S. Parks, R. L. Perlman, and I. Pastan. 1971. Regulation of gal messenger ribonucleic acid synthesis in Escherichia coli by 3′,5′-cyclic adenosine monophosphate. J. Biol. Chem. 2462898-2903. [PubMed] [Google Scholar]
- 28.Morell, P., and N. S. Radin. 1969. Synthesis of cerebroside by brain from uridine diphosphate galactose and ceramide containing hydroxy fatty acid. Biochemistry 8506-512. [DOI] [PubMed] [Google Scholar]
- 29.Musso, R. E., R. Di Lauro, S. Adhya, and B. de Crombrugghe. 1977. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell 12847-854. [DOI] [PubMed] [Google Scholar]
- 30.Pai, T., Q. Chen, Y. Zhang, R. Zolfaghari, and A. C. Ross. 2007. Galactomutarotase and other galactose-related genes are rapidly induced by retinoic acid in human myeloid cells. Biochemistry 4615198-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, D., K. S. Ryu, D. Choi, J. Kwak, and C. Park. 2007. Characterization and role of fucose mutarotase in mammalian cells. Glycobiology 17955-962. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson, H., and G. Pettersson. 2001. Kinetics of the coupled reaction catalysed by a fusion protein of β-galactosidase and galactose dehydrogenase. Biochim. Biophys. Acta 1549155-160. [DOI] [PubMed] [Google Scholar]
- 33.Roy, S., S. Semsey, M. Liu, G. N. Gussin, and S. Adhya. 2004. GalR represses galP1 by inhibiting the rate-determining open complex formation through RNA polymerase contact: a GalR negative control mutant. J. Mol. Biol. 344609-618. [DOI] [PubMed] [Google Scholar]
- 34.Ryu, K. S., C. Kim, I. Kim, S. Yoo, B. S. Choi, and C. Park. 2004. NMR application probes a novel and ubiquitous family of enzymes that alter monosaccharide configuration. J. Biol. Chem. 27925544-25548. [DOI] [PubMed] [Google Scholar]
- 35.Semsey, S., K. Virnik, and S. Adhya. 2006. Three-stage regulation of the amphibolic gal operon: from repressosome to GalR-free DNA. J. Mol. Biol. 358355-363. [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick, B. 1961. Crystallographic examination of the alpha- and beta-anomers of d-galactose. J. Chem. Soc., p. 3157-3158.
- 37.Thoden, J. B., J. Kim, F. M. Raushel, and H. M. Holden. 2003. The catalytic mechanism of galactose mutarotase. Protein Sci. 121051-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomizawa, J., T. Itoh, G. Selzer, and T. Som. 1981. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. USA 781421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]