Abstract
In the facultatively phototrophic proteobacterium Rhodobacter sphaeroides, formation of the photosynthetic apparatus is oxygen dependent. When oxygen tension decreases, the response regulator PrrA of the global two-component PrrBA system is believed to directly activate transcription of the puf, puh, and puc operons, encoding structural proteins of the photosynthetic complexes, and to indirectly upregulate the photopigment biosynthesis genes bch and crt. Decreased oxygen also results in inactivation of the photosynthesis-specific repressor PpsR, bringing about derepression of the puc, bch, and crt operons. We uncovered a hierarchical relationship between these two regulatory systems, earlier thought to function independently. We also more accurately assessed the spectrum of gene targets of the PrrBA system. First, expression of the appA gene, encoding the PpsR antirepressor, is PrrA dependent, which establishes one level of hierarchical dominance of the PrrBA system over AppA-PpsR. Second, restoration of the appA transcript to the wild-type level is insufficient for rescuing phototrophic growth impairment of the prrA mutant, whereas inactivation of ppsR is sufficient. This suggests that in addition to controlling appA transcription, PrrA affects the activity of the AppA-PpsR system via an as yet unidentified mechanism(s). Third, PrrA directly activates several bch and crt genes, traditionally considered to be the PpsR targets. Therefore, in R. sphaeroides, the global PrrBA system regulates photosynthesis gene expression (i) by rigorous control over the photosynthesis-specific AppA-PpsR regulatory system and (ii) by extensive direct transcription activation of genes encoding structural proteins of photosynthetic complexes as well as genes encoding photopigment biosynthesis enzymes.
In the facultatively phototrophic alphaproteobacterium Rhodobacter sphaeroides, photosynthesis (PS) operates under anoxic conditions. A decrease in oxygen tension triggers significant upregulation of PS gene transcription (35, 38). The photosynthetic apparatus is comprised of the reaction center, encoded by the puh and puf operons, and two light-harvesting complexes, encoded by the puf and puc operons. Enzymes involved in the biosynthesis of photosynthetic pigments, i.e., bacteriochlorophyll a and carotenoids, are encoded by the bch and crt genes, respectively. Most PS-specific genes are located in the R. sphaeroides PS gene cluster, whereas the puc operons are located separately (4, 49). Three major regulatory systems control oxygen-dependent transcription of PS genes. One of these is composed of the antirepressor AppA (15, 17) and the repressor PpsR (7, 36) and is primarily responsible for the regulation of PS genes (30). Two other systems are global regulatory systems, i.e., the redox-responsive two-component system PrrBA (2, 47) and the anaerobic activator FnrL (48). Several additional factors are involved in PS gene expression (12, 46), but their roles are more limited and less precisely defined (reviewed in references 27 and 47).
The R. sphaeroides PpsR protein is a master repressor of PS genes (30). Inactivation of the ppsR gene is sufficient to turn on PS gene expression and formation of the photosynthetic apparatus even at a high oxygen concentration, whereas ppsR overexpression is sufficient to block PS development even in the absence of oxygen. PpsR directly represses transcription of most bch and crt genes, the puc operons, and the hemC and hemE genes involved in the early steps of tetrapyrrole biosynthesis (14, 30). The upstream regions of these genes contain two PpsR binding sites, TGTcN10gACA. PpsR also indirectly affects expression of the puh and puf operons by an as yet poorly understood mechanism(s) (30).
The antirepressor protein AppA binds PpsR and conveys oxygen and light signals to the repressor. Genetic inactivation of appA results in phototrophically impaired cells that fail to derepress PS genes in response to oxygen deprivation, which is similar to PpsR overexpression (17). AppA is a unique dual sensor of oxygen and light (3, 20, 29), sensing oxygen via its SCHIC (sensor containing heme instead of cobalamin) domain (31) and light via the BLUF (sensor of blue light using flavin adenine dinucleotide) domain (13). Both oxygen and light are anticipated to disrupt the AppA-PpsR interaction and thus result in increased DNA binding by PpsR (29, 31). However, until now, only light has been shown to disrupt AppA-PpsR heteromers in vitro (29).
The PrrBA two-component regulatory system (9, 10) (also known as RegBA) has a global range of targets that includes PS genes as well as genes involved in nitrogen, hydrogen, sulfur, and carbon metabolism, heme biosynthesis, respiration, and other processes (8, 11). The histidine kinase PrrB is believed to sense redox poise of the cell by monitoring electron flow to the cytochrome c oxidase cbb3 (34). It is controversial at this point, however, whether or not PrrB monitors electron flow via a ratio of oxidized/reduced membrane quinones (23, 43). Under conditions of low oxygen tension, when the electron flow is reduced, the kinase activity of PrrB is stimulated, which increases phosphorylation of the response regulator PrrA. PrrA is a transcription factor which binds DNA as a dimer and activates or represses gene expression. The phosphorylated form of PrrA, PrrA∼P, has increased DNA binding capacity (25, 37).
The prrA null mutant is incapable of phototrophic growth (9). Interestingly, phototrophic growth of this mutant can be rescued by inactivating ppsR (30). This suggests that the photosynthesis growth defect of the prrA mutant stems primarily from its inability to release repression imposed by PpsR. Therefore, PrrA must control either the expression or activity of the components of the AppA-PpsR system (30).
In this study, we began to test this possibility. We found that PrrA controls the AppA-PpsR system on two levels, by affecting appA (but not ppsR) transcription and by posttranscriptional regulation. In addition, we learned that PrrA directly activates the transcription of numerous bch and crt genes, which have been considered exclusive targets of PpsR (7). The hierarchical relationship between the major regulatory systems and extensive overlaps in their gene targets expose a previously unanticipated complexity in PS gene regulation in R. sphaeroides and possibly other anoxygenic phototrophic bacteria.
(The preliminary findings reported here were presented at the 105th General Meeting of the American Society for Microbiology, Atlanta, GA, 2005 [12a].)
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. R. sphaeroides strains were grown at 30°C in Sistrom's minimal medium A with succinate as a carbon source (5). Unless specified otherwise, 60-ml cultures were grown in 100-ml glass culture tubes under continuous vigorous sparging with the following defined gas mixtures: 20% O2, 79% N2, and 1% CO2 (high oxygen) and 0.5% O2, 98.5% N2, and 1% CO2 (low oxygen). Phototrophic cultures were grown in completely filled, tightly capped 13-ml tubes exposed to white light at 30 W m−2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| R. sphaeroides strains | ||
| 2.4.1 | Wild type | Laboratory collection |
| APP11 | 2.4.1 (appA)::Tpr | 15 |
| PRRBCA2 | 2.4.1 (prrBAC)::Tpr | 33 |
| RPS1 | PRRBCA2 ppsR::Kmr | 30 |
| P. denitrificans strain | ||
| ATCC 17741 | Wild type | American Type Culture Collection |
| E. coli strains | ||
| DH5α phe | Strain used for cloning | 9 |
| S17-1 | Tra+ strain used for plasmid mobilization | 41 |
| Plasmids | ||
| pRK415 | IncP Tcr; vector | 22 |
| pECapp | pRK415::PdorC::appA | This study |
| p484-Nco1 | pRK415::appA (native PappA) | 15 |
| pRK415prrA | pRK415::prrA | This study |
| pSmNs | pRK415::ppsR | 14 |
| pSmNsABC | pSmNs::prrBAC | This study |
| pLXapp | IncQ Smr SprappA::lacZYA | This study |
| pCF200Km | IncQ Smr SprpucB1::lacZYA | 26 |
| pLX14 | IncQ Smr SprbchF::lacZYA | 17 |
| pUI2711 | IncQ Smr SprcrtA::lacZYA | 45 |
| pUC19spf′ | Vector for in vitro transcription | 1 |
| pUC19spf::bchC | pUC19spf′ plus upstream region of bchC | This study |
| pUC19spf::bchE | pUC19spf′ plus upstream region of bchE | This study |
| pUC19spf::bchF | pUC19spf′ plus upstream region of bchF | This study |
| pUC19spf::crtA | pUC19spf′ plus upstream region of crtA | This study |
| pJC412 | pUC19spf′ plus upstream region of pucB1 | 1 |
| pRKK146 | pUC19spf′ plus upstream region of cycA P2 promoter | 22 |
Paracoccus denitrificans ATCC 17741 was grown in the same medium and at the same temperature as R. sphaeroides. Ten-milliliter cultures were grown in 125-ml flasks on a rotating shaker. Antibiotics were used for R. sphaeroides and P. denitrificans, where necessary, at the following concentrations: tetracycline, 1 μg ml−1; and streptomycin and spectinomycin, 50 μg ml−1 each.
Escherichia coli strains were grown at 30°C in LB medium supplemented with the following antibiotics: ampicillin, 100 μg ml−1; tetracycline, 10 μg ml−1; and streptomycin and spectinomycin, 50 μg ml−1 each for strain S17-1 and 25 μg ml−1 each for strain DH5α.
Isolation of spontaneous phototrophically competent suppressors of the prrA null mutant.
Several independent cultures of the prrA null mutant, PRRBCA2 (Table 1), were grown under low oxygen. Approximately 5 × 108 cells from each independent culture were transferred to 13-ml tubes, which were completely filled with new medium, tightly capped, and exposed to light. After 7 to 10 days, when visible phototrophic growth was observed, the tubes were opened and aliquots were plated on Sistrom's medium plates. Single colonies from each culture were subsequently streak purified, and their phototrophic competence was verified. The ppsR genes from the colonies were PCR amplified using high-fidelity Hotstart Pfu DNA polymerase (Stratagene) and sequenced with ppsR-specific primers at the University of Wyoming NAE facility.
Plasmid construction.
Plasmid pLXapp containing an appA-lacZ transcriptional fusion was constructed as follows. The 0.28-kb upstream region of appA was PCR amplified from the R. sphaeroides genomic DNA by use of appA-specific primers APP-P (5′-AACTGCAGTGCTGTCAACCATCGTG) and APPA-Xb (5′-GCTCTAGAGTGTTGCATCCTTCGCC) (underlined sequences were added). The amplified fragment was digested with PstI and XbaI and cloned into vector pLX1 (16) upstream of the promoterless lacZ gene.
The pECapp plasmid was constructed by cloning the appA gene in vector pRK415 (Table 1) downstream of the dorC promoter, which in turn was PCR amplified as a 0.25-kb fragment from pNMT68 (32).
Plasmids for in vitro transcription assays were constructed by cloning the upstream regions of the desired genes into the PstI-XbaI or HindIII-XbaI sites of vector pUC19spf′ upstream of a strong transcription terminator. The following primers were used for gene amplification (sequences added for cloning purposes are underlined): for pUC19spf:bchC, primers BCHCup-H (5′-CAGAAGCTTCTTCTTTTCCGAGATCAAGATC) and BCHCup-Xb (5′-GCTCTAGACGGCGGTCGTTCTCACAG); for pUC19spf:bchE, primers BCHEup-H (5′-CAGAAGCTTGCTCGATCGCCGATTTCCTC) and BCHEup-Xb (5′-GCTCTAGAGTGAACGAATACGATACGC); for pUC19spf:bchF, primers BCHF-H (5′-CCCAAGCTTGGAGGAACGCATGTCAAAG) and BCHF-Xb (5′-GCTCTAGATCTTGAGGTTCGCCTTC); and for pUC19spf:crtA, primers CRTA-H (5′-CCCAAGCTTCTGCAGTGCTTGTCAAC) and CRTA-Xb (5′-GCTCTAGATGTCATTCCTCCTGCAG).
Plasmid mobilization into R. sphaeroides and P. denitrificans was performed using E. coli S17-1 as a donor, as described earlier (14).
RNA extraction and qPCR.
RNAs were extracted from exponentially grown R. sphaeroides cultures at an A600 of 0.18 ± 0.02, using an RNeasy Midi kit (Qiagen) according to a previously described protocol (12, 35). cDNA was prepared from total RNA by use of SuperScript II (Stratagene) and random hexamers as described earlier (12, 35). cDNA samples were used for quantitative reverse transcription-mediated real-time PCR (qPCR). The iCycler iQ real-time PCR detection system (Bio-Rad) with SYBR green chemistry was used to monitor amplification and to quantify PCR products. The primers and conditions used for probe amplification were described earlier; expression levels of the rpoZ gene were used for normalization (12). Each qPCR reaction was performed at least in triplicate; average data from two to four biological replicates are reported.
Spectroscopy of photosynthetic complexes.
R. sphaeroides strains were grown at low oxygen to an A600 of 0.3 ± 0.05, pelleted, resuspended in 10 mM phosphate buffer (pH 7.2), and passed through a French pressure cell, followed by removal of cell debris. Soluble cell extracts were adjusted to identical A660 values (which correspond to absorption that is independent of photosynthetic pigment content), and UV-visible spectra were recorded using a Shimadzu UV-1601PC spectrophotometer as previously described (15).
β-Galactosidase assays.
Assays of β-galactosidase activity in P. denitrificans were performed using o-nitrophenyl-β-d-galactopyranoside (ONPG) on cells permeabilized with chloroform and sodium dodecyl sulfate as described earlier (14).
In vitro transcription assays.
In vitro transcription experiments were performed as previously described (1). Briefly, crude R. sphaeroides RNA polymerase (RNA Pol) preparations were used to drive transcription from the circular plasmid DNA templates based on pUC19spf′ in the presence of 32P-labeled nucleotides. The transcripts were analyzed following their separation by polyacrylamide gel electrophoresis. The R. sphaeroides PrrA protein was purified and phosphorylated in vitro by use of acetyl phosphate to obtain PrrA∼P as described earlier (6). Various concentrations of PrrA or PrrA∼P were added to in vitro transcription reaction mixtures. Transcript levels were calculated based on radiation intensities of the resulting transcripts measured by phosphorimaging. All in vitro transcripts were normalized to the amount of RNA1 transcript in each reaction mix, which is PrrA independent (1).
Primer extension and manual DNA sequencing.
The gene-specific primers designed for cloning into pUC19spf′ were used for primer extension, which was performed essentially as described elsewhere (39). Briefly, R. sphaeroides RNA was annealed with individual gene primers and extended in the presence of deoxynucleotides, including [32P]dATP, using the reverse transcriptase Superscript II at 45°C following the manufacturer's protocol (Stratagene).
DNA sequencing was performed using a Thermo Sequenase cycle sequencing kit (USB Corp.) according to the specifications of the manufacturer, employing the same gene-specific primers and [32P]dATP. The products of primer extension and sequencing reactions were resolved using denaturing polyacrylamide gel electrophoresis as described elsewhere (39).
RESULTS
Inactivation of ppsR is sufficient for restoring phototrophic growth of the prrA null mutant.
The premise of this study, i.e., that the global PrrBA system affects the activity of the AppA-PpsR system, is based on our earlier observation that inactivation of the ppsR gene restores phototrophic growth of the prrA mutant (30). However, the constructed prrA ppsR double mutant, RPS1 (Table 1), grows slowly and requires relatively high light intensity, which necessitates the use of liquid, as opposed to agar, medium and makes it difficult to ensure strain clonality (30). Therefore, we could not exclude the possibility that an additional spontaneous mutation accumulated in the constructed double mutant that contributed to its phototrophic capacity. To test whether ppsR inactivation is sufficient for restoring phototrophic growth of the prrA mutant, we decided to select spontaneous phototrophically competent prrA suppressor mutants. We reasoned that if such suppressor mutations can be isolated and proven to reside in ppsR, this would establish that ppsR inactivation is sufficient. If suppressor mutations were located elsewhere, we would identify new factors mediating interactions between the PrrBA and AppA-PpsR regulatory systems.
We placed the prrA mutant strain PRRBCA2 under anaerobic conditions in front of light and observed phototrophic growth following a 5- to 7-day incubation. We analyzed six isolates capable of phototrophic growth from independent PRRBCA2 cultures. Each contained a mutation in ppsR. A total of four different point mutations and one insertion were identified (Table 2). The insertion resulted in a frameshift in the ppsR gene, thus resulting in a truncated protein. All point mutations were T→C transversions resulting in Leu→Pro substitutions. The Pro residues are expected to impair secondary structures and to interfere with PpsR tetramerization and/or DNA binding (19). For example, the 459L→P mutation, located in the helix-turn-helix motif of PpsR, is likely to prevent DNA binding. The 248L→P mutation has been isolated previously (19) and shown to inactivate PpsR.
TABLE 2.
Spontaneous suppressor mutations in ppsR rescuing phototrophic growth of the prrA null mutant, PRRBCA2a
| Nucleotide mutation | Corresponding amino acid mutation |
|---|---|
| 311T→C | 104L→P |
| 524T→C | 175L→P |
| 743T→C | 248L→P |
| 1376T→C | 459L→P |
| 723CCGCGTGTCTT (insertion) | Frameshift |
Numbers correspond to the nucleotide and amino acid sequences of ppsR (GenBank accession number L37197).
Our ability to readily isolate suppressor mutations in ppsR confirms that ppsR inactivation is sufficient for rescuing phototrophic growth of the prrA null mutant and that the phototrophic growth impairment of this mutant stems primarily from its inability to release repression of the PpsR regulon (30). Therefore, PrrA must control, directly or indirectly, the expression and/or activity of the components of the AppA-PpsR system.
PrrA is required for appA expression.
To test whether PrrA affects the expression of the appA or ppsR gene, we compared appA and ppsR transcript levels (measured by qPCR) in the prrA mutant, PRRBCA2, with those in the wild type. We found that mRNA levels of ppsR were unchanged (not shown), whereas mRNA levels of appA were decreased in PRRBCA2, to approximately 13% of the wild-type level [Fig. 1A, compare PRRBCA2(pRK415) and 2.4.1(pRK415)]. It is worth mentioning that a decreased appA mRNA level in the prrA null mutant was recently independently observed by DNA microarray analysis (11).
FIG. 1.
Relative transcript abundance measured by qPCR. (A) Restoration of the appA transcript in the prrA null mutant, PRRBCA2, grown under low-oxygen conditions. The expression of appA in PRRBCA2 is assigned to 1 arbitrary unit. (B) Effect of appA level on transcript abundance of PS genes measured in cultures grown under low-oxygen conditions. Expression values are averages from three to four independent experiments, for each of which qPCR was done in triplicate. Error bars represent standard deviations.
The greatly decreased appA transcript level would prevent AppA from inactivating the PpsR repressor and thus make the prrA mutant in effect a double PrrA− AppA− mutant. The PpsR regulon in such a mutant must be repressed even under low- or no-oxygen conditions, which could explain the low expression of the PpsR-dependent bch and crt genes. Since we earlier showed that PpsR overexpression is sufficient to prevent phototrophic growth (14), we assumed that a decreased appA transcript level was responsible for the phototrophic growth defect of the prrA mutant.
PrrA activates appA transcription indirectly.
To determine whether PrrA affects appA gene expression directly or indirectly, we tested the effect of PrrA on appA transcription in vitro, using R. sphaeroides RNA Pol in the presence or absence of PrrA or its phosphorylated form, PrrA∼P. However, the level of appA transcription in vitro was below the level of detection (not shown).
We further tested the effect of PrrA on appA transcription in the heterologous host, Paracoccus denitrificans, which is a close relative of R. sphaeroides that lacks PS genes and PS-specific regulators. We and others have successfully used this bacterium as a model to investigate the functionality of the transcription factors involved in PS gene expression (12, 14, 17, 36). We showed earlier that the R. sphaeroides prrA gene expressed in P. denitrificans efficiently activates the expression of PrrA-dependent genes (16).
To explore whether PrrA affects appA gene transcription directly, we constructed an appA-lacZ transcriptional fusion (plasmid pLXapp) (Table 1) and introduced it into P. denitrificans harboring either the vector pRK415 or pRK415prrA (Table 1), expressing the R. sphaeroides prrA gene from its native promoter as well as from the lac promoter. We observed no significant difference in appA-lacZ expression between the two strains (not shown). This suggests that PrrA does not activate appA transcription directly. Therefore, an additional regulatory factor must exist in R. sphaeroides that mediates the effect of PrrA on appA transcription. This as yet unidentified factor is expected to be an important link between the PrrBA and AppA-PpsR regulatory pathways.
Restoration of appA transcript levels in the prrA mutant.
To test the hypothesis that low appA expression is responsible for the phototrophic growth defect of the prrA mutant, we brought the level of the appA transcript in the prrA mutant to the wild-type level by expressing appA from a plasmid. To do this, we placed appA downstream of several promoters of various strengths in a four- to six-copy vector, pRK415 (Table 1). The appA-expressing plasmids were transferred into strain PRRBCA2, and the appA mRNA levels in these strains were measured by qPCR. mRNA levels of appA expressed under the control of its native promoter, plasmid p484-Nco1, were approximately 3.5-fold higher than needed [Fig. 1A, compare PRBCA2(p484-Nco1) and 2.4.1(pRK415)]. Fortuitously, one of the tested plasmids, pECapp, restored the level of appA transcript to that of the wild type, under both high (not shown)- and low-oxygen conditions [Fig. 1A, compare PRBCA2(pECapp) and 2.4.1(pRK415)].
We tested the expression of selected PS genes in PRRBCA2(pECapp) and PRRBCA2(pRK415) under low-oxygen conditions, anticipating that the wild-type level of the appA transcript in the prrA mutant would restore expression of the crt and bch genes to the wild-type levels (30). We also anticipated that expression of pucB1 would be restored only partially, because pucB1 is under dual regulation, i.e., repressed by PpsR as well as activated by PrrA (9, 14). Expression of pucB1 in strain PRRBCA2(pECapp) was indeed restored only partially (Fig. 1B), whereas expression of bchF and bchC was restored to close to the wild-type levels, or somewhat higher.
However, in contrast to our expectations, expression of the bchE and crtA genes in PRRBCA2(pECapp) was not fully restored and remained significantly below the wild-type levels (Fig. 1B). Several possibilities may account for incomplete restoration of the bchE and crtA transcript levels in PRRBCA2(pECapp). (i) PrrA may activate expression of these genes not only through the AppA-PpsR pathway but also directly. (ii) The AppA-PpsR pathway in PRRBCA2(pECapp) may not be fully functional in the prrA mutant, despite wild-type levels of appA mRNA, because in addition to regulating appA transcription, PrrA may affect the activity of the AppA-PpsR system at a posttranscriptional level. Below we show that both possibilities appear to be true.
PrrA directly activates transcription of crt and bch genes in vitro.
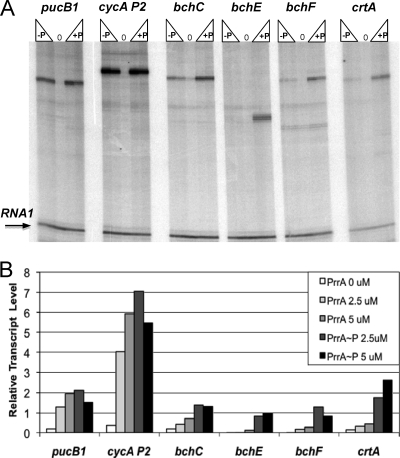
To test the possibility that PrrA directly activates transcription of bchE and crtA, we used in vitro transcription assays. To this end, we employed the R. sphaeroides RNA Pol (1) in the absence or presence of PrrA or PrrA∼P. As templates for in vitro transcription, we used pUC19spf′-derived plasmids containing upstream regions of selected bch and crt genes placed upstream of a strong transcription terminator (Table 1). pUC19spf′ contains the RNA1 gene, whose transcription is PrrA independent, which allowed us to normalize the amounts of generated bch and crt transcripts to the amount of the RNA1 transcript generated in each assay. As positive controls, we used known PrrA-dependent promoters, cycA P2 and pucB1 (1, 21) (Fig. 2A).
FIG. 2.
Transcription activation of the crt and bch genes by PrrA in vitro. (A) Typical autoradiographs of transcripts generated in vitro by the R. sphaeroides RNA Pol holoenzyme from the pUC19spf-derived templates in the absence (0) or presence of PrrA (−P) or PrrA∼P (+P), at concentrations of 2.5 and 5 μM. PrrA-dependent transcription of the cycA P2 (plasmid pRKK146) and pucB1 (plasmid pJC412) promoters was used as a positive control. (B) Relative transcript abundance. The RNA1 transcript generated in each reaction was used to normalize the transcript abundance in each reaction mix.
We found that unphosphorylated PrrA and, more so, PrrA∼P activated transcription of bchE and crtA (Fig. 2A). This is consistent with the notion that PrrA directly activates transcription of these genes in vivo. We extended our analysis to the bchF and bchC genes. Surprisingly, we found that PrrA activated transcription of these genes in vitro as well (Fig. 2A). The quantification of PrrA activation revealed that the extent of activation was high for all tested genes (Fig. 2B). This suggests that many bch and crt genes are likely to be activated directly by PrrA in vivo.
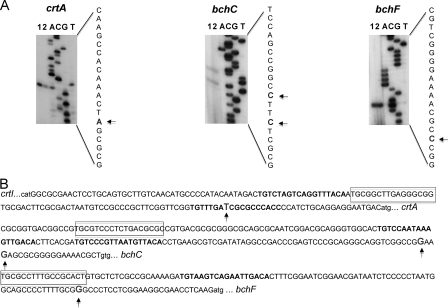
The sizes of the crtA and bch transcripts observed in the in vitro transcription assays were in agreement with what was expected based on the assumption that the promoters of these genes are located some 15 to 50 nucleotides (nt) upstream of their translational start codons (Fig. 2A). To verify that the transcripts obtained in vitro corresponded to the transcripts in vivo, we determined the sequences of the 5′ ends of the major crtA, bchC, and bchF transcripts (Fig. 3A). mRNAs were purified from the wild-type strain grown at low oxygen tension, when expression of the crt and bch genes is high, and used in primer extension experiments. For each tested gene, one or two closely located major transcription start sites were identified (Fig. 3A) that corresponded to the transcript sizes seen by in vitro transcription assays. The observed 5′ ends were consistent with the transcriptional activation by PrrA observed in the in vitro transcription assays.
FIG. 3.
(A) Transcription start sites for the PrrA-activated crtA, bchC, and bchF genes. Primers used for cloning into pUC19spf′ were used for extension as described in Materials and Methods. Lanes 1 and 2, primer extension reactions; lanes A, C, G, and T, nucleotides corresponding to sequencing of the pUC19spf′-based templates. Arrows show transcription start sites. (B) Upstream regions of the crtA, bchC, and bchF genes. Putative PrrA binding sites are boxed, residues corresponding to the PrrA consensus sequence (24) are shown in gray, the PpsR binding sites (30) are shown in bold, and nucleotides corresponding to transcription start sites are enlarged.
Based on the experimentally determined transcription start sites, we localized putative PrrA binding sites for the tested genes (Fig. 3B). PrrA binds to a degenerate sequence and DNA curvature may play a role in determining its binding affinity, and therefore precise site identification has proved to be complicated. The most conserved part of the binding sequence is believed to be the GCG-Nx-CGC palindrome involved in PrrA dimer binding, where N is any nucleotide and x is variable but often equals nine (24, 28). The upstream region of each tested gene contained at least one putative PrrA binding site. Note that the putative PrrA binding site upstream of crtA is located in the intergenic region between crtA and the divergently transcribed crtIB operon (Fig. 3B) and therefore that PrrA likely activates both crtA and crtIB.
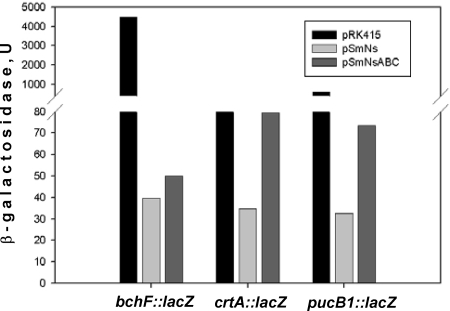
PrrA activates transcription of selected crt and bch genes in a heterologous host.
To verify the possibility that PrrA activates crt and bch gene transcription, we compared the expression of two transcriptional fusions, crtA-lacZ and bchF-lacZ, in P. denitrificans expressing either R. sphaeroides PpsR alone (plasmid pSmNs) (Table 1) or both PpsR and PrrA (plasmid pSmNsABC) (Table 1). As a positive control, we used the pucB1-lacZ transcriptional fusion. As expected from our earlier work (14), in the presence of PpsR, expression of pucB1-lacZ was low compared to that in cells containing the control vector (Fig. 4, compare pSmNs and pRK415). The presence of PrrA in addition to PpsR resulted in the upregulation of pucB1-lacZ expression by approximately twofold compared to the expression in the presence of PpsR alone (Fig. 4, compare pSmNsABC and pSmNs). This gave us a measure of the activation potential of PrrA in this setup.
FIG. 4.
Expression of lacZ transcriptional fusions in P. denitrificans. The effects of prrA and ppsR on the bchF-lacZ, crtA-lacZ, and pucB1-lacZ transcriptional fusions were determined. Black bars, vector pRK415; light gray bars, pSmNs (pRK415:ppsR); dark gray bars, pSmNsABC (pRK415:ppsR:prrBAC). One unit of β-galactosidase activity equals 1 nmol ONPG A600 unit−1 ml culture−1 min−1. Average data from three independent experiments are shown, with standard deviations not exceeding 15% of the averages.
When PrrA activation was tested using the crtA-lacZ transcriptional fusion, we observed a similar, approximately twofold activation (Fig. 4). However, the extent of activation of the bchF-lacZ expression by PrrA was only approximately 1.25-fold, which is borderline significant. These results, together with the in vitro transcription data, suggest that PrrA can activate transcription of the crt and bch genes, but the relative contributions of PrrA in vivo vary for different bch and crt genes. PrrA appears to activate transcription of bchE and crtA but to contribute less significantly or insignificantly to transcription of bchF and bchC.
The PrrBA system affects the AppA-PpsR pathway via multiple mechanisms.
To explore whether the PrrBA system affects the AppA-PpsR pathway exclusively via appA transcription, we tested phototrophic growth of the prrA mutant containing wild-type levels of the appA transcript, i.e., PRRBCA2(pECapp). We found that this strain could not grow phototrophically (not shown). Furthermore, photosynthetic complexes in PRRBCA2(pECapp) grown at low oxygen were significantly less abundant than the complexes in the prrA ppsR double mutant RPS1 measured under the same conditions (Fig. 5). Note that the lack of direct activation by PrrA of bch and crt genes described above in itself does not account for the phototrophic incompetence of PRRBCA2(pECapp) because PrrA is absent from both the phototrophic growth-impaired strain PRRBCA2(pECapp) and the phototrophically competent strain RPS1.
FIG. 5.
Photosynthetic complexes of strains grown under low-oxygen conditions. Cultures of all strains were adjusted to the same optical density (A660) prior to absorption spectroscopy. Spectra were shifted vertically to ease their comparisons.
The phototrophic impairment of PRRBCA2(pECapp) suggests that while PrrA-dependent appA expression is important, it is not the only mechanism through which PrrBA affects the AppA-PpsR pathway. The additional mechanism(s) remains to be uncovered. Since ppsR expression is PrrA independent, this mechanism must be posttranscriptional. Consistent with this interpretation is our observation that overexpression of appA beyond the wild-type transcript level, using plasmid p484-Nco1 (Fig. 1A), results in a further derepression of the PpsR regulon in PRRBCA2 (Fig. 1B) and brings the level of photosynthetic spectral complexes in PRRBCA2(p484-Nco1) closer to that of the prrA ppsR double mutant RPS1 (Fig. 5).
DISCUSSION
In R. sphaeroides, the major oxygen-dependent regulatory systems, PrrBA and AppA-PpsR, were believed to work in parallel, essentially independently of each other. However, in our previous study (30), we uncovered an apparent paradox. If the AppA-PpsR system were to function independently of PrrBA, then a lack of oxygen, which is sensed by the AppA antirepressor (31), would be expected to result in PpsR inactivation and derepression of the PpsR-dependent genes. However, in the prrA null mutant, PS genes are only marginally upregulated under anoxic conditions, and the prrA mutant cannot grow phototrophically (9). If PpsR were to respond to oxygen independently of PrrBA, then the phenotype of the prrA ppsR double mutant under anoxic conditions would be similar to that of the prrA null mutant, yet the double mutant can grow phototrophically (30). The observed discrepancy suggested to us that the PrrBA two-component system affects the activity of the AppA-PpsR pathway (30). In the beginning of this study, we verified our key assumption that ppsR inactivation is sufficient for restoring phototrophic growth of the prrA mutant. We readily isolated several independent spontaneous mutations in ppsR that restored phototrophic growth of the prrA mutant, which confirms that ppsR inactivation in itself is sufficient for restoring phototrophic growth of the prrA mutant.
In the process of investigating why the AppA-PpsR system in the prrA mutant does not properly respond to decreased oxygen, we revealed that oxygen-dependent PS gene regulation in R. sphaeroides is hierarchical and that the PrrBA system is positioned upstream of AppA-PpsR in this hierarchy. The dominance of the PrrBA system is exerted at two (or more) levels. One level involves PrrA-dependent appA gene expression. The second level involves posttranscriptional control by PrrBA over the AppA and/or PpsR activity. In this study, we explored the former mechanism in more detail.
The greatly decreased (13% of the wild-type level) appA mRNA level observed in the prrA null mutant makes this mutant in essence a PrrA− AppA− double mutant (Fig. 1A). In such a mutant, PpsR cannot adequately respond to oxygen changes because of an insufficient amount of the AppA antirepressor, and therefore the PpsR regulon remains overrepressed even under low- or no-oxygen conditions (Fig. 6). This interpretation is consistent with our earlier observations that a decreased AppA/PpsR ratio brought about by ppsR overexpression is qualitatively similar to appA inactivation (17, 30). Neglecting the importance of maintaining a wild-type AppA to PpsR ratio, e.g., using heterologous expression of AppA and PpsR from plasmid-borne systems, may easily compromise conclusions regarding the ability of the AppA-PpsR system to respond to environmental stimuli (20). The fact uncovered here that in the prrA null mutant, the AppA/PpsR ratio is greatly increased offers a simple explanation for the observation that light does not affect PS gene expression in this mutant, as observed by Jäger et al. (20), who presented a different interpretation.
FIG. 6.
Complex, hierarchical relationships between the major pathways controlling oxygen-dependent PS gene expression, i.e., the PrrBA and AppA-PpsR pathways. Thick arrows indicate regulatory genes (prrA, appA, and ppsR) or categories of PS genes (bch, crt, puc, puf, and puh), and geometrical shapes correspond to regulatory proteins (PrrA, AppA, and PpsR). +, activation; −, inhibition; ?, proposed unknown regulator. See the text for details.
Interestingly, our data suggest that PrrA does not activate appA transcription directly (Fig. 4). Therefore, an important regulatory component linking the PrrBA and AppA-PpsR systems remains to be identified (Fig. 6). The factor(s) that mediates the second, posttranscriptional level of control by PrrBA over the AppA-PpsR system activity is also unknown (Fig. 6). The existence of such a posttranscriptional level stems from our observation that restoring the appA transcript to the wild-type level is insufficient for rescuing phototrophic growth of the prrA null mutant (Fig. 5).
In addition to discovering the hierarchical relationship between the major oxygen-responsive regulatory pathways in R. sphaeroides, our work has led to a better understanding of the scope of involvement of the PrrBA pathway in PS gene expression in R. sphaeroides. PrrA has been known to affect bch and crt gene expression (9). However, its role was presumed to be indirect. Our data provide an explanation for the indirect role of PrrA, i.e., PrrA controls, or “macromanages,” the AppA-PpsR regulatory system, which directly represses most bch and crt genes (30). Unexpectedly, we found that PrrA also “micromanages” selected bch and crt genes (Fig. 2 and 4) by directly activating their transcription (Fig. 6). Oh et al. (33) proposed earlier that bchE transcription is regulated by PrrA, but they stopped short of showing that activation is direct. From this and other studies, a better picture of interactions between the two major regulatory pathways controlling oxygen-dependent PS gene expression is emerging (Fig. 6).
The hierarchical regulation of PS gene expression uncovered here is consistent with the position of PrrBA as a dominant regulatory system in relationship to other regulatory systems controlling carbon, nitrogen, sulfur, and hydrogen metabolism (8, 27). The peculiarity of the PrrBA-AppA-PpsR interactions is that both regulatory systems depend on the same environmental stimulus (oxygen) and that both control energy generation processes (respiration and PS for PrrBA and PS for AppA-PpsR). Therefore, a higher extent of their interdependence may be expected. In line with this expectation is our observation that the relationship between PrrBA and AppA-PpsR is nonlinear. AppA-PpsR seem to exhibit a feedback regulation on PrrBA. This feedback regulation manifests itself in the PpsR-dependent pattern of prrA transcription (30), which must have an effect on expression of PrrA-dependent genes (30) (also see pufB mRNA in Fig. 2B). Since PpsR does not appear to directly affect prrA transcription (30), how AppA-PpsR exert feedback regulation remains uncertain and will require further investigation (Fig. 6). Now that we have highlighted uncharacterized links between the PrrBA and AppA-PpsR regulatory systems in R. sphaeroides (Fig. 6), the goal is to elucidate molecular mechanisms underlying these links.
How common is such complex regulation of PS gene expression, and hence photosystem formation, in anoxygenic photosynthetic bacteria? While the precise answer to this question will have to await molecular analysis of regulation involving more species, it is already clear that the observed complexity is not unique to R. sphaeroides. By analyzing the literature on PS gene regulation in the related organism Rhodobacter capsulatus, we observed the same regulatory principles. (i) The PrrA homolog of R. capsulatus, RegA (40), directly activates bch and crt genes, as shown recently by Willett et al. (44). This is similar to the micromanagement role of PrrA in R. sphaeroides. (ii) A hierarchical relationship between RegBA and CrtJ has not been shown but can be predicted based on the ability of the regA (prrA) mutant of R. capsulatus to grow anaerobically phototrophically at high light but not at lower light. A possible explanation would be that the R. capsulatus CrtJ (PpsR) repressor is not fully inactivated in the regA mutant even under anoxic conditions. Note that since R. capsulatus lacks an AppA homolog, the nature of RegBA-CrtJ interactions may not be identical to that of PrrBA-AppA-PpsR interactions. (iii) In R. capsulatus, CrtJ directly represses transcription of the ubiquinol oxidase genes, encoding a low-oxygen terminal oxidase in this species (42). This may affect electron flow to the cbb3 cytochrome c oxidase and/or the quinone/quinol ratio (43), therefore providing a mechanism of feedback regulation by CrtJ of the RegBA system. We suggest that while “wiring schemes” vary even in relatively closely related species of anoxygenic phototrophs, regulation of PS gene expression in this group probably involves the same key elements, i.e., extensive PS gene target overlap between the global PrrBA/RegBA two-component system and the PS-specific AppA-PpsR/CrtJ system, hierarchical dominance of PrrBA/RegBA, and feedback regulation from AppA-PpsR/CrtJ.
Acknowledgments
This work was supported by NIH grants NCRR P20 RR15640 (M.G.), GM075273 (T.J.D.), and DOE DE-FG02-01ER63232 (T.J.D. and M.G.).
We are grateful to Samuel Kaplan for sharing unpublished results and to Jill Zeilstra-Ryalls for participating in the early stages of the in vitro transcription assays and for stimulating discussions.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Anthony, R. J., H. A. Green, and T. J. Donohue. 2003. Rhodobacter sphaeroides RNA polymerase and its sigma factors. Methods Enzymol. 37054-65. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, C., S. Elsen, L. R. Swem, D. L. Swem, and S. Masuda. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos. Trans. R. Soc. Lond. B 358147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. The single flavoprotein, AppA, from Rhodobacter sphaeroides integrates both redox and light signals. Mol. Microbiol. 45827-836. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary, M., and S. Kaplan. 2000. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 28862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 4925-68. [DOI] [PubMed] [Google Scholar]
- 6.Comolli, J. C., A. J. Carl, C. Hall, and T. Donohue. 2002. Transcriptional activation of the Rhodobacter sphaeroides cytochrome c2 gene P2 promoter by the response regulator PrrA. J. Bacteriol. 184390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsen, S., M. Jaubert, D. Pignol, and E. Giraud. 2005. PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol. Microbiol. 5717-26. [DOI] [PubMed] [Google Scholar]
- 8.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 17632-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eraso, J. M., and S. Kaplan. 1996. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1787037-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eraso, J. M., J. H. Roh, X. Zeng, S. J. Callister, M. S. Lipton, and S. Kaplan. 2008. Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J. Bacteriol. 1904831-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149377-388. [DOI] [PubMed] [Google Scholar]
- 12a.Gomelsky, L., O. V. Moskvin, R. A. Stenzel, D. F. Jones, T. J. Donohue, and M. Gomelsky. 2005. Abstr. 105th Gen. Meet. Am. Soc. Microbiol., abstr. H-103. American Society for Microbiology, Washington, DC.
- 13.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27497-500. [DOI] [PubMed] [Google Scholar]
- 14.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 1771634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1774609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomelsky, M., and S. Kaplan. 1996. The Rhodobacter sphaeroides 2.4.1 rho gene: expression and genetic analysis of structure and function. J. Bacteriol. 1781946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic evidence suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 27335319-35325. [DOI] [PubMed] [Google Scholar]
- 19.Gomelsky, M., I. M. Horne, H. J. Lee, J. M. Pemberton, A. G. McEwan, and S. Kaplan. 2000. Domain structure, oligomeric state, and mutational analysis of PpsR, the Rhodobacter sphaeroides repressor of photosystem gene expression. J. Bacteriol. 1822253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jäger, A., S. Braatsch, K. Haberzettl, S. Metz, L. Osterloh, Y. Han, and G. Klug. 2007. The AppA and PpsR proteins from Rhodobacter sphaeroides can establish a redox-dependent signal chain but fail to transmit blue-light signals in other bacteria. J. Bacteriol. 1892274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karls, R. K., J. R. Wolf, and T. J. Donohue. 1999. Activation of the cycA P2 promoter for the Rhodobacter sphaeroides cytochrome c2 gene by the photosynthesis response regulator. Mol. Microbiol. 34822-835. [DOI] [PubMed] [Google Scholar]
- 22.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. J., I. J. Ko, J. M. Lee, H. Y. Kang, Y. M. Kim, S. Kaplan, and J. I. Oh. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1895617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laguri, C., M. K. Phillips-Jones, and M. P. Williamson. 2003. Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity. Nucleic Acids Res. 316778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laguri, C., R. A. Stenzel, T. J. Donohue, M. K. Phillips-Jones, and M. P. Williamson. 2006. Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides. Biochemistry 457872-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. K., and S. Kaplan. 1992. cis-Acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J. Bacteriol. 1741146-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackenzie, C., J. M. Eraso, M. Choudhary, J. H. Roh, X. Zeng, P. Bruscella, A. Puskás, and S. Kaplan. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61283-307. [DOI] [PubMed] [Google Scholar]
- 28.Mao, L., C. Mackenzie, J. H. Roh, J. M. Eraso, S. Kaplan, and H. Resat. 2005. Combining microarray and genomic data to predict DNA binding motifs. Microbiology 1513197-3213. [DOI] [PubMed] [Google Scholar]
- 29.Masuda, S., and C. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110613-623. [DOI] [PubMed] [Google Scholar]
- 30.Moskvin, O. V., L. Gomelsky, and M. Gomelsky. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J. Bacteriol. 1872148-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moskvin, O. V., S. Kaplan, M. A. Gilles-Gonzalez, and M. Gomelsky. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 28228740-28748. [DOI] [PubMed] [Google Scholar]
- 32.Mouncey, N. J., and S. Kaplan. 1998. Cascade regulation of dimethyl sulfoxide reductase (dor) gene expression in the facultative phototroph Rhodobacter sphaeroides 2.4.1T. J. Bacteriol. 1802924-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, J. I., J. M. Eraso, and S. Kaplan. 2000. Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1823081-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 194237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 1864748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 1762869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranson-Olson, B., D. F. Jones, T. J. Donohue, and J. H. Zeilstra-Ryalls. 2006. In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J. Bacteriol. 1883208-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roh, J. H., W. E. Smith, and S. Kaplan. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J. Biol. Chem. 2799146-9155. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sganga, M. W., and C. E. Bauer. 1992. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell 68945-954. [DOI] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 42.Swem, D. L., and C. E. Bauer. 2002. Coordination of ubiquinol oxidase and cytochrome cbb(3) oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 1842815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swem, L. R., X. Gong, C. A. Yu, and C. E. Bauer. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 2816768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willett, J., J. L. Smart, and C. E. Bauer. 2007. RegA control of bacteriochlorophyll and carotenoid synthesis in Rhodobacter capsulatus. J. Bacteriol. 1897765-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeliseev, A. A., and S. Kaplan. 1995. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27021167-21175. [DOI] [PubMed] [Google Scholar]
- 46.Yeliseev, A. A., and S. Kaplan. 1999. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27421234-21243. [DOI] [PubMed] [Google Scholar]
- 47.Zeilstra-Ryalls, J. H., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell. Mol. Life Sci. 61417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeilstra-Ryalls, J. H., and S. Kaplan. 1995. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J. Bacteriol. 1776422-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 1856171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]