Abstract
The ubiquitous algal metabolite dimethylsulfoniopropionate (DMSP) is a major source of carbon and reduced sulfur for marine bacteria. Recently, the enzyme responsible for the demethylation of DMSP, designated DmdA, was identified, and homologs were found to be common in marine bacterioplankton cells. The recombinant DmdA proteins from the cultured marine bacteria Pelagibacter ubique HTCC1062 and Silicibacter pomeroyi DSS-3 were purified with a three-step procedure using anion-exchange, hydrophobic interaction, and hydroxyapatite chromatographies. The P. ubique enzyme possessed an Mr on sodium dodecyl sulfate-polyacrylamide gel electrophoresis of 38,500. Under nondenaturing conditions, the Mr was 68,000, suggesting that the enzyme was likely to be a dimer. The purified enzyme exhibited strict substrate specificity for DMSP, as DmdA from both S. pomeroyi and P. ubique possessed no detectable demethylase activity with glycine betaine, dimethyl glycine, methylmercaptopropionate, methionine, or dimethylsulfonioacetate. Less than 1% activity was found with dimethylsulfoniobutanoate and dimethylsulfoniopentanoate. The apparent Kms for DMSP were 13.2 ± 2.0 and 5.4 ± 2.3 mM for the P. ubique and S. pomeroyi enzymes, respectively. In cell extracts of S. pomeroyi DSS-3, the apparent Km for DMSP was 8.6 ± 1.2 mM, similar to that of purified recombinant DmdA. The intracellular concentration of DMSP in chemostat-grown S. pomeroyi DSS-3 was 70 mM. These results suggest that marine bacterioplankton may actively accumulate DMSP to osmotically significant concentrations that favor near-maximal rates of DMSP demethylation activity.
Dimethylsulfoniopropionate (DMSP) is synthesized by marine phytoplankton primarily for use as an intracellular osmolyte, although the compound has also been recognized as an antioxidant and predator deterrent (26, 39, 46). DMSP production by phytoplankton may account for 10% of the total carbon fixation in parts of the ocean (3, 37). Once released from phytoplankton cells, the carbon and sulfur in DMSP are both rapidly transformed in the marine microbial food web. While some phytoplankton are capable of degrading DMSP to dimethyl sulfide (DMS), marine bacteria are considered the primary mediators of DMSP transformation in seawater (36). DMSP consumption by marine bacteria satisfies 1 to 15% of their carbon demand and most, if not all, of the bacterial sulfur demand (20, 37).
Bacterial degradation of DMSP occurs through two competing pathways, known as the cleavage and the demethylation pathways. The cleavage pathway results in the formation of DMS, the major natural source of sulfur to the atmosphere (2, 36). In the atmosphere, oxidation of DMS produces aerosols that can influence global climate by causing solar radiation backscatter and acting as cloud condensation nuclei (5, 29). A majority of DMSP, however, is degraded through the demethylation pathway (19). The initial step of the demethylation pathway is removal of a methyl group from DMSP to form methylmercaptopropionate (MMPA). Subsequently, MMPA can be demethylated to form mercaptopropionate or can be demethiolated to form methanethiol and acrylate or another three-carbon compound (19, 43). Methanethiol is rapidly taken up by marine bacteria and incorporated into proteins (19).
The initial demethylation of DMSP is critical to oceanic sulfur flux because it precludes the possibility of DMS emission (15). Recently, the enzyme responsible for the demethylation of DMSP, designated DmdA, was discovered in two marine isolates from the roseobacter and SAR11 taxa, Silicibacter pomeroyi DSS-3 and Pelagibacter ubique HTCC1062, respectively. These taxa are among the most abundant bacterial groups found in ocean surface waters (13, 31). Based on metagenomic sequence data in the Sorcerer II Global Ocean Sampling Expedition, it has been estimated that 58% of marine bacteria possess the gene encoding DmdA (16).
Phylogenetic analyses of all identified DmdA sequences reveal a diverse set of proteins that form five evolutionarily distinct clades, known as clades A, B, C, D, and E (16). However, the bacterioplankton that harbor these DmdA orthologs remain largely unknown. At present, clades B and C lack genes from any cultured organisms. Having no taxonomic anchors, it is not possible to make comparisons of DmdA phylogeny to organismal phylogeny or to determine if the clades of DmdA are a result of organismal evolution or ecological adaptation. To examine this question and gain insights into biological controls on DMSP degradation, the recombinant DmdA enzymes from S. pomeroyi and P. ubique, representatives of clades A and D, respectively, were purified and characterized.
MATERIALS AND METHODS
Plasmid construction and expression of recombinant proteins.
The dmdA homologs in the Pelagibacter ubique genome (11) (SAR11_0246) and the Silicibacter pomeroyi DSS-3 genome (30) (SPO_1913) were synthesized commercially with Escherichia coli codon usage (GenScript Corporation). The synthesized genes were inserted into the expression vector pCYB1 (New England Biolabs) to generate pABX101 and pCRX1, respectively, which were transformed into E. coli Top10F′ and BL21(DE3), respectively. Plasmid-bearing cells were grown in Luria-Bertani broth at 37°C until cultures reached an optical density at 600 nm of 0.6, at which time the cultures were induced with 25 μM or 200 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for pABX101 and pCRX1, respectively, and transferred to 25°C for overnight incubation. Cells were harvested by centrifugation at 10,000 × g for 10 min. Pellets were resuspended in 50 mM Tris-HCl (pH 8.0) with 1 mM EDTA and placed on ice. Cells were then lysed by sonication, using three 30-s bursts. Lysed cells were centrifuged at 30,000 × g for 30 min to remove cell debris from the supernatant.
Preparation of Silicibacter pomeroyi DSS-3 extract.
Silicibacter pomeroyi DSS-3 was grown in batch or continuous culture using an artificial seawater medium consisting of 0.08 M HEPES (pH 6.9), 0.29 mM KH2PO4, 7.1 mM NH4Cl, 0.068 mM FeEDTA, 2% (wt/vol) sea salts (Sigma), and 0.1% (vol/vol) vitamin solution (12). Cell extract was prepared after 2 days of growth on DMSP as the sole carbon source in batch culture. Cells were harvested by centrifugation at 10,000 × g for 10 min and washed once in growth medium without DMSP. The pellet was resuspended in 400 mM HEPES (pH 7.5) with 1 mM EDTA and placed on ice. Cells were then lysed by sonication and centrifuged as described above.
Purification of DmdA.
The recombinant DmdA from P. ubique was purified from 8 ml of E. coli soluble extract after induction of the plasmid-borne gene. The extract was loaded onto a Q-Sepharose HP (GE Healthcare) column (1.6 by 10.0 cm) equilibrated with 50 mM Tris-HCl (pH 8.0) and 1 mM EDTA with a flow rate of 1 ml/min. The column was washed with the buffer at a flow rate of 1 ml/min. All enzyme activity was retained in 8 ml of flowthrough. Active fractions were pooled and brought to 1.7 M (NH4)2SO4 with the addition of the finely ground solid (NH4)2SO4. The sample was applied to a phenyl-Superose (GE Healthcare) column (1.0 by 10 cm) equilibrated with 50 mM Tris-HCl (pH 8.0) containing 1.7 M (NH4)2SO4 and 1 mM EDTA. Proteins were eluted with a linear gradient from 1.7 to 0 mM (NH4)2SO4 in a total volume of 80 ml. Activity eluted at about 0.8 M NH4SO4. Fractions containing activity were pooled and concentrated using a Centricon Ultracel YM-10 filter. Concentrated protein was diluted eight times with 10 mM Na2HPO4 (pH 7.0) and applied to a CHT ceramic hydroxyapatite type 1 (Bio-Rad) column (1.0 by 9 cm) at a flow rate of 0.5 ml/min. The column was washed with 10 ml of 10 mM Na2HPO4 (pH 7.0), and proteins were eluted with linear gradient of 10 to 500 mM Na2HPO4 (pH 7.0) in a total volume of 50 ml. Activity eluted at about 200 mM Na2HPO4. The active fractions were concentrated to 8 mg/ml using a Centricon Ultracel YM-10 filter.
The recombinant DmdA from S. pomeroyi was purified similarly to the P. ubique enzyme except for the following modifications. A linear gradient from 1.1 to 0 M (NH4)2SO4 was used to elute proteins from the phenyl-Superose column. Activity eluted at about 0.7 M (NH4)2SO4. Fractions containing activity were pooled and concentrated to about 0.1 ml using a Centricon Ultracel YM-10 filter. Concentrated protein was then diluted to 2 ml in 5 mM Na2HPO4 (pH 6.8) and again concentrated. Concentrated protein was diluted eight times with 5 mM Na2HPO4 (pH 6.8) and 0.03 mM CaCl2 and applied to a CHT ceramic hydroxyapatite type 1 (Bio-Rad) column (1.0 by 9 cm) equilibrated with the same buffer at a flow rate of 0.5 ml/min. The column was then washed with 10 ml of 5 mM Na2HPO4 (pH 6.8) and 0.03 mM CaCl2 at a flow rate of 0.5 ml/min. Enzyme activity eluted in the flowthrough. The active fraction was concentrated to 0.3 mg/ml using a Centricon Ultracel YM-10 filter.
Protein concentration.
The concentration of purified DmdA from P. ubique was determined by the biuret method using bovine serum albumin as a standard (14). The mass extinction coefficient at 280 nm was determined to be 10.72 g−1 liter cm−1. The protein concentration was then routinely determined using the mass extinction coefficient. Concentrations of other proteins were determined using the Bio-Rad Bradford reagent with bovine serum albumin as the standard.
Enzyme assays.
To minimize the effect of DMSP on reaction pH, a stock of buffered DMSP was prepared. A 600 mM solution of DMSP in 10 ml of 400 mM HEPES with 1 mM EDTA was brought to pH 6.5 with the dropwise addition of about 2 ml of 0.5 M NaOH with constant stirring. The final volume was then brought to 20 ml with 400 mM HEPES and 1 mM EDTA (pH 7.5). High-pressure liquid chromatography (HPLC) analysis of the buffered DMSP stock showed the presence of about 1 mM acrylate, indicating that only a small portion of DMSP was hydrolyzed during the neutralization procedure.
Unless specified differently, assays were performed using 400 mM HEPES (pH 7.5), 1 mM EDTA, and 2 mM dithiothreitol in 0.1 ml by combining 20 μl of 300 mM buffered DMSP stock solution and 0.685 mM tetrahydrofolate (THF). Due to the sensitivity of THF to O2, assays were performed in an anaerobic chamber under an N2-H2 (95:5, vol/vol) atmosphere. Reactions were initiated with the addition of enzyme, and the mixtures were incubated for 10 min. Reactions were quenched by addition of 20 μl of 50% H3PO4, and mixtures were briefly centrifuged to remove denatured proteins. Formation of either 5-methyl-THF or MMPA was determined by HPLC.
To determine the pH optimum, 400 mM of the following buffers were used at the indicated pH values: sodium 1,3-bis(Tris)propane (Bis-Tris propane) (6.5, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5), sodium 2-(N-morpholino)ethanesulfonic acid (MES) (5.5, 6.0, and 6.5), sodium 3-(N-morpholino)propanesulfonic acid (MOPS) (6.5, 7.0, and 7.5), sodium HEPES (7.0, 7.5, and 8.0), and Tris-HCl (7.5, 8.0, 8.5, and 9.0). Activity was determined with 10 mM DMSP.
Assays to determine substrate specificity were performed using either 4 mM or 10 mM of analog. Reaction mixtures contained 2 μg of DmdA, and reactions were performed for 3 hours.
Kinetic analyses were performed as described above except that the DMSP concentrations were 2.5, 5.0, 10, 20, 40, and 60 mM and the THF concentrations were 0.042, 0.085, 0.17, 0.34, and 0.68 mM. Assay mixtures with S. pomeroyi DSS-3 extract were incubated for 35 min before quenching. Inhibition kinetics were determined using 5, 20, and 60 mM DMSP with 0, 1, 5, 10, and 20 mM inhibitor. Kinetic data were analyzed using SigmaPlot 10.0 with the Enzyme Kinetics module (Systat Software Inc.).
HPLC analysis was performed using a Waters Alliance 4600 instrument with a reverse-phase SB-AQ column (2.6 by 250 mm; Agilent). The running buffer consisted of 6% (vol/vol) acetonitrile, 25 mM NaH2PO4, and 0.8% (vol/vol) H3PO4 with a flow rate of 0.75 ml/min. MMPA and acrylate were detected by UV absorption at 214 nm, and 5-methyl-THF was detected at 280 nm.
To confirm the reaction products, 2 μg of P. ubique DmdA was incubated with 60 mM DMSP and 0.17 mM THF for 2 h, at which time most THF was consumed, as determined by HPLC analysis. The reaction mixture was diluted 1:20 in 100 mM phosphate buffer (pH 7.0), and a UV spectrum of the reaction mixture was taken.
Determination of native molecular weight by gel filtration.
Purified DmdA from P. ubique was applied to a Sephacryl S200 HR (GE Healthcare) column (1.6 by 32 cm) equilibrated with 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 200 mM NaCl at a flow rate of 0.5 ml/min. Under the same conditions, the following molecular mass standards were chromatographed: carbonic anhydrase (29 kDa), albumin from chicken egg white (46 kDa), and bovine serum albumin monomer (66 kDa) and dimer (132 kDa).
PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the Next-Gel system (Amresco) with 12.5% polyacrylamide minigels. Gels were stained with 0.1% Coomassie blue in 50% methanol-10% acetic acid.
Substrate synthesis.
MMPA was synthesized by alkaline hydrolysis of its methyl ester, methyl-3-(methylthio)propionate (44). Dimethylsulfonioacetate and DMSP were synthesized from bromoacetic acid and DMS or acrylic acid and DMS as described previously (4, 38). Dimethylsulfoniobutanoate and dimethylsulfoniopentanoate were synthesized by mixing 2 g of bromobutyric acid or bromovaleric acid, respectively, with 10 ml of DMS in a round-bottom flask and heating the flask briefly in warm water to initiate the reaction. Vessels were covered and allowed to incubate at room temperature for 5 days. The remaining DMS was then evaporated, the product was dissolved in 5 ml of distilled water, and an equal volume of diethyl ether was added. The solution was mixed thoroughly for several minutes, and the aqueous layer containing the product was removed and evaporated. The last step was repeated, and the products were confirmed using 1H nuclear magnetic resonance (NMR).
To determine if the analogs were contaminated with DMSP, small amounts of DMSP were added to the analogs. The 1H NMR peaks for DMSP were then compared to the background in the analog spectrum to determine the sensitivity for small amounts of contamination.
Intracellular DMSP.
S. pomeroyi DSS-3 was grown at 30°C in a carbon-limited chemostat using 1 mM DMSP at a flow rate of 0.1 ml/min and a dilution rate of 0.0416 per hour. After five volumetric exchanges, the chemostat outflow was collected on ice for 5 min, and 0.4 ml of outflow was immediately centrifuged for 1 min to pellet cells. The cells were then washed in 1 ml of medium without DMSP, and the supernatant was transferred into a 9.8-ml vial. Finally, the cells were resuspended in medium without DMSP and transferred to a 9.8-ml vial. The vials were sealed with Teflon-coated stoppers, and 1 ml of 5 M NaOH was added with a syringe. Alkaline hydrolysis of DMSP yields equimolar concentrations of DMS and acrylate. The vials were vortexed briefly and incubated for 15 min at 30°C. The headspace was then analyzed for DMS by gas chromatography on an SRI 8610-C gas chromatograph with a Chromosil 330 column with nitrogen carrier gas at a flow rate of 60 ml min−1, an oven temperature of 60°C, and a flame photometric detector (9). Cell volume was calculated by first converting absorbance at 660 nm to dry weight using the following equation: dry weight (μg ml−1) = 364.74A660 + 6.7A660 (27). Dry weight was converted to wet weight by assuming that 30% of the cell's total weight was dry weight and then subtracting the dry weight (32). The volume was then calculated by using the density of water to convert from the water weight to volume. To measure the amount of DMSP remaining in spent medium, 1.5 ml of culture was collected on ice and immediately centrifuged for 1 min. The supernatant was decanted and placed into a 9.8-ml vial. The medium was then purged with N2 for 5 min to remove dissolved DMS. The vial was then sealed and analyzed for DMSP as described above.
Phylogenetic analysis.
DmdA homologs were found using BLASTp searches against selected genomes. Sequences were aligned, edited, and analyzed using MEGA version 4.0 (40). Sequences were manually trimmed to 261 amino acids. A phylogenetic tree was constructed using the minimum evolution method, including 1,000 bootstrap replicates. The GenBank accession number for all sequences used in the phylogenetic analysis are as follows: Homo, P48728; Pisum, P49364; Escherichia, P27248; Saccharomyces, P48015; Silicibacter V, AAV97197; Bacillus, P54378; Desulfovibrio, ABB38793; Pelagibacter IV, AAZ21486; Pyrococcus, O58888; Silicibacter I, AAV95190; Pelagibacter I, AAZ21068; clade B, ECV34452; clade C, EDI11852; clade E, EAW42451; Rattus I, Q64380; Rattus II, Q63342; Haloarcula, AAV45054; Rhizobium II, AAQ87218; Silicibacter VI, AAV96623; Roseovarius, EAP75561; Burkholderia, EDT09370; Silicibacter III, AAV94849; Mycobacterium, ABP47600; Silicibacter IV, AAV94866; Rhizobium, EDR75388; Pelagibacter III, AAZ22069; Marinobacter, EDM49742; Pelagibacter II, EAS84178; and Silicibacter II, AAV94935.
RESULTS
Purification of DmdA.
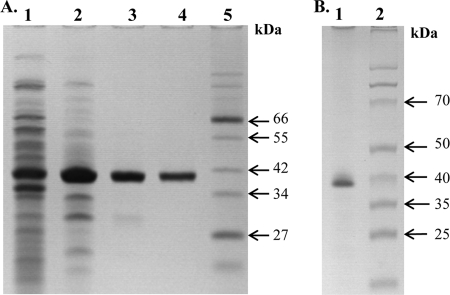
Recombinant DmdA from P. ubique was purified to electrophoretic homogeneity using a three-step chromatographic purification as shown in Fig. 1A. SDS-PAGE of the purified protein failed to detect contaminating proteins, and comparison to a standard indicated a purity of >92%. Initial purifications yielded a labile enzyme which lost activity in a matter of days. Addition of EDTA to buffers greatly improved enzyme stability. In 50 mM Tris-HCl (pH 8.0) with 1 mM EDTA, the enzyme was stable for more than 6 months when stored at either −20° or 4°C. The enzyme did not bind to anion- or cation-exchange resins within ranges of pH 6 to 9. This inability to bind anion-exchange resin was also observed for the recombinant glycine cleavage T-protein, which is homologous to DmdA (33). Nevertheless, substantial purification was obtained by passing the extracts through either anion- or cation-exchange resin.
FIG. 1.
SDS-PAGE of purified recombinant P. ubique DmdA (A) and S. pomeroyi DmdA (B). The proteins from various purification steps were separated by SDS-PAGE on 12.5% polyacrylamide and stained with Coomassie blue. (A) Crude soluble cell extract of recombinant E. coli (lane 1), Q-Sepharose flowthrough (lane 2), phenyl-Superose eluate (lane 3), hydroxyapatite eluate (lane 4), and broad-range protein marker (New England Biolabs) (lane 5). (B) Hydroxyapatite flowthrough (lane 1) and broad-range protein marker (Fermentas) (lane 2).
Similarly, the recombinant DmdA from S. pomeroyi did not bind anion-exchange resin at pH 8.0. Moreover, the enzyme also failed to bind CHT ceramic hydroxyapatite type 1, even at very low salt concentrations. Thus, it was not possible to purify this protein to electrophoretic homogeneity by these methods. Integration of the bands identified on SDS-PAGE, as shown in Fig. 1B, indicated a purity of 70%.
Properties of DmdA.
Upon SDS-PAGE, the Mrs of the denatured enzymes were 38,500 to 39,500, which was consistent with the predicted molecular mass, based on the amino acid sequence, of 39.5 to 41.5 kDa. The native molecular mass of the DmdA from P. ubique was investigated by gel filtration. In duplicate experiments, DmdA eluted as a single peak with an Mr of 66,000 to 69,000, indicating that the enzyme may exist as a dimer. Because the enzyme did not enter native polyacrylamide gels, it was not possible to confirm the native molecular mass by this method. The inability of DmdA to bind to ion-exchange resins or migrate into native polyacrylamide gels suggests that the recombinant enzyme was neutral in charge. However, the predicted isoelectric points were 6.5 and 5.3 for DmdA from P. ubique and S. pomeroyi, respectively. Therefore, the absence of an observed charge with ion-exchange resins and native polyacrylamide gels suggests that the charge on DmdA was hidden.
The activity of DmdA in several buffers was examined to find the optimum reaction conditions. Bis-Tris propane, which has a wide buffering range, was inhibitory at pHs of above 7.5 and was not used further. The DmdA from P. ubique was consistently more active in HEPES buffer than in MOPS or Tris-HCl buffer at the same pH. Similarly, the DmdA from S. pomeroyi was more active in HEPES than in MOPS buffer, but the activities in HEPES and Tris-HCl buffers were the same. These differences in activities are not due to differences in the counterions or ionic strengths of the buffers. For instance, 400 mM of (NH4)2SO4, K2HPO4, MgSO4, Na-acetate, NaCl, and KCl had no effect on the enzyme activity. In contrast, 100 mM and 400 mM MgCl2 inhibited activity by 20 and 80%, respectively. Therefore, the different activities observed in the buffers were due to direct interaction of DmdA with the buffers and not the counterions.
The optimum pHs for the DmdA enzymes from P. ubique and S. pomeroyi were similar. Maximum activity was observed at pH 7.0 to 8.0 for both enzymes. The P. ubique DmdA possessed about 50% activity at pH 6.5 and 8.3. The DmdA from S. pomeroyi possessed 50% activity at pH 6.0 and 8.8. Thus, all subsequent assays were carried out at pH 7.5 using 400 mM sodium HEPES buffer.
Product confirmation.
To confirm that the reaction transfers a methyl group from DMSP to THF, the products of the enzyme reaction were examined by UV absorption spectroscopy and chromatographic analysis. An enzyme reaction was run to completion so that most of the THF was consumed as determined by UV spectra of the products. The product possessed an absorption maximum at 290 nm and a minimum at 245 nm, identical to those of authentic 5-methyl-THF in the same buffer (18). The formation of 5-methyl-THF was further confirmed because the product coeluted with the authentic 5-methyl-THF on HPLC (data not shown). The production of MMPA was also confirmed by comparison of the elution time of the reaction product with that of the authentic compound upon HPLC (data not shown).
Substrate specificity.
The methyl donor specificity of DmdA from both P. ubique and S. pomeroyi was investigated using a number of substrate analogs (Table 1). No activity was observed with dimethylsulfonioacetate or the nitrogen-containing compounds tested. Very low rates of methyl-THF formation were observed from the DMSP analogs dimethylsulfoniobutanoate and dimethylsulfoniopentanoate. This activity could be due to either the low activities of DmdA for these substrates or contamination of the analogs by DMSP. Examination of the 1H NMR spectra indicated that the analogs contained less than 1% DMSP. Next, enzyme assays were performed with low concentrations of the analogs. Under these conditions, >3% of the substrates were consumed. Since the amount of substrate demethylated was greater than the maximum amount of DMSP contamination, the activity could not be attributed solely to DMSP contamination.
TABLE 1.
Rates of demethylation for DMSP and analogs
| Substrate | Sp act (nmol min−1 mg−1)a for recombinant DmdA from:
|
|
|---|---|---|
| P. ubique | S. pomeroyi | |
| Dimethyl glycine | <0.6 | <0.6 |
| Glycine betaine | <0.6 | <0.6 |
| Methionine | <0.6 | <0.6 |
| Dimethylsulfonioacetate | <0.6 | <0.6 |
| DMSP | 2,490 | 649 |
| Dimethylsulfoniobutanoate | 7 | 2 |
| Dimethylsulfoniopentanoate | 11 | 8 |
Values are averages from duplicate experiments using 4 mM substrate.
Inhibition.
Inhibition of DMSP demethylation by the purified DmdA from P. ubique was studied using the substrate analogs and the product of DMSP demethylation, MMPA. DMSP demethylation was strongly inhibited by MMPA, where Lineweaver-Burk plots showed a series of nonparallel lines intersecting to the left of the origin, indicative of noncompetitive inhibition (see Fig. S1 in the supplemental material). The data fit best to a noncompetitive partial model of inhibition with a Ki of 2.1 ± 0.4 mM. The substrate analogs dimethylsulfoniobutanoate and dimethylsulfoniopentanoate were weak inhibitors of DMSP demethylation. Kinetic analysis of dimethylsulfoniobutanoate and dimethylsulfoniopentanoate inhibition yielded a best fit to the mixed partial model, with Kis of 47 and 19 mM, respectively.
Kinetics.
The Michaelis-Menten constants for purified DmdA from P. ubique and S. pomeroyi were determined. Lineweaver-Burk plots showed a series of intersecting lines (Fig. 2), indicative of a sequential mechanism in which both DMSP and THF must bind to the enzyme before catalysis. A random bi-bi mechanism, where the order of substrate binding and product release occurs randomly, yielded the best fit to each data set (Table 2). The high Km values of the purified enzyme for DMSP suggested that the recombinant enzymes might not be in their physiological conformations. To test this hypothesis, the kinetic constants for the native enzyme in cell extracts of S. pomeroyi were determined (Table 2 and Fig. 2). The agreement of the Km values for the recombinant and native activities confirmed that the recombinant enzymes were in their physiologically active forms.
FIG. 2.
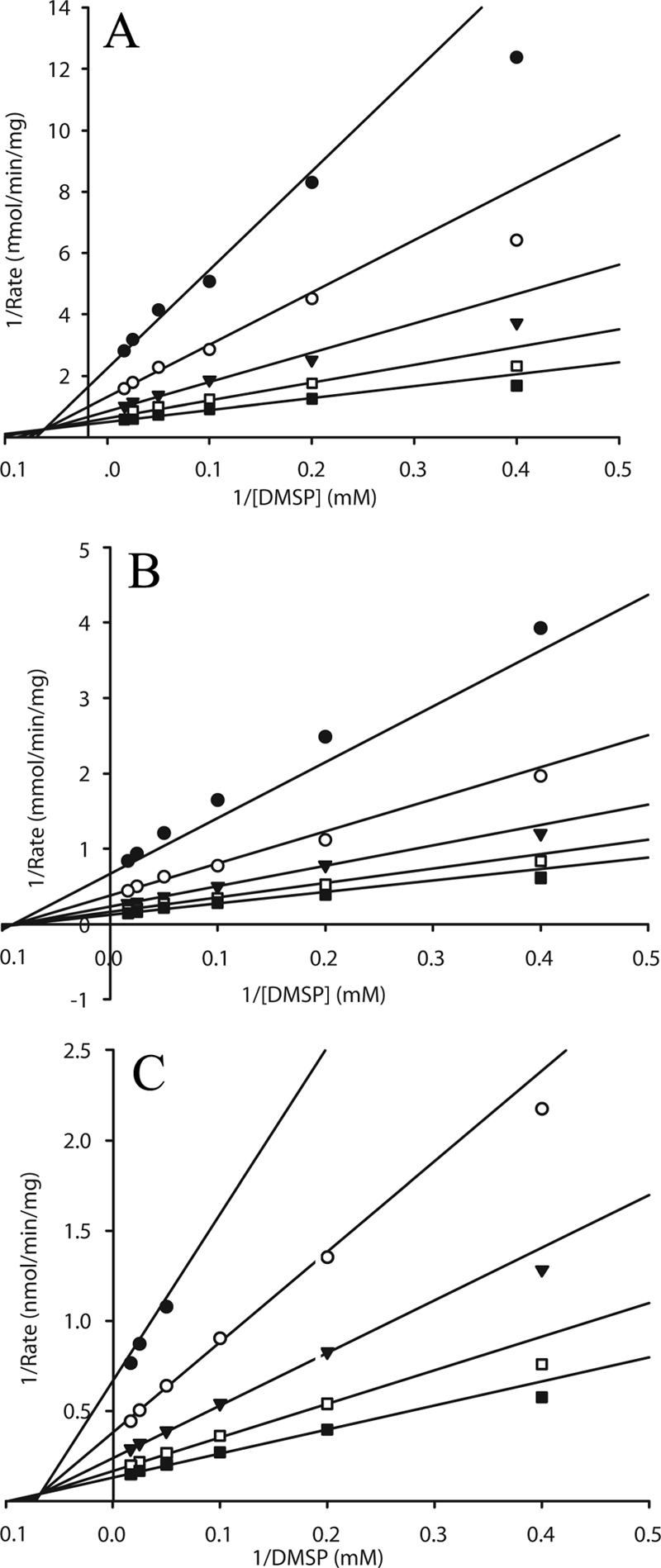
Lineweaver-Burk plots of DMSP demethylation. (A) DmdA from S. pomeroyi. (B) DmdA from P. ubique. (C) Crude cell extract from S. pomeroyi DSS-3. Lines are best fit to the overall data set of each graph using the random bi-bi model. Each plot indicates a different concentration of THF, as follows: •, 0.042 mM; ○, 0.085 mM; ▾, 0.17 mM; □, 0.34 mM; ▪, 0.68 mM.
TABLE 2.
Kinetic constants of DmdA
| Enzyme source | r2 (random bi-bi model) |
Km (mM, mean ± SD) for:
|
Vmax (μmol min−1 mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) | |
|---|---|---|---|---|---|---|
| THF | DMSP | |||||
| P. ubique | 0.994 | 0.29 ± 0.03 | 13.2 ± 2.0 | 11.7 | 8.1 | 618 |
| S. pomeroyi | 0.976 | 0.21 ± 0.04 | 5.4 ± 2.3 | 3.7a | 2.4 | 450 |
| S. pomeroyi extract | 0.997 | 0.26 ± 0.02 | 8.6 ± 1.2 | 0.0105 | NAb | NA |
The Vmax for the recombinant S. pomeroyi enzyme was corrected for a protein purity of 70%.
NA, not applicable.
Intracellular DMSP.
The high Km values observed and low concentration of DMSP typically found in the environment (nM range) suggested that cells might be accumulating DMSP intracellularly. To test this hypothesis, the intracellular concentration of DMSP in S. pomeroyi DSS-3 was measured by growing cells in a chemostat with DMSP as the limiting nutrient. The initial concentration of DMSP in the medium was 1 mM. The spent medium contained 2 μM, indicating that the cells consumed 99.8% of the available DMSP. In contrast, the intracellular DMSP concentrations were determined to be 152 and 157 μmol (g [dry weight] of cells)−1) in duplicate measurements, implying that the intracellular DMSP concentration was about 70 mM. DMSP was not detected in the supernatant of the single wash step, indicating that the intracellular measurement was not affected by DMSP carryover and that cells did not readily lose significant amounts of DMSP during the washing. Thus, chemostat-grown S. pomeroyi cells accumulated very high levels of DMSP despite the very low concentrations in the culture medium.
Phylogenetic analyses.
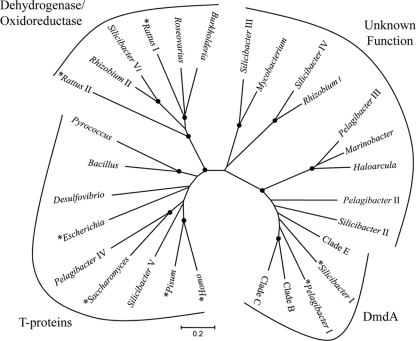
DmdA is a member of a diverse enzyme family that includes the glycine cleavage T-protein and domains of dimethylglycine oxidase and sarcosine dehydrogenase (Fig. 3). Phylogenetic analyses showed that proteins confirmed as glycine cleavage T-proteins from prokaryotes, eukaryotes, and archaea clustered together, distinct from proteins with DMSP demethylase activity. The carboxy termini of the dimethylglycine oxidase (EC 1.5.3.10) and sarcosine dehydrogenase (EC 1.5.99.1) are homologous to that of the T-protein (28). The sarcosine dehydrogenase and dimethylglycine oxidase enzymes from Rattus and several open reading frames from proteobacteria also form an independent cluster. Several open reading frames with unknown function but annotated as aminomethyl transferase proteins from both bacteria and archaea form additional clusters. Presumably, these represent novel enzymes within this family.
FIG. 3.
Phylogenetic tree of homologs to T-proteins and DmdA. The tree was constructed using the minimum evolution method with MEGA4 software. Enzymes with confirmed activity are indicated by an asterisk. Five DmdA sequences were used, representative of the five known phylogenetic clades, including clades A and D (from Silicibacter pomeroyi and Pelagibacter ubique, respectively [16]) and clades B, C, and E (from uncultured marine bacterioplankton). Closed circles at the branch points indicate ≥90% replication with 1,000 bootstraps. The scale bar represents 0.2 amino acid substitution per site. GenBank accession numbers of protein sequences are listed in Materials and Methods.
DISCUSSION
The properties of the purified DmdA enzymes from S. pomeroyi and P. ubique are consistent with their role in the THF-dependent demethylation of DMSP, as previously hypothesized (15). The protein sequence of DmdA places the enzyme in the aminomethyl transferase family (EC 2.1.2.10), which includes the well-characterized T-protein of the glycine cleavage system. The glycine cleavage system is comprised of four proteins which catalyze the conversion of glycine to 5,10-methylene-THF, CO2, and NH3. First, the P-protein catalyzes the decarboxylation of glycine and transfers the remaining aminomethylene group to the lipoic acid arm of the H-protein. Next, the T-protein liberates ammonia and transfers the methylene group to THF. Finally, the L-protein oxidizes the lipoic acid moiety of the H-protein (25). This complicated multiprotein system contrasts with the demethylation of DMSP, which is catalyzed by a single enzyme. Even though the glycine cleavage T-proteins and DmdA form separate groups within a diverse enzyme family, they possess similar Kms for THF, i.e., 0.17 mM for T-protein (34) and 0.21 to 0.29 mM for DmdA. In the glycine cleavage system, the carbon donor is covalently bound to the H-protein, so it is not possible to compare these kinetic constants.
A THF-dependent DMSP-demethylating enzyme was previously purified from a sulfate-reducing bacterium, but the identity of the gene encoding this protein was not reported (17). This enzyme possessed an Mr upon SDS-PAGE of 35,000 but was extremely labile and O2 sensitive. Despite the similarity in molecular weight to DmdA, the difference in O2 sensitivity suggests that these enzymes are not closely related. In addition, phylogenetic analyses of DmdA against all sequenced deltaproteobacteria, which include the sulfate-reducing bacteria, did not identify a potential ortholog of DmdA.
The Km of DmdA for DMSP is higher than those observed for bacterial DMSP cleavage enzymes that mediate the competing pathway to DMS. DMSP lyase purified from a facultatively anaerobic Alcaligenes species had a Km for DMSP of 1.4 mM (9), while the Km of a purified DMSP lyase from Desulfovibrio acrylicus was 0.45 mM (8). These values are close to those expected for enzymes active with common intracellular metabolites but are an order of magnitude lower than the values for DmdA. Whether similarly low Kms for DMSP cleavage enzymes will be found in planktonic marine bacteria is not yet known, but the answer may shed light on the routing of DMSP to the demethylation versus cleavage pathways in situ. If the intracellular concentrations of DMSP in S. pomeroyi and P. ubique are in the range typical for many other metabolites, DmdA would have only low activity in vivo. Instead, the kinetic constants for DmdA are consistent with the accumulation of DMSP to the high levels typical of osmoprotectants. For example, the common osmolyte glycine betaine is accumulated to intracellular concentrations of 130 to 170 mM by some bacteria (1, 35). Similarly, DMSP is a known osmoprotectant in marine and other bacteria (6, 10, 45). Natural populations of marine microorganisms taking up 35S-labeled DMSP can retain most of the compound untransformed for 30 h, suggesting intracellular accumulation (21, 22). The assimilation of both glycine betaine and DMSP in the marine environment may rely on the same high-affinity transport system capable of taking up the compounds at the low nM levels typical of seawater (24). The fact that S. pomeroyi accumulates DMSP to 70 mM from a medium concentration of 2 μM suggests that it may rely on DMSP as an osmolyte while metabolizing only that which is supplied in excess of the cells' requirement for osmoprotection.
Although measurements of intracellular DMSP concentration in P. ubique were not performed because of the challenges of maintaining cultures of this oligotrophic marine bacterium in the laboratory, the enzyme kinetics suggest that this microorganism must also accumulate high levels of DMSP for demethylation to occur. Recently, it was shown that growth of P. ubique is dependent on an exogenous source of reduced sulfur, such as methionine or DMSP (42). In radiotracer experiments using 35S-labeled DMSP, P. ubique took up 70% and incorporated 50% of DMSP sulfur into protein (42). For DMSP sulfur to be incorporated into cellular protein, DMSP must first be demethylated by DmdA (23). Therefore, the assimilation of DMSP sulfur suggests that P. ubique also accumulates DMSP intracellularly to high levels.
To investigate whether the phylogenetic diversity of DmdA has functional significance, the properties of the DmdA enzymes from S. pomeroyi and P. ubique, which represent phylogenetic clades A and D, were compared. The enzymes exhibited similar pH optima and kinetics properties, and both had strict substrate specificities. While the Kms for THF were very similar, the Kms for DMSP were somewhat different, i.e., 5.4 mM and 13.2 mM for S. pomeroyi and P. ubique, respectively. Likewise, the calculated turnover numbers were also different, 2.4 s−1 for S. pomeroyi and 8.1 s−1 for P. ubique. Despite these differences, the catalytic efficiencies (kcat/Km) were very similar. Whether or not these small differences in Km and Vmax reflect physiological adaptations of the DmdA enzymes harbored by S. pomeroyi and P. ubique is of ecological interest. Although both organisms probably use DMSP as an osmoprotectant, P. ubique requires DMSP or another exogenous source of reduced sulfur for growth (42), while S. pomeroyi is capable of assimilating sulfate. S. pomeroyi is also able to degrade DMSP through the cleavage pathway. While the ability of P. ubique to cleave DMSP has yet to be determined, homologs of recently identified genes involved in DMSP cleavage are absent in P. ubique (7, 41). Thus, it is possible that P. ubique lacks the cleavage pathway. These potential physiological differences may be reflected in the differing Kms for DMSP, which may be part of the strategies for controlling DmdA activity in situ. The characterization of additional DmdA enzymes, particularly from clades B, C, and E, is needed to further examine the kinetic diversity of DmdA. Nevertheless, the properties of DmdA reported here greatly expand our knowledge of the conditions under which DMSP demethylation can occur.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (OCE-0724017) and the Gordon and Betty Moore Foundation.
We thank Robert Phillips for advice on substrate synthesis, James Henriksen for technical advice, and Alison Buchan for providing plasmid pABX101.
Footnotes
Published ahead of print on 10 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abee, T., R. Palmen, K. J. Hellingwerf, and W. N. Konings. 1990. Osmoregulation in Rhodobacter sphaeroides. J. Bacteriol. 172149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreae, M. O., and H. Raemdonck. 1983. Dimethyl sulfide in the surface ocean and the marine atmosphere: a global view. Science 221744-747. [DOI] [PubMed] [Google Scholar]
- 3.Archer, S. D., C. E. Widdicombe, G. A. Tarran, A. P. Rees, and P. H. Burkill. 2001. Production and turnover of particulate dimethylsulphoniopropionate during a coccolithophore bloom in the northern North Sea. Aquat. Microb. Ecol. 24225-241. [Google Scholar]
- 4.Chambers, S. T., C. M. Kunin, D. Miller, and A. Hamada. 1987. Dimethylthetin can substitute for glycine betaine as an osmoprotectant molecule for Escherichia coli. J. Bacteriol. 1694845-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlson, R. J., J. E. Lovelock, M. O. Andreae, and S. G. Warren. 1987. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature 326655-661. [Google Scholar]
- 6.Cosquer, A., V. Pichereau, J. A. Pocard, J. Minet, M. Cormier, and T. Bernard. 1999. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl. Environ. Microbiol. 653304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curson, A. R., R. Rogers, J. D. Todd, C. A. Brearley, and A. W. Johnston. 2008. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine alpha-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10757-767. [DOI] [PubMed] [Google Scholar]
- 8.der Maarel, M. J. E. C., W. Aukema, and T. A. Hansen. 1996. Purification and characterization of a dimethylsulfoniopropionate cleaving enzyme from Desulfovibrio acrylicus. FEMS Microbiol. Lett. 143241-245. [Google Scholar]
- 9.de Souza, M. P., and D. C. Yoch. 1995. Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl. Environ. Microbiol. 6121-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz, M. R., P. T. Visscher, and B. F. Taylor. 1992. Metabolism of dimethylsulfoniopropionate and glycine betaine by a marine bacterium. FEMS Microbiol. Lett. 9661-65. [Google Scholar]
- 11.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 3091242-1245. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, J. M., F. Mayer, M. A. Moran, R. E. Hodson, and W. B. Whitman. 1997. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int. J. Syst. Bacteriol. 47369-376. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 634237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gornall, A. G., C. J. Bardawill, and M. M. David. 1949. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 177751-766. [PubMed] [Google Scholar]
- 15.Howard, E. C., J. R. Henriksen, A. Buchan, C. R. Reisch, H. Burgmann, R. Welsh, W. Ye, J. M. Gonzalez, K. Mace, S. B. Joye, R. P. Kiene, W. B. Whitman, and M. A. Moran. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314649-652. [DOI] [PubMed] [Google Scholar]
- 16.Howard, E. C., S. Sun, E. J. Biers, and M. A. Moran. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 102397-2410. [DOI] [PubMed] [Google Scholar]
- 17.Jansen, M., and T. A. Hansen. 2000. DMSP: tetrahydrofolate methyltransferase from the marine sulfate-reducing bacterium strain WN. J. Sea Res. 43225-231. [Google Scholar]
- 18.Kaufman, B. T., K. O. Donaldson, and J. C. Keresztesy. 1963. Chromatographic separation of the diastereoisomers of dl, L-5, 10-methylenetetrahydrofolate. J. Biol. Chem. 2381498-1500. [PubMed] [Google Scholar]
- 19.Kiene, R. P. 1996. Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 5469-83. [Google Scholar]
- 20.Kiene, R. P., and L. J. Linn. 2000. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45849-861. [Google Scholar]
- 21.Kiene, R. P., and L. J. Linn. 2000. The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: tracer studies using 35S-DMSP. Geochim. Cosmochim. Acta 642797-2810. [Google Scholar]
- 22.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43209-224. [Google Scholar]
- 23.Kiene, R. P., L. J. Linn, J. Gonzalez, M. A. Moran, and J. A. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 654549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiene, R. P., L. P. H. Williams, and J. E. Walker. 1998. Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aquat. Microb. Ecol. 1539-51. [Google Scholar]
- 25.Kikuchi, G. 1973. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol. Cell Biochem. 1169-187. [DOI] [PubMed] [Google Scholar]
- 26.Kirst, G. O. 1996. Osmotic adjustment in phytoplankton and macroalgae: the use of dimethylsulfoniopropionate (DMSP), p. 121-129. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, NY.
- 27.Koch, A. L. 1994. Growth measurement, p. 248-277. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 28.Leys, D., J. Basran, and N. S. Scrutton. 2003. Channelling and formation of ‘active’ formaldehyde in dimethylglycine oxidase. EMBO J. 224038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovelock, J. E., R. J. Maggs, and R. A. Rasmussen. 1972. Atmospheric dimethyl sulphide and the natural sulphur cycle. Nature 237452-453. [Google Scholar]
- 30.Moran, M. A., A. Buchan, J. M. Gonzalez, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432910-913. [DOI] [PubMed] [Google Scholar]
- 31.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420806-810. [DOI] [PubMed] [Google Scholar]
- 32.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA.
- 33.Okamura-Ikeda, K., K. Fujiwara, and Y. Motokawa. 1992. Molecular cloning of a cDNA encoding chicken T-protein of the glycine cleavage system and expression of the functional protein in Escherichia coli. Effect of mRNA secondary structure in the translational initiation region on expression. J. Biol. Chem. 26718284-18290. [PubMed] [Google Scholar]
- 34.Okamura-Ikeda, K., K. Fujiwara, and Y. Motokawa. 1982. Purification and characterization of chicken liver T-protein, a component of the glycine cleavage system. J. Biol. Chem. 257135-139. [PubMed] [Google Scholar]
- 35.Perroud, B., and D. Le Rudulier. 1985. Glycine betaine transport in Escherichia coli: osmotic modulation. J. Bacteriol. 161393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simo, R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16287-294. [DOI] [PubMed] [Google Scholar]
- 37.Simo, R., S. D. Archer, C. Pedros-Alio, L. Gilpin, and C. E. Stelfox-Widdicombe. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 4753-61. [Google Scholar]
- 38.Slow, S., M. Vasudevamurthy, R. Fraser, C. McEntyre, M. Lever, S. Chambers, and P. George. 2007. Dimethylthetin treatment causes diffuse alveolar lung damage: a pilot study in a sheep model of continuous ambulatory peritoneal dialysis (CAPD). Exp. Toxicol. Pathol. 58285-290. [DOI] [PubMed] [Google Scholar]
- 39.Sunda, W., D. J. Kieber, R. P. Kiene, and S. Huntsman. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418317-320. [DOI] [PubMed] [Google Scholar]
- 40.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 41.Todd, J. D., R. Rogers, Y. G. Li, M. Wexler, P. L. Bond, L. Sun, A. R. Curson, G. Malin, M. Steinke, and A. W. Johnston. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315666-669. [DOI] [PubMed] [Google Scholar]
- 42.Tripp, H. J., J. B. Kitner, M. S. Schwalbach, J. W. Dacey, L. J. Wilhelm, and S. J. Giovannoni. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452741-744. [DOI] [PubMed] [Google Scholar]
- 43.Visscher, P. T., M. R. Diaz, and B. F. Taylor. 1992. Enumeration of bacteria which cleave or demethylate dimethylsulfoniopropionate in the Caribbean Sea. Mar. Ecol. Prog. Ser. 89293-296. [Google Scholar]
- 44.Wackett, L. P., J. F. Honek, T. P. Begley, V. Wallace, W. H. Orme-Johnson, and C. T. Walsh. 1987. Substrate analogues as mechanistic probes of methyl-S-coenzyme M reductase. Biochemistry 266012-6018. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe, G. V. 1996. Accumulation of dissolved DMSP by marine bacteria and its degradation via bacterivory, p. 277-291. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, NY.
- 46.Wolfe, G. V., and M. Steinke. 1997. Grazing-activated chemical defence in a unicellular marine alga. Nature 387894-897. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.