Abstract
There are mutants of Salmonella enterica (with mutations in fliF and fliL) that shed flagella when they are swimming in a viscous medium or on the surface of soft agar. Filaments with hooks and the distal rod segment FlgG are recovered. We tried to extract flagellar filaments from such cells by pulling on them with an optical trap but failed, even when we used forces large enough to straighten the filaments. Thus, flagella are firmly anchored.
The flagellar filaments of swimming bacteria are subjected to forces strong enough to generate changes in polymorphic form (e.g., from normal to semicoiled to curly) (5, 8, 10). There are defects in the hook-associated protein at the base of the filament that allow transformations that otherwise are forbidden (e.g., from normal or curly to straight) (6). Also, there are defects in the MS ring (in FliF) that cause cells to shed filaments when the are swimming in gel-like media (4% gelatin), provided that their motors switch (i.e., are not biased strongly clockwise or counterclockwise) (9). The flagella appear to break at a point proximal to the distal rod protein, FlgG. Recently, R. M. Harshey's group (1) found that fliL mutants, when attempting to swarm (on 0.6% agar), shed flagella in a similar way, although switching was not required. Might it be possible to extract such flagella by pulling on them with an optical trap? We generated polymorphic transformations by attaching reconstituted filaments to glass at their proximal ends (they attach spontaneously [7]) and to latex beads near their distal ends by pulling on the beads with an optical trap (4). So we thought that it would be interesting to try the same thing with filaments attached to intact cells to see whether filaments of the fliF mutant SJW3060 (9) or of the fliL null mutant UA74 obtained from R. M. Harshey or the null mutants SJW2295 and SJW2296 obtained from S. Yamaguchi are less robust than those of wild-type strain SJW1103 (11).
Cells were grown to saturation overnight in LB broth (1% Bacto tryptone [Difco], 0.5% yeast extract, 0.5%NaCl) at 37°C and then diluted 1:100 in the same medium and grown again for 2 h. Cells were pelleted by centrifugation for 15 min at 8,000 × g and washed once in motility buffer (10 mM potassium phosphate [pH 7], 67 mM NaCl, 0.1 mM EDTA). Cells were labeled in this medium with an amine-reactive Cy3 dye (catalog no. PA23001; Amersham Biosciences, Piscataway, NJ) for 1.5 h, using the procedures of Turner et al. (10). Then the cells were pelleted, washed once, and added to anti-Cy3 antibody-coated beads in motility buffer containing 0.1% bovine serum albumin (BSA).
The antibody-coated beads were prepared by adsorbing anti-Cy3 antibody (catalog no. ab6902-1; Abcam, Cambridge, MA) onto 1.4-μm-diameter latex beads (catalog no. 17133; Polysciences, Warrington, PA) by mixing 10 μl BSA (10% in water), 20 μl antibody (from a 1-mg/ml aliquot prepared previously and kept at −20°C), 10 μl beads (as supplied), and 70 μl motility buffer. The mixture was rotated overnight in a cold room and rinsed into motility buffer containing 0.001% Tween 20.
The cell-bead mixture was placed on a microscope slide within a grease ring, covered with a no. 1 coverslip, and viewed with the optical trap used previously to measure filament force-extension curves (4). The experiments were done at room temperature (∼22°C). The BSA prevented the beads from sticking irreversibly to the coverslip but also reduced the number of stuck cells, so we had to hunt a bit to find stuck cells with accessible filaments. A bead was plucked from the coverslip or out of the suspension and brushed near the end of an exposed filament. Most filaments were rotating and reversing, and actively spinning filaments were difficult to catch with a bead, so we selected filaments that had stopped spinning. In about one-half of such encounters, the attachment of the bead to the filament was so strong that the bead pulled out of the trap rather than detached from the filament.
Calibration of the trap at maximum power with a free bead gave a Lorentzian spectrum with a roll-off of ∼1.8 kHz, corresponding to a stiffness of ∼150 pN/μm (3). Calibration of the quadrant photodiode by moving a stuck bead gave a value of ∼12.9 V/μm. Several pulls were made for each tagged flagellum, and the escape voltages were recorded. Pulling was done by slowly translating the stage over a period of about 1 min, which moved the cell body directly away from the trapped bead. As noted above, the bead either detached from the filament or was pulled out of the trap. In the latter case, the bead was returned to the trap and the filament was pulled again, typically from 5 to 20 more times. The peak voltage was consistent from pull to pull, and most tethered beads escaped from the trap at ∼5 V, corresponding to a displacement of ∼0.39 μm or a force of ∼60 pN. Figure 1 shows the results of a typical experiment. Based on our previous measurement of filament stiffness (3.5 pN/μm2) (4), we expected this force to change the filament shape roughly as observed. Polymorphic transformations were not apparent; the filaments merely stretched until they were nearly straight, and the flagellum assumed a uniform long-pitch helical form, just as one would observe when pulling on a helical spring.
FIG. 1.
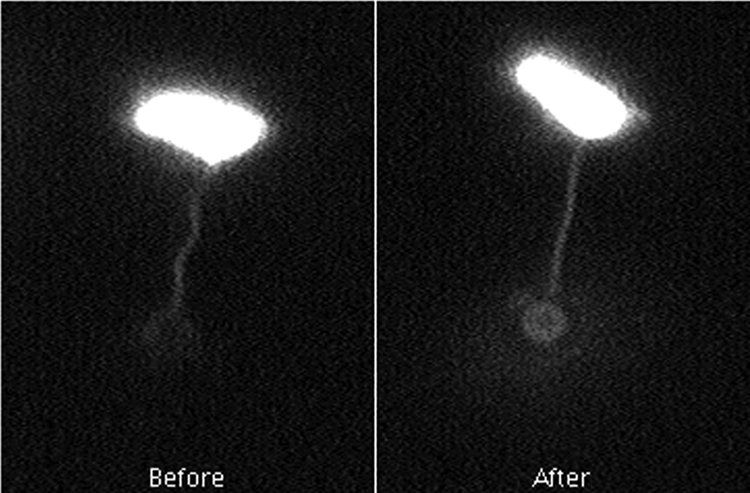
Shape change during stretching. The images show a cell before the stretch and a cell after the stretch, just before the bead escaped from the trap. The bead is in better focus in the latter image, where the pitch of the helix is ∼1.35 times normal and the radius of the helix is ∼0.33 times normal. The images are single frames from a video recording.
In no case were we able to extract a filament from a cell, regardless of the direction of pull, whether we used wild-type strain SJW1103, fliF mutant SJW3060, or fliL mutant UA74, SJW2295, or SJW2296. As determined using Stokes’ law, the force required to pull a sphere with a radius 1 μm through water at a speed of 30 μm/s is ∼0.6 pN (2), so the tensile stress that filaments of Salmonella might experience during swimming is at least 100 times less than the maximum force that we applied. We concluded that the Salmonella flagellar filaments are firmly anchored.
Okino et al. (9) found that when cells of strain SJW3060 swam in 4% gelatin, large bundles (much larger than the cell bodies) were released into the viscous medium, but only if the flagellar motors switched frequently from clockwise to counterclockwise. The filaments appeared to break at the proximal end of the outermost rod protein, FlgG; only this protein was released with the hook and the filament, even though the mutation was in the MS ring, to which the inner rod proteins are thought to attach. But the process appears to be different with fliL null mutants, because shedding required that motors rotate but did not require that motors switch (1); once again, the filaments appeared to break at the proximal end of FlgG. In either case, the structural failure might have been due to torsional rather than tensile stress. We did move flagella around cells in an attempt to mimic torsional stress, but since symmetric beads can rotate freely within the optical trap, we could not apply much torque in this way; we probably only flexed the flagellar hook.
It would be interesting to apply larger tensile stress using the probe of a scanning force microscope or a flexible quartz fiber, although it might be necessary to covalently link filaments to such probes. Superparamagnetic beads could be employed with a magnetic trap to exert large torques. The shedding of filaments observed in gelatin or on agar may involve the interactions of many cells, since in gelatin very large bundles of filaments were shed and on agar cells live in a crowded environment. And why do the filaments appear to break at the distal rod protein, given that the defect in the fliF mutant must be at the point of attachment of a proximal rod protein to the MS ring? Perhaps the proximal rod proteins, once free of the cell wall, depolymerize. Finally, why is the attachment of the filament so much more robust than appears to be required for freely swimming cells? This is presumably because life in the world of crowds, gels, or multiple interfaces is more demanding than swimming in bulk fluid. A cell's flagella are engineered to resist externally imposed forces that are at least 2 orders of magnitude greater than those produced by an isolated swimming cell.
Acknowledgments
We thank Rasika Harshey and Shigeru Yamaguchi for supplying the bacterial strains used in this study and the referees for useful comments.
This work was funded by grant AI066540 from the National Institutes of Health.
Footnotes
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Attmannspacher, U., B. E. Scharf, and R. M. Harshey. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68328-341. [DOI] [PubMed] [Google Scholar]
- 2.Berg, H. C. 1993. Random walks in biology. Princeton University Press, Princeton, NJ.
- 3.Berg-Sørensen, K., and H. Flyvbjerg. 2004. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 75594-612. [DOI] [PubMed] [Google Scholar]
- 4.Darnton, N. C., and H. C. Berg. 2007. Force-extension measurements on bacterial flagella: triggering polymorphic transformations. Biophys. J. 922230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnton, N. C., L. Turner, S. Rojevsky, and H. C. Berg. 2007. On torque and tumbling in swimming Escherichia coli. J. Bacteriol. 1891756-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahrner, K. A., S. M. Block, S. Krishnaswamy, J. S. Parkinson, and H. C. Berg. 1994. A mutant hook-associated protein (HAP3) facilitates torsionally-induced transformations of the flagellar filament of Escherichia coli. J. Mol. Biol. 238173-186. [DOI] [PubMed] [Google Scholar]
- 7.Kamiya, R., and S. Asakura. 1976. Flagellar transformations of Salmonella flagella in vitro. J. Mol. Biol. 106167-186. [DOI] [PubMed] [Google Scholar]
- 8.Macnab, R. M., and M. K. Ornston. 1977. Normal-to-curly flagellar transitions and their role in bacterial tumbling: stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 1121-30. [DOI] [PubMed] [Google Scholar]
- 9.Okino, H., M. Isomura, S. Yamaguchi, Y. Magariyama, S. Kudo, and S.-I. Aizawa. 1989. Release of flagellar filament-hook-rod complex by a Salmonella typhimurium mutant defective in the M ring of the basal body. J. Bacteriol. 1712075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner, L., W. S. Ryu, and H. C. Berg. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 1822793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130255-265. [DOI] [PubMed] [Google Scholar]


