Abstract
Quinolone-resistant and CTX-M-15-producing Escherichia coli isolates belonging to clone ST131 have been reported in the community. This study was designed to identify these E. coli isolates in the stools of 332 independent healthy subjects living in the area of Paris, France. Stools were plated on media without antibiotics, in order to obtain the dominant (Dm) fecal E. coli strain, and with nalidixic acid (NAL) and cefotaxime. Quinolone susceptibility, phylogenetic groups, and molecular profiles, including multilocus sequence types (ST), were determined for all NAL-resistant (NAL-R) isolates. Groups were also determined for the Dm strains from participants with NAL-R isolates and from a subgroup without NAL-R isolates. All B2 isolates were typed; pulsed-field gel electrophoresis was performed for the ST131 isolates, and the results were compared with those for intercontinental clone ST131. Two participants (0.6%) had extended-spectrum β-lactamase-producing (SHV-2, TEM-52) fecal E. coli isolates, and 51 (15%) had NAL-R isolates; 51% of NAL-R isolates belonged to phylogenetic group A, 31% to group D, 16% to group B2, and 2% to group B1. The Dm strain was NAL-R in 3.3% of the 332 subjects. Forty-nine percent of the NAL-R isolates belonged to clones: ST10 and ST606 for group A isolates, ST117 and ST393 for group D isolates. Of all B2 isolates studied from 100 subjects (8 NAL-R strains; 19 NAL-susceptible dominant strains), 52% belonged to three clones: ST131 (n = 7), ST95 (n = 4), and ST141 (n = 3). This is the first study to show the presence of fecal E. coli isolates of clone ST131 in 7% of independent healthy subjects not colonized by CTX-M-15-producing isolates.
Escherichia coli, a universal commensal of humans and several animal species, is also one of the most common enterobacterial species to cause extraintestinal infections in their hosts (30). According to several recent publications, E. coli isolates producing extended-spectrum β-lactamases (ESBL) of the CTX-M type have emerged in the community in numerous countries (25-28). These isolates resist extended-spectrum cephalosporins because of CTX-M production and are often resistant to other antibiotic families, in particular fluoroquinolones and/or cotrimoxazole (28). This pattern of multidrug resistance is dangerous for the treatment of community-acquired infections, because these drugs are often prescribed by general practitioners, especially for urinary tract infections (UTIs). Moreover, a clonal group producing CTX-M-15 has been identified among fluoroquinolone-resistant and CTX-M-producing E. coli isolates (phylogenetic group B2, O25:H4, ST131) in both inpatients and outpatients all over the world, strongly suggesting that this clone can disseminate widely (20).
Because of these recent data and because the main reservoir of extraintestinal pathogenic E. coli isolates is the human digestive tract (8), the aim of this study was to identify CTX-M-15-producing E. coli isolates and E. coli clone ST131 in the guts of healthy adult subjects living in the area of Paris, France.
MATERIALS AND METHODS
Participants.
Fresh stool specimens were provided by 332 healthy adult volunteers who visited the Medical Center IPC (located in Paris) for a checkup between 14 and 27 February 2006. All participants provided informed consent, and the protocol was reviewed and approved by the relevant institutional review board of the medical center.
The physician who examined the participants collected demographic data (age, gender, weight, and address) and clinical data concerning hospital stays and antibiotic intake in the past month, as well as any occupational activity in a health care setting. Information about hospital stays and work in a health care setting was also obtained for people living with the participants, as well as information about any visits made to the IPC center at the same time or during the study period. Stools and participant files were transferred anonymously to the microbiological department of Hôpital Beaujon every day, and stools were stored at −80°C until use.
Sample processing and bacterial strains.
Aliquots (50 mg) of each stool specimen were spread onto two Drigalski agar plates (Bio-Rad, Marnes la Coquette, France) supplemented with either 0.5 mg/liter of cefotaxime (CTX) or 20 mg/liter of nalidixic acid (NAL) and were incubated for 48 h at 37°C. Putative E. coli colonies were selected arbitrarily from each plate. Biochemical identification was performed by using the API 20E system (bioMérieux, Marcy l'Etoile, France).
The dominant (Dm) fecal E. coli strain was obtained from subjects with fecal E. coli isolates resistant (R) to NAL and from a randomly chosen subgroup of subjects without NAL-R isolates, as follows. An aliquot (50 mg) of stools was spread onto a Drigalski agar plate and incubated for 48 h at 37°C. From the terminal streak area of each plate, an isolated colony suspected to be E. coli was chosen. If multiple morphologies were noted, all unique morphotypes were sampled. Each colony selected was identified using the API 20E system (bioMérieux). Since statistically a strain needs to be fairly prevalent within the fecal flora to be recovered from the terminal streak area, the colony selected was considered to represent the Dm E. coli strain (18).
Strain T1, a representative strain of clone ST131 (20), was used as a control strain for the enterobacterial repetitive intergenic consensus 2 (ERIC-2) PCR typing method.
ESBL detection and characterization.
E. coli isolates from CTX-supplemented plates were tested for ESBL production by using the double-disk synergy test described by Jarlier et al. (13). The type of ESBL produced by E. coli isolates with a positive double-disk synergy test was determined by using bla gene-specific primers and the PCR conditions described previously (15).
Antibiotic susceptibility testing.
E. coli isolates from NAL-supplemented plates were tested by disk diffusion for susceptibility to NAL and ciprofloxacin (CIP) by using the method and interpretative criteria recommended by the French Antibiogram Committee (1).
Phylogenetic classification.
Phylogenetic groups were determined by a multiplex PCR assay (6).
ERIC-2 PCR typing.
The genetic diversity of the NAL-R isolates within each phylogenetic group was determined by the ERIC-2 PCR typing method as previously described (20). This method was also applied to Dm strains when the NAL-R isolate and the Dm strain of a subject belonged to the same phylogenetic group, and it was also applied to all isolates found to belong to group B2 in this study.
MLST.
The NAL-R isolates and all group B2 isolates with identical ERIC-2 PCR profiles upon visual comparison were further analyzed by multilocus sequence typing (MLST) as previously described (20).
Serotyping and PFGE typing of isolates identified as E. coli ST131 by MLST.
O and H antigen determination was performed as previously described (24). XbaI PFGE analysis was performed as previously described (2). Profiles were compared digitally using BioNumerics software (Applied Maths). Cluster analysis of Dice similarity indices based on the unweighted-pair group method using average linkages (UPGMA) was used to generate a dendrogram describing the relationships among PFGE profiles. Isolates were considered to belong to the same PFGE group if their Dice similarity index was ≥85% (5). The PFGE profiles of isolates identified as E. coli ST131 in the present study were compared with those displayed by the 36 intercontinental ST131 isolates producing CTX-M-15, and the Spanish ST648 strain FV7591, producing CTX-M-1, that we previously studied and published (20).
Statistical analysis.
Continuous variables were compared by Fisher's exact test, while the t test was used to compare age and weight. A P value of ≤0.05 was considered statistically significant.
RESULTS
Prevalence of ESBL-producing and quinolone-resistant fecal E. coli isolates.
Fresh stools were obtained from 332 healthy adult subjects. Demographic data were available for 329. All subjects lived in houses or flats in the Paris area. None of the participants lived in the same house as any other participant in the study. There were 187 men and 142 women, with a mean age of 60 years (range, 24 to 88 years). Globally, 2% of the participants had been hospitalized and 8.5% had been given antibiotics within the month prior to stool sampling.
Two of the 332 subjects (0.6%) had fecal E. coli isolates from the CTX-supplemented plates that were positive by the double-disk synergy test. These two isolates harbored genes encoding ESBL SHV-2 and TEM-52, respectively.
Fecal E. coli isolates from 51 (15%) of the 332 subjects were selected on NAL-supplemented plates. Forty-nine percent of these NAL-R isolates were classified as resistant to CIP, resulting in a 7% prevalence of CIP-R fecal E. coli isolates among the 332 healthy subjects studied.
As shown in Table 1, the 51 subjects with NAL-R isolates did not differ from the subjects without NAL-R isolates with respect to demographic or clinical criteria, particularly hospital stays and antibiotic intake in the month before stool sampling.
TABLE 1.
Demographic and clinical data for 329 healthy subjects according to NAL-R E. coli digestive carriage status
| Parameter | Value for subjects:
|
|
|---|---|---|
| With NAL-R E. coli | Without NAL-R E. coli | |
| No. of subjects | 51 | 278 |
| Male | 33 | 154 |
| Female | 18 | 124 |
| Mean age (yr) | 61 | 60 |
| Male | 62 | 58 |
| Female | 59 | 61 |
| Mean wt (kg) | 71 | 71 |
| Male | 76 | 79 |
| Female | 65 | 63 |
| No. working in health care settings | 1 | 2 |
| No. with prior hospitalization | 0 | 7 |
| No. with prior antibiotherapy | 5 | 23 |
| No. of households with: | ||
| Work in health care settings | 0 | 5 |
| Prior hospitalization | 3 | 12 |
Phylogenetic groups and ERIC-2 PCR typing of E. coli NAL-R isolates.
The phylogenetic groups of the 51 NAL-R isolates were determined in order to know whether E. coli isolates of group B2, clone ST131, were present among the quinolone-resistant fecal isolates, even though these isolates did not produce CTX-M-15. Sixteen percent of these isolates belonged to group B2, whereas 51% belonged to group A, 31% to group D, and 2% to group B1 (Table 2). The eight NAL-R group B2 isolates were typed by the ERIC-2 PCR method and compared to strain T1, the representative strain of clone ST131 in this study. Four unique profiles were found in these eight isolates; two profiles (B2.I and B2.II) were each found in a single isolate, and two other profiles, B2a and B2b, were found in four and two isolates, respectively (Table 3). Moreover, profile B2a was absolutely identical to the profile of strain T1 (data not shown).
TABLE 2.
Phylogenetic groups of the isolates of three E. coli populations
| Populationa | Number (%) of isolates in group:
|
Total no. of participants studied | |||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| NAL-R E. coli | 26 (51) | 1 (2) | 8 (16) | 16 (31) | 51 |
| Dm strains of subjects: | |||||
| With NAL-R E. coli | 22 (43)b | 6 (12) | 5 (10) | 18 (36)c | 51 |
| Without NAL-R E. coli | 23 (47) | 4 (8) | 14 (28) | 8 (16) | 49 |
| Total | 71 | 11 | 27 | 42 | 100 |
The NAL-R E. coli population and the population of Dm strains of subjects with NAL-R E. coli come from the same 51 participants.
Seven of these isolates were NAL-R.
Four of these isolates were NAL-R.
TABLE 3.
Phylogenetic groups and ERIC-2 PCR and ST profiles of 51 NAL-R isolates
| Participant code(s) | Phylogenetic group | ERIC-2 PCR profile | ST profile |
|---|---|---|---|
| 105a | A | A.I | |
| 114 | A | A.II | |
| 115 | A | A.III | |
| 136 | A | A.IV | |
| 172a | A | A.V | |
| 175 | A | A.VI | |
| 228a | A | A.VII | |
| 247b | A | A.VIII | |
| 282 | A | A.IX | |
| 286 | A | A.X | |
| 289 | A | A.XI | |
| 291a | A | A.XII | |
| 312 | A | A.XIII | |
| 60,a 79, 17, 184,a 267 | A | Aa | ST10 |
| 159, 272, 305,a 311 | A | Aa | ST167 |
| 47 | A | Aa | ST709 |
| 203 | A | Aa | ST744 |
| 295, 296 | A | Ab | ST606 |
| 224 | B1 | NAc | |
| 36 | B2 | B2.I | |
| 22 | B2 | B2.IId | |
| 2, 183, 39, 187 | B2 | B2a | ST131 |
| 74, 190 | B2 | B2b | ST95 |
| 8 | D | D.I | |
| 12a | D | D.II | |
| 24 | D | D.III | |
| 34 | D | D.IV | |
| 53 | D | D.V | |
| 84 | D | D.VI | |
| 90 | D | D.VII | |
| 151a | D | D.VIII | |
| 156a | D | D.IX | |
| 327 | D | D.X | |
| 11,a 208, 325 | D | Da | ST117 |
| 223, 313, 314 | D | Db | ST393 |
The NAL-R isolate was the Dm fecal E. coli strain in the subject.
Subject carrying the SHV-2-producing isolate.
NA, not available.
This ERIC-2 PCR profile was also found in two Dm susceptible group B2 strains.
To ascertain whether clonal strains also existed among isolates from other phylogenetic groups, ERIC-2 PCR profiles were determined for isolates from groups A and D. As shown in Table 3, the 26 NAL-R group A isolates displayed 15 profiles, of which 13 (A.I to A.XIII) were specific for 1 isolate each, 1 (Aa) was specific for 11 isolates, and 1 (Ab) was specific for 2 isolates. The 16 NAL-R group D isolates displayed 12 profiles, of which 2, Da and Db, were found in 3 isolates each and 10 (D.I to D.X) were each specific for 1 isolate (Table 3). Thus, ERIC-2 PCR showed that 25 (49%) of the 51 NAL-R fecal E. coli isolates identified in the 332 healthy subjects were clonal strains.
MLST of the NAL-R isolates identified as clones by ERIC-2 PCR typing.
MLST was used to confirm the clonal relatedness of the NAL-R isolates with identical ERIC-2 PCR profiles and to characterize the different clones. Identical sequence types (ST) were found for the group B2 isolates: ST131 for the four isolates with the B2a profile and ST95 for the two isolates with the B2b profile (Table 3). The 11 group A isolates with the ERIC-2 PCR profile Aa showed four ST: ST10, ST167, ST709, and ST744 (Table 3). However, since they differed from each other by a single allelic sequence, these four ST belonged to clone complex ST10. The two group A isolates with the Ab profile by ERIC-2 PCR had identical ST (ST606). The three group D isolates with the Da profile by ERIC-2 PCR displayed ST117, whereas those with the Db profile displayed ST393 (Table 3). Overall, the MLST method confirmed the ERIC-2 PCR-based clonal relatedness of 49% of the NAL-R isolates detected in 51 independent healthy subjects and characterized nine clones, including clone ST131 (Table 3).
Rates of NAL-R isolates and NAL-R clonal strains among the Dm strains.
The phylogenetic groups of the Dm strains of the 51 participants with NAL-R isolates were determined. When the Dm strain and the NAL-R isolate in a given participant belonged to the same phylogenetic group, their ERIC-2 PCR profiles were determined. Thus, the phylogenetic group distribution of the Dm strains of subjects with NAL-R fecal isolates did not differ significantly from that of the NAL-R isolates (Table 2). In 11 (21.5%) cases, the NAL-R isolate and the Dm strain belonged to the same group and had identical ERIC-2 PCR profiles. These cases included seven group A and four group D isolates, four (36%) of which were clonal strains (two ST10, one ST167, and one ST117 strain). As a result, the NAL-R isolates were more often subdominant (n = 40) than Dm (n = 11) strains. Five (45%) of the 11 Dm NAL-R strains were CIP-R.
Overall, 11 (3.3%) of the 332 subjects in the study had a NAL-R Dm strain; 4 had (1.2%) a clonal NAL-R Dm strain; 5 (1.5%) had a CIP-R Dm strain; and none had a NAL-R group B2 Dm strain.
Identifying clones in Dm strains of group B2.
Table 2 shows that only 10% (n = 5) of the Dm strains of the 51 participants with NAL-R isolates belonged to group B2 and that all five of these strains were susceptible (S) to NAL. The Dm strains of a subgroup of 49 randomly chosen participants without NAL-R isolates were studied in order to determine (i) whether the rate of group B2 isolates in participants without NAL-R isolates was different from that in participants with NAL-R isolates and (ii) whether clones existed among the NAL-S group B2 Dm strains. Analysis of the phylogenetic group distribution of the Dm strains of participants without NAL-R isolates (Table 2) showed that it differed, but not significantly, from that of the Dm strains of participants with NAL-R isolates. The 14 NAL-S group B2 Dm strains of participants without NAL-R isolates plus the 5 NAL-S group B2 Dm strains of participants with NAL-R isolates were analyzed by the ERIC-2 PCR and MLST methods and compared to the 8 NAL-R group B2 subdominant isolates (Table 4). Profiles B2a and B2b, identified in four and two subdominant NAL-R isolates (Table 3), respectively, were also found in three and two NAL-S Dm strains, respectively (Table 4). As expected, Dm strains with profile B2a had ST131 and those with profile B2b had ST95. A new clonal B2 strain (B2c) was identified by comparing the ERIC-2 PCR profiles of the 8 NAL-R and 19 NAL-S isolates. The NAL-R isolate of subject 22 (profile B2.II) (Table 3) and two NAL-S Dm strains (Table 4) belong to this clone, whose ST was shown to be ST141.
TABLE 4.
Distribution of group B2 isolates into different clones according to their susceptibility to NAL
| Strain | No. of isolates in clone:
|
Total no. of:
|
|||
|---|---|---|---|---|---|
| B2a (ST131) | B2b (ST95) | B2c (ST141) | Clonal isolates | Group B2 isolates | |
| NAL-R | 4 | 2 | 1 | 7 | 8 |
| NAL-S Dm | 3 | 2 | 2 | 7 | 19 |
| Total | 7 | 4 | 3 | 14 | 27 |
Overall (Table 4), 7 (37%) of the 19 NAL-S Dm strains belonging to group B2 (from the group of 100 healthy subjects whose Dm strains were studied) were clonal strains. In contrast, clonal strains constituted 87% (7/8) of the group B2 isolates among the NAL-R subdominant isolates detected in 51 of the 100 healthy subjects studied. Thus, 14 (52%) of the 27 group B2 fecal isolates studied were clonal strains. Fifty percent of the B2 clonal strains belonged to clone ST131 (Table 4), identified in 7 of the 100 healthy subjects.
Serotyping and PFGE of seven ST131 isolates.
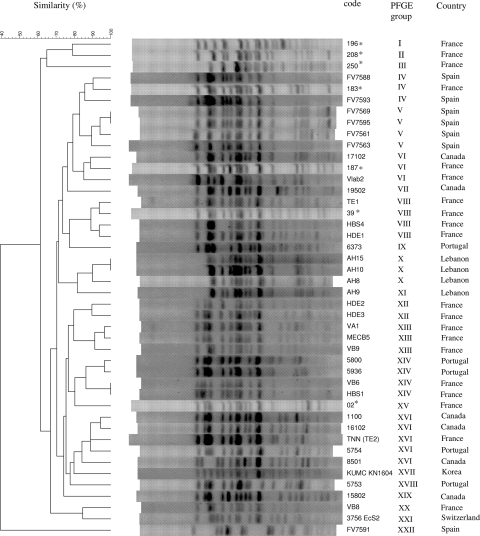
The seven B2 isolates belonging to ST131 (Table 4) were serotyped and shown to belong to serotype O25:H4. Their XbaI PFGE profiles were compared to those of 36 intercontinental CTX-M-15-producing E. coli clinical isolates previously shown to belong to clone ST131 (20). Figure 1 shows the dendrogram generated from the PFGE profiles of the 43 ST131 isolates. Overall, 21 PFGE groups with a similarity index of ≥85% were identified. Four of the seven isolates (Dm strains from subjects 196, 208, and 250 and the NAL-R strain from subject 02) each belonged to a unique PFGE group. In contrast, the three remaining NAL-R isolates belonged to three different PFGE groups, which included other, previously described ST131 isolates (Fig. 1). Thus, the NAL-R isolate of subject 183 was in the same PFGE group as two Spanish strains; that of subject 187 was in the same PFGE group as a Canadian and a French strain; and that of subject 39 was in the same PFGE group as three French strains.
FIG. 1.
XbaI PFGE dendrogram of 43 Escherichia coli ST131 isolates and a Spanish ST648 strain. The 7 ST131 fecal isolates analyzed in the present study (asterisked) were compared with 36 intercontinental ST131 isolates and the Spanish ST648 strain, which were published previously (24). The dendrogram for the 44 isolates, produced by the UPGMA algorithm based on Dice similarity coefficients, included 22 PFGE groups defined on the basis of ≥85% similarity of PFGE profiles.
Overall, the 43 ST131 strains constituted one large cluster (defined by a 62% similarity level), which was associated with the “outgroup” ST648 strain FV7591 (PFGE profile XXII) by <40% similarity (Fig. 1).
DISCUSSION
Recent studies have shown a significant increase in the number of CTX-M-producing E. coli isolates, including CTX-M-15-producing E. coli clone ST131, in the community (19, 22, 25, 26, 28). Assessment of this increase has been based on the increase in the number of CTX-M-positive clinical samples obtained from outpatients and inpatients at hospital admission. However, appropriate studies to effectively measure the prevalence of CTX-M-positive E. coli isolates in the community—for example, prospective studies evaluating the fecal carriage of ESBL-producing E. coli isolates in healthy volunteers without any recent contact with health care settings and without recent exposure to antibiotics—remain scarce. One study performed in 2005 by Palecchi et al. on healthy children in South America reported an ESBL-producing fecal E. coli prevalence of 1.7%, with a predominance of CTX-M enzymes (23). A second study, by Valverde et al., in 2003, reported a higher prevalence of ESBL-positive E. coli isolates (3.7%) but a lower proportion of CTX-M enzymes (50% CTX-M and 50% SHV-12) in 108 independent healthy Spanish (Madrid) volunteers (32). Despite the apparent similarity of our subjects to those of Valverde et al., our results were quite different: 0.6% of our 332 healthy subjects had E. coli fecal isolates producing ESBL that were not CTX-M enzymes. The difference in these results may seem surprising, since both studies were performed several years after the emergence and dissemination of CTX-M enzymes in the respective countries (9, 15, 21, 29), and both were performed on healthy subjects. However, there are no data on the risk factors of fecal carriage of CTX-M enzymes in healthy adult subjects, living in large urban areas of developed countries, who have had no recent contact with health care settings and no recent exposure to antibiotics. Thus, it is difficult to explain the conflicting results between the Valverde study and ours. No information was given on the mean age of the 108 healthy participants in the former study. In that study, the four subjects with ESBL-producing E. coli isolates were 23 to 25 years old, so our population seems to have been older (mean age, 60). If age affects the prevalence of CTX-M-positive fecal E. coli isolates, the difference between the Spanish and French studies could be related to this factor. Moreover, no details were given about how the healthy Spanish volunteers were chosen and their potential relationships. Our 332 subjects belonged to distinct households, and most probably had no relationship with each other, because their residences extended over an area of 12,000 km2 with 11,500,000 inhabitants. If social and geographical links between healthy subjects have an impact on the prevalence of CTX-M-positive fecal E. coli isolates, more data about these parameters in the Spanish study would be interesting.
The source of CTX-M-producing fecal isolates in independent healthy subjects without direct or indirect (via households) contact with any health care setting could be food. Although food-producing animals have been shown to be infected or colonized by E. coli strains producing CTX-M-1 and -15 in France (11, 17), food-borne CTX-M enzymes do not seem to be a significant health concern, since none of our independent healthy subjects was found to be positive for CTX-M enzymes in fecal E. coli isolates.
In contrast, food could be a significant source of human fecal NAL-R E. coli strains, as previously suggested (14), since 15% of our independent healthy subjects harbored such strains in the gut. Once again, comparing our results to previous results is difficult, because the populations studied are often not comparable. Thus, Garau et al. found a 24% prevalence of NAL-R fecal E. coli in 104 Spanish (Barcelona) adults resembling our participants except that they were visiting the hospital emergency room for a noninfectious disease when they were included in the study (10). Bruinsma et al. found a prevalence of 1% to 12% depending on the country (Canada, Greece, or The Netherlands) where healthy volunteers in this study lived (4), while Grenet et al. found a prevalence of 8% in the Wayampis Amerindians, an isolated community in French Guyana (12). Globally, our results are similar to those previously published. However, we showed that only 20% of the NAL-R fecal isolates were Dm strains. Distinguishing between Dm and subdominant fecal strains seems to be relevant with regard to E. coli extraintestinal pathogenesis. Indeed, Moreno et al. showed that the prevalence of fecal isolates, notably those less virulent, is an important determinant for UTI pathogenesis in women (18). This finding suggests that the risk of having a UTI caused by an antibiotic-resistant E. coli isolate could be higher in subjects in whom the Dm strain is resistant to antibiotics than in subjects in whom the antibiotic-resistant isolates are subdominant.
Another important result was the presence of clonal fecal E. coli isolates in independent healthy subjects. Almost 50% of the NAL-R isolates were clonal strains, and the clonal structure was independent of the phylogenetic groups. The use of the MLST method made it possible to characterize these clones (n = 9) and identify the ST131 clone (group B2), which had been reported, until this study, for isolates that produced the ESBL CTX-M-15 and were resistant to quinolones (20). The discovery of clone ST131 in NAL-R isolates was followed by the discovery of this clone in NAL-S Dm strains. All these results showed that clone ST131 was the dominant clone in the isolates of group B2.
The presence of fecal E. coli clones in apparently epidemiologically unrelated subjects has already been suggested by profile comparison methods (16, 24). The present study clearly identifies the clones (ST) that were suggested by ERIC-2 PCR, which we have confirmed to be an accurate method of detecting clone complexes (31). Thus, clone ST69 (also called CgA), which is widely disseminated in North America (3, 16, 31), was shown to be absent in our healthy subjects by MLST, while clone ST131, which is present on three continents in the form of isolates producing CTX-M-15 (20), was present in the form of isolates free of CTX-M enzymes and S or R to quinolones in 7% of healthy subjects.
PGFE typing showed that the seven ST131 isolates of this study formed a PFGE-based cluster (defined at the 62% similarity level) with the 36 previously published international ST131 isolates (20). This strongly suggests recent divergence from a common ancestor. Furthermore, the marked similarity of the PFGE profiles of some of our fecal E. coli isolates with those of certain clinical isolates producing CTX-M-15 from either France, Spain, or Canada strongly suggests a recent acquisition of CTX-M-15-mediating-plasmids by clone ST131. Although very few studies on the intercontinental clone ST131 are available, it has been shown to be a virulent clone that produces biofilms (7, 20). Finding this clone in the guts of independent healthy subjects at a prevalence of 7% could indicate that it has particular, as yet undefined properties.
In conclusion, this study showed that CTX-M-producing fecal isolates were absent, while quinolone-resistant subdominant fecal isolates were highly prevalent, in healthy adults living in the Paris area. We also found that 50% of the NAL-R fecal isolates and 50% of the group B2 fecal isolates were clonal isolates. Finally, this is the first study to show the presence of fecal E. coli isolates of clone ST131 that do not produce CTX-M-15 in 7% of independent healthy subjects.
Acknowledgments
This work was supported by grants from the Fondo de Investigación Sanitaria (FIS-REIPI-RD06/008/1018) and the Xunta de Galicia (07MRU036261PR). A. Mora acknowledges the Ramón y Cajal program of the Spanish Ministry of Education and Science.
We thank F. Bert for advice and help in the writing of this article.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Anonymous. 2003. Comité de l'Antibiogramme de la Société Française de Microbiologie Report 2003. Int. J. Antimicrob. Agents 21364-391. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, M., J. E. Blanco, G. Dahbi, A. Mora, M. P. Alonso, G. Varela, M. P. Gadea, F. Schelotto, E. A. Gonzalez, and J. Blanco. 2006. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (μΒ and ξR/β2Β). J. Med. Microbiol. 551165-1174. [DOI] [PubMed] [Google Scholar]
- 3.Boczek, L. A., E. W. Rice, B. Johnston, and J. R. Johnson. 2007. Occurrence of antibiotic-resistant uropathogenic Escherichia coli clonal group A in wastewater effluents. Appl. Environ. Microbiol. 734180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruinsma, N., J. M. Hutchinson, A. E. van den Bogaard, H. Giamarellou, J. Degener, and E. E. Stobberingh. 2003. Influence of population density on antibiotic resistance. J. Antimicrob. Chemother. 51385-390. [DOI] [PubMed] [Google Scholar]
- 5.Carrico, J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazao, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 435483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 664555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont, O., M. Lavollay, S. Vimont, C. Deschamps, C. Forestier, C. Branger, E. Denamur, and G. Arlet. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 611024-1028. [DOI] [PubMed] [Google Scholar]
- 8.Donskey, C. J. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39219-226. [DOI] [PubMed] [Google Scholar]
- 9.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 481249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garau, J., M. Xercavins, M. Rodriguez-Carballeira, J. R. Gomez-Vera, I. Coll, D. Vidal, T. Llovet, and A. Ruiz-Bremon. 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 432736-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girlich, D., L. Poirel, A. Carattoli, I. Kempf, M. F. Lartigue, A. Bertini, and P. Nordmann. 2007. Extended-spectrum β-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 734681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenet, K., D. Guillemot, V. Jarlier, B. Moreau, S. Dubourdieu, R. Ruimy, L. Armand-Lefèvre, P. Bau, and A. Andremont. 2004. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg. Infect. Dis. 101150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10867-878. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J. R., M. R. Sannes, C. Croy, B. Johnston, C. Clapots, M. A. Kuskowski, J. Bender, K. E. Smith, P. L. Winokur, and E. A. Belongia. 2007. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002-2004. Emerg. Infect. Dis. 13838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leflon-Guibout, V., C. Jurand, S. Bonacorsi, F. Espinasse, M.-C. Guelfi, F. Duportail, B. Heym, E. Bingen, and M.-H. Nicolas-Chanoine. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 483736-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 3451007-1013. [DOI] [PubMed] [Google Scholar]
- 17.Meunier, D., E. Jouy, C. Lazizzera, M. Kobisch, and J. Y. Madec. 2006. CTX-M-1- and CTX-M-15 type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28402-407. [DOI] [PubMed] [Google Scholar]
- 18.Moreno, E., A. Andreu, T. Perez, M. Sabate, J. R. Johnson, and G. Prats. 2006. Relationship between Escherichia coli strains causing urinary tract infection in women and the dominant faecal flora of the same hosts. Epidemiol. Infect. 1341015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, M. Perilli, G. Amicosante, S. Stefani, A. Toniolo, and G. M. Rossolini. 2006. CTX-M-type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 502700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61273-281. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas-Chanoine, M. H., V. Jarlier, and “La Collégiale” de Bactériologie-Virologie-Hygiène Hospitalière de l'Assistance Publique, Hôpitaux de Paris, France. 2008. Extended-spectrum β-lactamases in long-term-care facilities. Clin. Microbiol. Infect. 14(Suppl. 1)111-116. [DOI] [PubMed] [Google Scholar]
- 22.Oteo, J., C. Navarro, E. Cercenado, A. Delgado-Iribarren, I. Wilhelmi, B. Orden, C. Garcia, S. Miguelanez, M. Perez-Vazquez, S. Garcia-Cobos, B. Aracil, V. Bautista, and J. Campos. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 442359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallecchi, L., A. Bartoloni, C. Fiorelli, A. Mantella, T. Di Maggio, H. Gamboa, E. Gotuzzo, G. Kronvall, F. Paradisi, and G. M. Rossolini. 2007. Rapid dissemination and diversity of CTX-M extended-spectrum β-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrob. Agents Chemother. 512720-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallecchi, L., C. Lucchetti, A. Bartoloni, F. Bartalesi, A. Mantella, H. Gamboa, A. Carattoli, F. Paradisi, and G. M. Rossolini. 2007. Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 511179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallecchi, L., M. Malossi, A. Mantella, E. Gotuzzo, C. Trigoso, A. Bartoloni, F. Paradisi, G. Kronval, and G. M. Rossolini. 2004. Detection of CTX-M-type β-lactamase genes in fecal Escherichia coli isolates from healthy children in Bolivia and Peru. Antimicrob. Agents Chemother. 484556-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitout, J. D., D. L. Church, D. B. Gregson, B. L. Chow, M. McCracken, M. R. Mulvey, and K. B. Laupland. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary health region: the emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 511281-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitout, J. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 5652-59. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Bano, J., M. D. Navarrro, L. Romero, L. Martinez-Martinez, M. A. Muniain, E. J. Perea, R. Perez-Cano, and A. Pascual. 2004. Epidemiology and clinical features of infections caused by extended-spectrum β-lactamase-producing Escherichia coli in nonhospitalized patients. J. Clin. Microbiol. 421089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero, L., L. Lopez, J. Rodriguez-Bano, J. Ramon-Hernandez, L. Martinez-Martinez, and A. Pascual. 2005. Long-term study of the frequency of Escherichia coli and Klebsiella pneumoniae isolates producing extended-spectrum β-lactamases. Clin. Microbiol. Infect. 11625-631. [DOI] [PubMed] [Google Scholar]
- 30.Smith, J. L., P. M. Fratamico, and N. W. Gunther. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4134-163. [DOI] [PubMed] [Google Scholar]
- 31.Tartof, S. Y., O. D. Solberg, A. R. Manges, and L. W. Riley. 2005. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J. Clin. Microbiol. 435860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valverde, A., T. M. Coque, M. Sanchez-Moreno, A. Rollan, F. Baquero, and R. Canton. 2004. Dramatic increase in prevalence of fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J. Clin. Microbiol. 424769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]