Abstract
The new Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test offers advanced automation for the detection of human immunodeficiency virus type 1 (HIV-1) RNA and DNA in dried blood spots (DBS) and whole blood. An analytical evaluation using an HIV-1 secondary standard yielded limits of detection of 514, 710, and 1,090 HIV RNA copies/ml for EDTA plasma, whole blood, and DBS, respectively. The precision and reproducibility of HIV-1 detection was equivalent for DBS and whole blood. Inclusivity was demonstrated for a reference panel of HIV-1 subtypes A to N. A clinical evaluation of the Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test was performed at a center for routine diagnostics in Johannesburg, South Africa, using 1,013 clinical specimens from HIV-1 exposed children. The Amplicor HIV-1 DNA test v1.5 with the MagNApure DNA isolation procedure was used as the reference method. A total of 995 valid results for whole blood with both methods yielded 691 and 303 concordant negative and positive results for the Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test, respectively. For the 800 valid DBS specimen results, 495 and 300 concordant negative and positive results were obtained, respectively. The resulting clinical specificities and sensitivities of the new test were 100% and 99.7% for whole blood and DBS, respectively. The new test was characterized by its robustness, enhanced automation, and improved sample throughput. The Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test will support early, reliable diagnosis of HIV in children in routine laboratory settings.
Globally at least 33.2 million individuals were estimated to be infected with human immunodeficiency virus (HIV) in 2007 (8). Two-thirds of HIV-infected adults and 90% of infected children reside in sub-Saharan Africa. Of those infected, 2.5 million represent new infections and more than 420,000 are infants (8). AIDS thus represents the most serious threat to the public health systems of many African countries.
HIV is transmitted largely by sexual contact, exposure to infected blood products (5), and vertical mother-to-child transmission (10). Vertical transmission can occur during pregnancy, childbirth, and postnatally via breastfeeding (1, 10, 14). For many infected infants, the progression of AIDS is rapid (20), with more than 50% of infants dying before the age of 2 years in the absence of antiretroviral (ARV) and/or prophylactic antibiotic treatment (2, 15). In a randomized, controlled clinical trial in South Africa, it was demonstrated that there was an increase in survival among infants who received immediate ARV therapy (96%) than among infants who received ARVs following significant immunological deterioration (84%) (13). Early confirmation of a pediatric infection is thus critical to enable immediate referral to care programs and will go a long way to reducing maternal anxiety (26). In addition, the identification of HIV-1-negative children would save follow-up clinic costs and reduce the need for antibiotic prophylaxis. Finally, the early diagnosis of HIV infection would provide a reliable measure of the efficacy of mother-to-child-transmission prevention programs.
Passively transferred maternal HIV type 1 (HIV-1) antibodies may be detectable in the HIV-exposed infant up to 18 months of life. Routine serological antibody testing is thus not informative of the true virologic status of HIV-exposed infants. In contrast, the direct detection of the virus or viral particles allows for the early diagnosis of pediatric HIV-1 infection (22). Thus, assays detecting HIV-1 nucleic acid are generally used for HIV-1 diagnosis in infants younger than 18 months. In South Africa, national guidelines recommend a PCR-based assay for HIV diagnosis at 6 weeks of age in all HIV-exposed infants unless they are ill, in which case the test is done earlier (12). The difficulties of implementing nucleic acid testing strategies should not be underestimated in resource-limited settings. The complexity of testing, costs of laboratory equipment, lack of skilled technicians, and difficulties of sample cold-chain transfer and blood collection from infants may limit the availability for routine diagnosis (25). The use of dried whole-blood spots (DBS) address the sample collection and transport difficulties and have been validated for the neonatal screening of many different disorders, not the least of which is the reliable PCR-based determination of HIV infection (3, 4, 18).
Currently, many routine diagnostic laboratories in southern Africa use the Amplicor HIV-1 DNA test v1.5 system (Roche Molecular Systems, Inc., Branchburg, NJ) for the qualitative detection of HIV-1 infection in infants (4, 6, 18, 25). This test uses the endpoint determination of PCR-amplified products and can be performed using either liquid blood or DBS. DBS can be prepared by manual excision or, as recently introduced, by automated punching with the BSD1000 gene punch instrument (BSD Robotics; Queensland, Australia). In the current format, sample preparation and detection require significant manual intervention.
In order to minimize user interventions and improve sample throughput, a new assay was designed by Roche for the Cobas AmpliPrep/Cobas TaqMan (CAP/CTM) system, making use of automated sample extraction connected to real-time PCR technology. The CAP/CTM system was initially developed for viral-load determination and is currently used to quantify HIV-1 RNA in EDTA plasma for monitoring the response to ARVs (21). In contrast, the new CAP/CTM HIV-1 Qual test enables the qualitative detection of HIV-1 RNA and proviral DNA in whole blood and in DBS. The purpose of this study was to evaluate the analytical performance of the CAP/CTM assay as well as to investigate its clinical performance compared to the Roche Amplicor HIV-1 DNA test v1.5 system. To this end, both tests were evaluated under routine diagnostic laboratory conditions in a high-throughput evaluation center in South Africa using clinical specimens from HIV-exposed children.
MATERIALS AND METHODS
Sample collection and preparation.
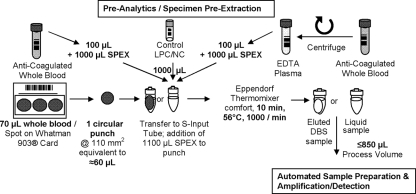
For both the analytical and clinical studies, fresh whole blood was collected in EDTA tubes. DBS were prepared from whole-blood samples within 5 days of the blood draw. Seventy microliters of whole blood was spotted into three circles on the DBS Whatman 903 cards (Fig. 1). The DBS cards were dried for at least 3 h and stored at room temperature in individual ziplock bags containing a desiccant for up to 4 months until analysis. Within 6 h of collection, EDTA plasma was obtained following centrifugation at 800 to 1,000 × g for 15 min and then stored at −80°C. The specimen analysis was conducted as per ethics clearance number M00-01-07 from the University of the Witwatersrand Human Ethics Committee, Johannesburg, South Africa.
FIG. 1.
Workflow of the CAP/CTM HIV-1 Qual test for preextraction processing of the three different specimen types—whole blood, DBS, and EDTA plasma—to support advanced automation and high sample throughput.
Workflow for the CAP/CTM HIV-1 Qual test and Amplicor HIV-1 DNA test v1.5.
The samples were processed first using the Amplicor HIV-1 DNA test v1.5 and then using the CAP/CTM assay. One hundred microliters of blood was extracted using the MagNApure LC DNA isolation kit I according to the high-performance protocol. Fifty microliters of the extracted sample was then used for amplification and detection on the Amplicor HIV-1 DNA test v1.5 as per the manufacturer's instructions. For the CAP/CTM assay, 100 μl blood was added to an S-input tube together with 1,000 μl of specimen preextraction (SPEX) reagent (containing sodium citrate dehydrate, 42.5% guanidine thiocyanate, <5% polydocanol, 1.8% dithiothreitol, and 0.01% citric acid) (see Fig. 1 for a graphical representation). Low positive and negative controls (1 ml of each) were added into separate input S-tubes and incubated in an Eppendorf Thermomixer comfort at 56°C for 30 min with no shaking. The input S-tubes were mixed for 15 s at 1,100 × g, placed on the SK24 sample racks, and then loaded on the CAP instrument. Eight hundred and fifty microliters of liquid sample was then used for amplification and detection on the CTM analyzer. The HIV-1 status of the individual specimen is automatically reported as positive or negative by AmpliLink software. The identical procedure was followed for EDTA plasma analysis. (Fig. 1).
The CAP/CTM HIV-1 Qual test uses the same HIV genome target sequences and reagents as the quantitative CAP/CTM HIV-1 test. In both assays, an HIV-1 internal control (IC) is added to each sample to serve as a control for both the extraction and amplification steps. This HIV-1 IC is a noninfectious armored RNA construct containing HIV sequences with primer binding sites identical to those of the wild-type HIV-1 target. A unique probe binding site allows for the differentiation of the IC-specific amplicon from the HIV-1 target amplicon.
Workflow for the CAP/CTM assay and the Amplicor HIV-1 DNA test v1.5 using DBS.
For the CAP/CTM assay, one 110 mm2 punch (Fig. 1) using either a prototype manual punch tool or manual excision of a spot, was generated. One DBS punch was transferred into each S-input tube followed by the manual addition of 1,100 μl SPEX reagent. Positive and negative controls were prepared as described above.
The input S-tubes were placed on an SK24 rack and processed using the CAP/CTM analyzer combination. Using the same samples, seven circular punches (3.2 mm in diameter, 56 mm2 in total) were prepared by the BSD1000 gene punch instrument for the Amplicor HIV-1 DNA test v1.5. For manual extraction, 500 μl of specimen blood wash reagent was added to the sample punches and incubated for 10 min at room temperature. After the supernatant was removed, the washing was repeated. Thereafter, 200 μl of freshly prepared extraction mix containing the IC was added, followed by consecutive 15-min incubation steps at 60°C and then at 100°C. The extracted sample was then subjected to amplification and detection as per the package insert of the Amplicor HIV-1 DNA test v1.5.
Limit of detection (LOD) of the CAP/CTM assay.
The HIV-1 secondary standard, which is traceable to the first World Health Organization international standard for HIV-1 RNA for nucleic-acid-testing assays (NIBSC code 97/656), was diluted in three independent dilution series with two separate HIV-negative specimen matrices. The first dilution series was conducted with an EDTA plasma pool from multiple donations (stored at −20°C) and the second using fresh, EDTA-anticoagulated whole blood from a single blood donation. The concentration levels were reported in copies/ml. The latter dilution series was then used to prepare DBS on Whatman 903 cards. The same technician executed all dilution series. The LOD evaluation was performed for the EDTA plasma, whole blood, and DBS using two different reagent lots. Each of the three dilution series was tested for each specimen, at five different concentration levels, with 12 replicates in each run. To estimate the LOD concentration, the hit rate profile from all valid replicates was combined and analyzed by applying a 95% probit model including a 95% confidence interval and by using a validated SAS software module.
Precision and reproducibility of the CAP/CTM test for whole blood and DBS.
HIV-negative fresh whole blood was used as a blank. In addition, a concentration level of 2.5× the LOD as determined for the respective specimen type was prepared by serial dilutions of spiked HIV-1 secondary standard. DBS were prepared immediately after the HIV-negative and 2.5× LOD-positive spiked panels were available. Single-day-use aliquots of the whole blood panels were stored frozen to prevent multiple freeze-thaw cycles. The assay reproducibility was tested for each of the two specimen types by measuring 11 replicates of each specimen over 10 working days. This study was completed using two different CAP/CTM reagent lots but the same SPEX reagent lot. Each run was performed with a new CAP/CTM reagent kit of the same lot. The positive results over the 10 performed runs were used to calculate the target hit rate.
Inclusivity of the CAP/CTM assay.
Subtype inclusivity was tested for one reagent lot using the NIBSC HIV subtype reference panel (NIBSC code 1/466), the subtype concentrations of which are certified according to bDNA (Bayer) assay determination. For each of the subtypes A to O, five replicates of 100-μl subtype sample were preincubated on the Eppendorf Thermomixer with 1,000 μl of SPEX reagent prior to processing with the CAP/CTM HIV-1 Qual test.
Clinical samples and study design.
The clinical study was conducted in the Department of Molecular Medicine and Hematology, University of the Witwatersrand, Johannesburg, South Africa. Approximately 1 ml of fresh EDTA whole blood was collected from children (6 weeks to 6 years of age) for routine diagnosis. A total of 1,013 random specimens were tested using the Amplicor HIV-1 DNA test v1.5, followed by the CAP/CTM assay. Three hundred and four were HIV-1 positive using the Amplicor HIV-1 DNA test v1.5. Any positive result with the Amplicor HIV-1 DNA test v1.5 on whole-blood specimens was repeated using the same assay for confirmation.
For DBS evaluation, six 70 μl DBS (two cards) were prepared from fresh whole blood. DBS cards were dried for at least 3 h and stored at room temperature in a bag containing a desiccant until further use. Based on the results for whole blood testing, DBS were analyzed with the identified 304 HIV-positive and an additional 500 randomly chosen HIV-negative specimens. Workflow and processing of the different specimen types are shown in Fig. 1.
Data analysis.
For the Amplicor HIV-1 DNA test v1.5, optical density readings at 450 nm were interpreted as follows: “not detected” (HIV-1 < 0.2, IC ≥ 0.2), negative for HIV-1 DNA; “inhibitory sample” (HIV-1 < 0.2, IC < 0.2), repeat testing once; “HIV-1 detected” (HIV-1 ≥ 1.5, any IC), positive for HIV-1 DNA and repeat testing once; and “equivocal” (0.2 ≤ HIV-1 < 1.5, IC ≥ 0.2), retest specimen in duplicate. If two out of three test results gave a positive result, the specimen was considered positive for HIV-1 DNA. For the CAP/CTM assay, “invalid” or positive valid results were not repeated. The results of the two tests were subjected to qualitative comparison. Samples were excluded from the evaluation when they were “invalid” in the CAP/CTM assay or “internal control inhibited” or “equivocal” or if they could not be repeated due to an insufficient sample in the Amplicor HIV-1 DNA test v1.5.
RESULTS
LOD for specimen types EDTA plasma, whole blood, and DBS.
The results from 72 replicates per concentration level were combined for the LOD calculation. Probit analysis for the EDTA plasma matrix resulted in an LOD of 514 copies/ml (95% confidence interval, 416 to 703 copies/ml) and for the fresh whole blood matrix and DBS, the LODs were 710 copies/ml (95% confidence interval, 571 to 982 copies/ml) and 1,090 copies/ml (95% confidence interval, 910 to 1,443 copies/ml), respectively. The LOD for DBS (60-μl blood equivalent) corresponding to the 110 mm2 of punch were comparable to the 100-μl sample input for EDTA plasma and whole blood.
Precision and reproducibility for DBS.
For 110 measurements, no false-positive or -negative results were observed. In addition, the mean target threshold cycle (CT) values displayed standard deviations of 0.42 and 0.65 for whole blood and DBS, respectively, demonstrating low intra-assay and interassay variability.
Inclusivity.
When testing five replicates per HIV-1 subtype from A to N, the CAP/CTM test displayed a target hit rate of 100% for each subtype. The only exception represented HIV-1 subtype O, which was not detected in any of the five replicates performed.
Clinical evaluation of the CAP/CTM assay using whole blood.
A total of 995 whole-blood specimens (98.7%) yielded valid results for both methods. Thirteen result pairs were excluded because the Amplicor HIV-1 DNA test v1.5 identified the sample result as either “equivocal” (5 of 13) or as “internal control inhibited” (8 of 13). Additionally, five result pairs were excluded because the CAP/CTM assay identified the results as “invalid.”
Both assays yielded 691 and 303 concordant negative and positive results, respectively. One specimen turned out to be discordant, yielding a positive result on the Amplicor HIV-1 DNA test v1.5, but a negative result for the CAP/CTM test, and was classified as false negative. For this specimen, there was no fresh whole blood left for repeat analysis. All other specimens, which were identified positive with the Amplicor HIV-1 DNA test v1.5 in the first run, could be confirmed by repeat analysis. In addition, 691 of the 995 specimens turned out to be concordant negative, and no confirmed false-positive result was obtained for whole blood specimens with either test. The resulting clinical sensitivities and clinical specificities for the two methods are summarized in Table 1.
TABLE 1.
Clinical parameters of the CAP/CTM HIV-1 Qual test and the Amplicor HIV-1 DNA test version 1.5 on clinical whole-blood specimens
| Clinical parameter | Amplicor test
|
CAP/CTM test
|
||||
|---|---|---|---|---|---|---|
| Results | Ratio (%) | 95% Confidence interval (%) | Results | Ratio (%) | 95% Confidence interval (%) | |
| Specificity | 691/691a | 100 | 99.6-100 | 691/691a | 100 | 99.6-100 |
| Sensitivity | 304/304a | 100 | 99.0-100 | 303/304a | 99.7 | 98.5-100 |
| Robustness | 1,000/1,013b | 98.7 | 98.0-99.3 | 1,008/1,013b | 99.5 | 99.0-99.8 |
Number of correct results/number of valid results.
Number of valid results/total number of specimens tested.
Clinical evaluation of the CAP/CTM test with DBS.
Eight hundred and twelve DBS specimens were available for analysis. Eleven sample result pairs were excluded because the Amplicor HIV-1 DNA test v1.5 identified the result either as indeterminate (8 of 11) or as “internal control inhibited” (3 of 11). One additional result pair was excluded from evaluation because the CAP/CTM Test identified the result as “invalid.”
Four hundred and ninety-five of the remaining 800 DBS specimens turned out to be concordant negative (Table 2), matching the results obtained from the same patient specimens on whole blood. No confirmed false-positive results were obtained by the CAP/CTM assay for DBS testing. The Amplicor HIV-1 DNA test v1.5, yielded one false-positive result, which was confirmed on repeat testing. Three hundred of the 800 specimens turned out to be concordant positive (Table 2). Two Amplicor HIV-1 DNA test v1.5 results that were initially equivocal were confirmed positive upon repeat analysis. The matching whole blood specimens’ results were concordant positive. There was one discordant DBS specimen result, for which the Amplicor HIV-1 DNA test v1.5 initially failed to detect HIV-1, but repeat testing yielded a positive result with DBS. However, the CAP/CTM test yielded a negative result in duplicate. This specimen was also discordant in whole blood testing, where it was repeatedly positive in the Amplicor HIV-1 DNA test v1.5 but negative in the CAP/CTM assay.
TABLE 2.
Clinical parameters of the CAP/CTM HIV-1 Qual test and the Amplicor HIV-1 DNA test version 1.5 on clinical DBS specimens
| Clinical parameter | Amplicor test
|
CAP/CTM test
|
||||
|---|---|---|---|---|---|---|
| Results | Ratio (%) | 95% Confidence interval (%) | Results | Ratio (%) | 95% Confidence interval (%) | |
| Specificity | 495/496a | 99.8% | 99.0-100 | 496/496a | 100 | 99.4-100 |
| Sensitivity | 301/304a | 99% | 97.1-99.8 | 303/304a | 99.7 | 98.5-100 |
| Robustness | 801/812b | 98.6 | 97.8-99.3 | 811/812b | 99.9 | 99.4-100 |
Number of correct results/number of valid results.
Number of valid results/total number of specimens tested.
In addition, the Amplicor HIV-1 DNA test v1.5 yielded another three initially false-negative results for DBS specimens, which were repeatedly negative and also classified as false negative since the matching whole-blood specimens were determined positive. The clinical specificities and sensitivities for both methods are summarized in Table 2.
Comparability of the CAP/CTM test results for matched whole blood and DBS specimens.
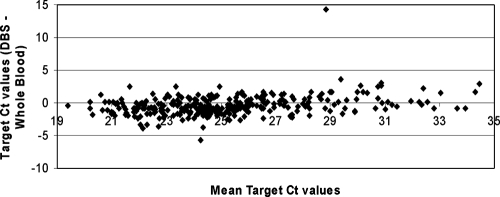
To further investigate the performance of the CAP/CTM test on whole blood and DBS, the 300 positive results, obtained from the clinical evaluation of the two matching specimen types, were compared with regard to their determined CT values. As illustrated by Bland Altman analysis in Fig. 2, target CT values of DBS and matched whole-blood specimens were very similar with minor scattering around −0.4 CT, except for one specimen. This outlier with a delayed DBS target CT value of 36 yielded a whole-blood CT value of 21.7. Repeat testing of this DBS specimen yielded a CT value of 21.5, indicating a DBS sample handling error in initial testing.
FIG. 2.
Comparison of the CAP/CTM HIV-1 Qual test target CT values for DBS and whole blood from 300 matched positive clinical specimens subjected to Bland Altman analysis. The CT value is defined as the number of PCR cycles needed to achieve a specified target threshold fluorescence signal required for reliable detection.
DISCUSSION
The CAP/CTM HIV-1 Qual test demonstrated improved automation for HIV-1 detection in children using whole blood and DBS. The clinical evaluation revealed sensitivities and specificities of 99.7% and 100% for the CAP/CTM assay, respectively, for whole blood and DBS. The performance is thus equivalent to data recently reported for the Amplicor HIV-1 DNA test v1.5 test using DBS (23). The CAP/CTM assay, however, displayed an improved assay robustness of 99.9% versus 98.6% over the gold standard assay demonstrating fewer results as “equivocal” or “internal control inhibited.”
DBS offer huge advantages for the diagnosis of pediatric HIV-1 infection in resource-limited settings (18, 23). DBS collection from infants by heel or finger stick is much easier to perform than liquid blood collection because it requires significantly less phlebotomy skills. Transport to centralized laboratories can be done ambiently with reduced biohazardous risk. Evaluation of DBS using the HIV-1 secondary standard showed an excellent LOD of 1,090 copies/ml with an equivalent input of 60 μl of whole blood, which would improve with higher input volumes. HIV-1 RNA is detected together with proviral HIV-1 DNA, thus potentially improving the clinical sensitivity over assays that only detect HIV-1 DNA. An advantage of combining HIV-1 RNA and DNA detection may be related to the fact that in perinatal infection, HIV-1 RNA is detectable earlier and more reliably than HIV-1 DNA (9, 19, 24). The slightly enhanced LOD observed for EDTA plasma compared to that for whole-blood specimens may indicate a lack of inhibitory effects on the reverse transcriptase PCR procedure. The new assay was able to detect HIV-1 subtypes A to N with an inclusivity equivalent to that of the CAP/CTM HIV-1 test used for viral-load monitoring (21).
The single big punch used for the CAP/CTM test proved to be advantageous over the need for seven smaller punches prepared for the Amplicor HIV-1 DNA test v1.5 as per laboratory protocol. This single punch introduced a greater volume of blood and reduced the impact of disturbing particles or cross-contaminating lint originating from the filter paper. Interestingly, the filter disk, once soaked with SPEX reagent, expands and adheres to the wall of the S-input tube, and no interference of the pipette tip with the filter disk was observed during the clinical trial. Introducing an automated punch instrument similar to the BSD1000 gene punch instrument already used with the Amplicor HIV-1 DNA test v1.5 may further advance the automation and convenience of the test procedure (6). The excision of punches from DBS cards with the BSD1000 gene punch instrument was shown to be associated with a very low risk of sample-to-sample cross-contamination, which represented an improvement over manual punching.
A further advantage of the CAP/CTM assay was that automation reduced the hands-on time and technical skill required to perform the assay by eliminating the extensive sample preparation procedure. Most PCR-based assays published to date for the detection of HIV in resource-limited settings while showing good sensitivity and specificity involve long, laborious DNA extraction protocols (7, 11, 16, 17). Specimen throughput is greatly facilitated using the CAP/CTM assay with 24 and 48 results available within a 5-h and an 8-h work shift, respectively. Combining an 8-h work shift with an unattended overnight run, the system provides ≥144 results by the next morning. For the MagNApure DNA isolation procedure combined with the Amplicor HIV-1 DNA test v1.5, 90 patient samples can be processed within an 8-h period, but constant user intervention is required. The automated workflow ensures the traceability of samples throughout the procedure, reducing the risks for sample mix-up errors and minimizing cross-contamination events.
In conclusion, the CAP/CTM HIV-1 Qual test yielded accurate and reliable results for the detection of HIV-1 infection in DBS and whole blood and displayed clinical specificities and sensitivities equal to that of the Amplicor HIV-1 DNA test v1.5. The results obtained in this study support the use of the CAP/CTM HIV-1 Qual test for the early detection of HIV-1 infection in high-volume, centralized laboratories in resource-limited countries.
Acknowledgments
We thank Wolfram Schumacher and Eveline Akkers, from Roche Diagnostics for their support and technical assistance in this study. We are grateful to Roche Diagnostics GmbH, Germany, who provided us with the reagent kits to perform this study. We thank Heidemarie Peuker, BISs Biomedical Investigation Services, for diligent preparation of the manuscript.
Footnotes
Published ahead of print on 15 October 2008.
REFERENCES
- 1.Bobat, R., D. Moodley, A. Coutsoudis, and H. Coovadia. 1997. Breastfeeding by HIV-1-infected women and outcome in their infants: a cohort study from Durban, South Africa. AIDS 111627-1633. [DOI] [PubMed] [Google Scholar]
- 2.Brahmbhatt, H., G. Kigozi, F. Wabwire-Mangen, D. Serwadda, T. Lutalo, F. Nalugoda, N. Sewankambo, M. Kiduggavu, M. Wawer, and R. Gray. 2006. Mortality in HIV-infected and uninfected children of HIV-infected and uninfected mothers in rural Uganda. J. Acquir. Immune Defic. Syndr. 41504-508. [DOI] [PubMed] [Google Scholar]
- 3.Cassol, S., T. Salas, M. J. Gill, M. Montpetit, J. Rudnik, C. T. Sy, and M. V. O'Shaughnessy. 1992. Stability of dried blood spot specimens for detection of human immunodeficiency virus DNA by polymerase chain reaction. J. Clin. Microbiol. 303039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creek, T., A. Tanuri, M. Smith, K. Seipone, M. Smit, K. Legwaila, C. Motswere, M. Maruping, T. Nkoane, R. Ntumy, E. Bile, M. Mine, L. Lu, G. Tebele, L. Mazhani, M. K. Davis, T. H. Roels, P. H. Kilmarx, and N. Shaffer. 2008. Early diagnosis of human immunodeficiency virus in infants using polymerase chain reaction on dried blood spots in Botswana's national program for prevention of mother-to-child transmission. Pediatr. Infect. Dis. J. 2722-26. [DOI] [PubMed] [Google Scholar]
- 5.Curran, J. W., H. W. Jaffe, A. M. Hardy, W. M. Morgan, R. M. Selik, and T. J. Dondero. 1988. Epidemiology of HIV infection and AIDS in the United States. Science 239610-616. [DOI] [PubMed] [Google Scholar]
- 6.Driver, G. A., J. C. Patton, J. Moloi, W. S. Stevens, and G. G. Sherman. 2007. Low risk of contamination with automated and manual excision of dried blood spots for HIV DNA PCR testing in the routine laboratory. J. Virol. Methods 146397-400. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, A., C. Lejczak, C. Lambert, J. Servais, N. Makombe, J. Rusine, T. Staub, R. Hemmer, F. Schneider, J. C. Schmit, and V. Arendt. 2004. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J. Clin. Microbiol. 4216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joint United Nations Programme on HIV/AIDS and World Health Organization. 2007. AIDS epidemic update, December 2007. Joint United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO), Geneva, Switzerland. http://www.unaids.org/en/KnowledgeCentre/HIVData/EpiUpdate/EpiUpdArchive/2007/.
- 9.Lambert, J. S., D. R. Harris, E. R. Stiehm, J. Moye, Jr., M. G. Fowler, W. A. Meyer, III, J. Bethel, and L. M. Mofenson. 2003. Performance characteristics of HIV-1 culture and HIV-1 DNA and RNA amplification assays for early diagnosis of perinatal HIV-1 infection. J. Acquir. Immune Defic. Syndr. 34512-519. [DOI] [PubMed] [Google Scholar]
- 10.Lehman, D. A., and C. Farquhar. 2007. Biological mechanisms of vertical human immunodeficiency virus (HIV-1) transmission. Rev. Med. Virol. 17381-403. [DOI] [PubMed] [Google Scholar]
- 11.Luo, W., H. Yang, K. Rathbun, C. P. Pau, and C. Y. Ou. 2005. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J. Clin. Microbiol. 431851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Department of Health, South Africa. 2004. National antiretroviral treatment guidelines, 1st ed. National Department of Health, Pretoria, South Africa. http://www.doh.gov.za/docs/factsheets/guidelines/artguide04-f.html.
- 13.National Institute of Allergy and Infectious Diseases. 2007. Questions and answers. Children with HIV early antiretroviral therapy (CHER) study: treating HIV-infected infants early helps them live longer. South African clinical trial modified because of initial data. National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD. http://www3.niaid.nih.gov/news/QA/CHER_QA.htm.
- 14.Nduati, R., G. John, D. Mbori-Ngacha, B. Richardson, J. Overbaugh, A. Mwatha, J. Ndinya-Achola, J. Bwayo, F. E. Onyango, J. Hughes, and J. Kreiss. 2000. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. JAMA 2831167-1174. [DOI] [PubMed] [Google Scholar]
- 15.Newell, M. L., H. Coovadia, M. Cortina-Borja, N. Rollins, P. Gaillard, and F. Dabis. 2004. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet 3641236-1243. [DOI] [PubMed] [Google Scholar]
- 16.Nyambi, P. N., K. Fransen, H. De Beenhouwer, E. N. Chomba, M. Temmerman, J. O. Ndinya-Achola, P. Piot, and G. van der Groen. 1994. Detection of human immunodeficiency virus type 1 (HIV-1) in heel prick blood on filter paper from children born to HIV-1-seropositive mothers. J. Clin. Microbiol. 322858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou, C. Y., H. Yang, S. Balinandi, S. Sawadogo, V. Shanmugam, P. M. Tih, C. Adje-Toure, S. Tancho, L. K. Ya, M. Bulterys, R. Downing, and J. N. Nkengasong. 2007. Identification of HIV-1 infected infants and young children using real-time RT PCR and dried blood spots from Uganda and Cameroon. J. Virol. Methods 144109-114. [DOI] [PubMed] [Google Scholar]
- 18.Patton, J. C., E. Akkers, A. H. Coovadia, T. M. Meyers, W. S. Stevens, and G. G. Sherman. 2007. Evaluation of dried whole blood spots obtained by heel or finger stick as an alternative to venous blood for diagnosis of human immunodeficiency virus type 1 infection in vertically exposed infants in the routine diagnostic laboratory. Clin. Vaccine Immunol. 14201-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reisler, R. B., D. M. Thea, V. Pliner, T. Green, F. Lee, S. Nesheim, T. Brown, M. Kalish, T. M. Folks, and W. Heneine. 2001. Early detection of reverse transcriptase activity in plasma of neonates infected with HIV-1: a comparative analysis with RNA-based and DNA-based testing using polymerase chain reaction. J. Acquir. Immune Defic. Syndr. 2693-102. [DOI] [PubMed] [Google Scholar]
- 20.Rouet, F., C. Sakarovitch, P. Msellati, N. Elenga, C. Montcho, I. Viho, S. Blanche, C. Rouzioux, F. Dabis, and V. Leroy. 2003. Pediatric viral human immunodeficiency virus type 1 RNA levels, timing of infection, and disease progression in African HIV-1-infected children. Pediatrics 112e289. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38304-312. [DOI] [PubMed] [Google Scholar]
- 22.Sherman, G., P. Cooper, A. Coovadia, A. Puren, S. Jones, M. Mokhachane, and K. Bolton. 2005. Polymerase chain reaction for diagnosis of human immunodeficiency virus infection in infancy in low resource settings. Pediatr. Infect. Dis. J. 24993-997. [DOI] [PubMed] [Google Scholar]
- 23.Sherman, G. G., G. Stevens, S. A. Jones, P. Horsfield, and W. S. Stevens. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J. Acquir. Immune Defic. Syndr. 38615-617. [DOI] [PubMed] [Google Scholar]
- 24.Steketee, R. W., E. J. Abrams, D. M. Thea, T. M. Brown, G. Lambert, S. Orloff, J. Weedon, M. Bamji, E. E. Schoenbaum, J. Rapier, M. L. Kalish, et al. 1997. Early detection of perinatal human immunodeficiency virus (HIV) type 1 infection using HIV RNA amplification and detection. J. Infect. Dis. 175707-711. [DOI] [PubMed] [Google Scholar]
- 25.Stevens, W., G. Sherman, R. Downing, L. Parsons, C. Ou, S. Crowley, G. Gershy-Damet, K. Fransen, M. Bulterys, J. Homsy, L. Lu, T. Finkbeiner, and J. Nkengasong. 2008. Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: consensus statement on the performance of laboratory assays for early infant diagnosis. Open AIDS J. 217-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varga, C., G. Sherman, J. Maphosa, and S. Jones. 2005. Psychosocial consequences of early diagnosis of HIV status in vertically exposed infants in Johannesburg, South Africa. Health Care Women Int. 26387-397. [DOI] [PubMed] [Google Scholar]