Abstract
Clostridium perfringens is an important pathogen of animals and humans and is the causative agent of necrotic enteritis (NE) in poultry. This study focuses on the typing of intestinal C. perfringens isolates (n = 61) from outbreaks of NE collected from several areas of Southern Ontario, using a recently developed multilocus sequence typing (MLST) technique. For comparison, C. perfringens isolates from healthy birds were also obtained and typed. An additional locus, the pfoS locus, was included in our analysis, in an attempt to increase the discriminatory ability of the method previously published. Birds were collected from two major poultry processors in Canada, and isolates from processor 2 formed a distinct MLST cluster. Isolates from healthy birds also collected from the outbreak flocks clustered together with isolates from the birds with NE. Although isolates from eight outbreaks clustered together, MLST types were also occasionally different between outbreaks. Strong linkage disequilibrium was observed between loci, suggesting a clonal C. perfringens population structure. Detection assays for toxin genes cpb2 (beta-2 toxin), tpeL, and the newly described netB (NetB toxin) were also performed. netB was almost always found in outbreak isolates, whereas cpb2 was found exclusively in healthy bird isolates. The toxin gene tpeL, which has not been previously identified in C. perfringens type A strains, was also found, but only in the presence of netB. Resistance to bacitracin was found in 34% of isolates from antimicrobial agent-free birds and in 100% of isolates from conventionally raised birds.
Clostridium perfringens is an important pathogen of many animals and can cause a disease known as necrotic enteritis (NE) in avian species. The disease results from extracellular toxin production by the bacterium in the intestinal tract and leads to tissue necrosis and malabsorption and is often fatal (29). Although the disease is generally controlled through the use of antimicrobial agents, it is reemerging in countries that are now banning the use of antimicrobial agents as growth promoters in poultry production (33). The disease generally occurs in outbreaks in which a small percentage of a flock can be affected but can lead to significant losses within a farm.
There are various molecular typing techniques used for assessing bacterial population diversity and for outbreak investigation of C. perfringens. These methods include, among others, pulsed-field gel electrophoresis (4, 7, 23), multilocus variable number of tandem repeats analysis (5, 25), and multilocus sequence typing (MLST) (16). MLST is a procedure for characterizing bacteria by using the sequences of several “house-keeping” genes (20). Generally, 400 to 500 base pairs of each gene are amplified by PCR and sequenced, and differences in the DNA sequences define the allelic profile for each isolate, known as the sequence type (ST). Genetic events such as point mutations or recombinations result in different alleles and subsequently different allele combinations (8, 32). Most bacterial species have sufficient variation within house-keeping genes to provide many alleles per locus, allowing multiple STs to be identified using a limited number of loci. An MLST protocol has been developed for C. perfringens by Jost and collaborators (16); however, it is not currently available as a standard protocol on MLST.net (http://www.mlst.net) or PubMLST (http://pubmlst.org). Although MLST is generally expensive, it can be set up as a high-throughput technique which generates data that can be shared between laboratories and allows for the creation of a global database of sequences. This global approach would be of great value for the study of NE epidemiology and of other diseases caused by C. perfringens.
In this study, our aim was to characterize the genetic diversity of C. perfringens isolates from outbreaks of NE in broiler chicken populations by using the MLST protocol described by Jost and collaborators (16). Our focus was on investigating the level of clustering of MLST types in NE outbreaks to assess the diversity of isolates within and between outbreaks. Others have shown that only a few pulsed-field gel electrophoresis types of C. perfringens can be isolated from field cases of NE, whereas a higher diversity can be found in the intestinal flora of healthy birds (7, 12, 23). With its population genetics implications, MLST data will help provide a clearer picture of the epidemiology of C. perfringens and of NE.
MATERIALS AND METHODS
Collection of Clostridium perfringens isolates.
Chickens from farms associated with two major poultry processors were collected in Southern Ontario between 2005 and 2007. Chickens which were sick or had died of NE were collected from processor 1, and whenever possible, healthy birds were collected at the same time from the same farm as the outbreak flock. These birds were raised without antimicrobial agents; the farms had employed antimicrobial agents in feed 2 weeks to 4 months prior to the sampling dates but did not use them thereafter. Birds from processor 2 were raised following conventional methods (non-antibiotic-free and nonorganic); from the information available, most of the flocks received coccidiostats (salinomycin, narasin, and nicarbazin) in feed, and at least some were given virginiamycin. Live animals were euthanized by following standard protocols approved by the Animal Care Committee of the University of Guelph, and necropsies were conducted on all birds by the Animal Health Laboratory of the University of Guelph to confirm the diagnosis of NE. Nonoutbreak, healthy birds were also included from processor 2. A total of 11 different outbreak flocks and seven different normal flocks were sampled.
From each bird, cecal, mid-small-intestinal, and duodenal samples were removed aseptically, and the contents were suspended in 5 ml Difco brain heart infusion base (Becton Dickinson Microbiology Systems, Sparks, MD) plus 20% glycerol (Fisher Scientific, Fair Lawn, NJ) and frozen at −70°C. Duodenal samples were used in NE cases, and mid-small-intestinal and cecal samples were used only when C. perfringens isolates could not be recovered from the duodenum. A 100-μl aliquot of each sample was plated onto Shahidi-Ferguson perfringens selective medium plates (Becton Dickinson Microbiology Systems). Plates were incubated anaerobically for 24 h at 37°C. Two single colonies with dark centers were subcultured on Shahidi-Ferguson perfringens plates and subcultured again for purity. At least three isolates collected from sick or dead birds from each outbreak were used for MLST analysis. Whenever possible, isolates from the matched healthy birds were obtained from the duodenum, but cecal isolates were used if no duodenal isolates could be successfully cultured.
A total of 61 C. perfringens isolates were cultured from 45 broiler chickens, and these isolates were used for MLST analysis. These included 41 isolates from birds with NE, 9 isolates from matched healthy birds, and 11 isolates from nonoutbreak birds (Table 1). An additional C. perfringens isolate, CP4, which is a known virulent strain isolated from an NE case in Ontario (30), was also included for MLST analysis and toxin typing.
TABLE 1.
Complete list of Clostridium perfringens isolates typed by MLST, including outbreak attributes, ST, bacitracin MIC determined by Etest, and presence or absence of netB, tpeL, and cpb2 genes detected by PCR
| Isolateb | Outbreak no. | Date received | Processora | ST | Bird health | Culture source | Bacitracin MIC (μg/ml) | Presence ofc:
|
||
|---|---|---|---|---|---|---|---|---|---|---|
| netB | tpeL | cpb2 | ||||||||
| 01 | 1 | Feb. 2005 | 1 | 01 | NE, dead | Duodenum | 3 | + | − | − |
| 02 | 1 | 01 | NE, dead | Duodenum | 3 | + | − | − | ||
| 03 | 1 | 01 | NE, dead | Duodenum | 3 | − | − | − | ||
| 04 | 1 | 10 | NE, dead | Duodenum | 3 | + | − | − | ||
| 05 | 2 | Mar. 2005 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − |
| 06 | 1 | 02 | NE, dead | Duodenum | >256 | + | − | − | ||
| 07 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − | ||
| 08 | 1 | 10 | NE, dead | Duodenum | >256 | + | − | − | ||
| 09 | 3 | July 2005 | 1 | 04 | NE, sick | Duodenum | 2 | + | + | − |
| 10 | 1 | 03 | NE, dead | Duodenum | 2 | + | − | − | ||
| 11 | 1 | 04 | NE, dead | Duodenum | 1.5 | + | − | − | ||
| 12 | 1 | 04 | NE, dead | Duodenum | 2 | + | − | − | ||
| 13 | 4 | Aug. 2005 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − |
| 14 | 1 | 05 | NE, dead | Duodenum | 2 | + | − | − | ||
| 15 | 1 | 06 | NE, dead | Duodenum | 3 | + | − | − | ||
| 16 | 5 | Aug. 2005 | 1 | 07 | Healthy | Cecum | 1.5 | − | − | − |
| 17 | 1 | 19 | Healthy | Cecum | 1.5 | − | − | − | ||
| 18 | 1 | 08 | Healthy | Cecum | 1.5 | + | − | − | ||
| 19 | 1 | 08 | NE, dead | Duodenum | 1.5 | + | − | − | ||
| 20 | 1 | 09 | NE, dead | Duodenum | 1.5 | + | − | − | ||
| 21 | 1 | 08 | NE, dead | Duodenum | 1.5 | + | − | − | ||
| 22 | 6 | Apr. 2006 | 1 | 01 | Healthy | Cecum | 3 | + | + | − |
| 23 | 1 | 10 | NE, dead | Duodenum | 3 | + | + | − | ||
| 24 | 1 | 01 | NE, dead | Duodenum | 3 | + | + | − | ||
| 25 | 1 | 01 | NE, dead | Duodenum | 2 | + | + | − | ||
| 26 | 7 | Sept. 2006 | 1 | 11 | Healthy | Cecum | 2 | + | + | − |
| 27 | 1 | 12 | Healthy | Duodenum | 2 | + | + | − | ||
| 28 | 1 | 13 | NE, dead | Duodenum | 2 | + | + | − | ||
| 29 | 1 | 13 | NE, dead | Duodenum | 2 | + | + | − | ||
| 30 | 1 | 14 | NE, dead | Duodenum | 2 | + | + | − | ||
| 31 | 8 | Nov. 2006 | 1 | 10 | NE, dead | Cecum | 2 | + | − | − |
| 32 | 1 | 15 | NE, dead | Duodenum | 3 | + | − | − | ||
| 33 | 1 | 01 | NE, dead | Duodenum | 2 | − | − | − | ||
| 34 | 1 | 10 | Healthy | Duodenum | 2 | + | + | − | ||
| 35 | 9 | Mar. 2007 | 1 | 01 | NE, dead | Cecum | >256 | + | − | − |
| 36 | 1 | 01 | NE, dead | Cecum | >256 | + | − | − | ||
| 37 | 1 | 01 | NE, dead | Mid-small intestine | >256 | + | − | − | ||
| 38 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − | ||
| 39 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − | ||
| 40 | 10 | Mar. 2007 | 1 | 10 | NE, sick | Cecum | >256 | + | − | − |
| 41 | 1 | 01 | NE, sick | Cecum | >256 | + | − | − | ||
| 42 | 1 | 16 | NE, dead | Cecum | >256 | + | − | − | ||
| 43 | 1 | 01 | NE, dead | Mid-small intestine | >256 | + | − | − | ||
| 44 | 1 | 10 | NE, dead | Duodenum | >256 | + | − | − | ||
| 45 | Nonoutbreak | July 2007 | 2, farm 1 | 17 | Healthy | Cecum | >256 | − | − | − |
| 46 | 2, farm 1 | 19 | Healthy | Cecum | >256 | − | − | − | ||
| 47 | 2, farm 2 | 18 | Healthy | Cecum | >256 | − | − | + | ||
| 48 | 2, farm 2 | 18 | Healthy | Cecum | >256 | − | − | + | ||
| 49 | 2, farm 3 | 19 | Healthy | Cecum | >256 | − | − | − | ||
| 50 | Nonoutbreak | July 2007 | 2, farm 4 | 18 | Healthy | Cecum | >256 | − | − | + |
| 51 | 2, farm 5 | 18 | Healthy | Cecum | >256 | − | − | + | ||
| 52 | 2, farm 6 | 18 | Healthy | Cecum | >256 | − | − | + | ||
| 53 | 2, farm 7 | 18 | Healthy | Cecum | >256 | − | − | + | ||
| 54 | 2, farm 1 | 20 | Healthy | Cecum | >256 | − | − | + | ||
| 55 | 2, farm 1 | 21 | Healthy | Cecum | >256 | − | − | + | ||
| 56 | 11 | Nov. 2007 | 1 | 06 | NE, dead | Cecum | 2 | + | − | − |
| 57 | 1 | 22 | NE, dead | Duodenum | 1.5 | + | − | − | ||
| 58 | 1 | 01 | NE, dead | Duodenum | >256 | + | − | − | ||
| 59 | 1 | 01 | NE, sick | Cecum | >256 | + | − | − | ||
| 60 | Nonoutbreak | Dec. 2007 | 1 | 06 | Healthy | Cecum | 2 | + | − | − |
| 61 | 1 | 06 | Healthy | Cecum | 2 | + | − | − | ||
Processor 1, no antimicrobial agent use; processor 2, conventional antimicrobial agent use.
Each pair of isolates 16 and 17, 23 and 24, 35 and 36, 40 and 41, and 60 and 61 was recovered from the same intestinal sample of a single bird within a given outbreak.
+, present; −, absent.
Multilocus sequence typing and analysis.
The eight loci described in the MLST protocol by Jost and collaborators (16) were used in this study, with an additional regulatory protein gene (pfoS) added in an attempt to increase the discriminatory ability of the method (Table 2). This gene is described in a publication by Rooney and collaborators (24). Initial pfoS sequencing results showed allelic diversity in some isolates, and it was therefore included in the method. Lysates for each isolate were prepared using an InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. Each 100-μl PCR mixture contained 1× PCR buffer, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 250 nM each primer, 5 U Taq polymerase (Promega, Madison, WI), and 2 μl of template DNA. The PCR conditions were the same as those used in the publication by Jost and collaborators (16), with only the final extension time modified to 7 min. A 96-well plate format was adopted for all manipulations to facilitate robot-handling of reagents using a Biomek NX automation workstation (Beckman Coulter, Fullerton, CA). PCR product cleanup was performed using a MinElute 96 UF PCR purification kit (Qiagen Inc., Valencia, CA) by following the manufacturer's instructions. Sequencing was performed using an ABI Prism 3730 sequencer (Applied Biosystems, Foster City, CA), and sequences were analyzed using Sequencher version 4.5 software (Gene Codes Corporation, Ann Arbor, MI).
TABLE 2.
MLST locus properties
| Gene | Primer name and sequence | Primer reference | Amplicon size(s) (bp) | Location on genomea (3.1 Mbp) | No. of alleles per locus in 61 isolatesb |
|---|---|---|---|---|---|
| plc | CPplcR; AGTTTTTCCATCCTTTGTTTTG | 16 | 544 | 49,000 | 6 |
| CPplcF; ATATGAATGGCAAAGAGGAAAC | 16 | ||||
| ddlA | CPddlA2Rb; TTGTCATACCAGGTAATGTATTT | This study | 397 | 1,004,000 | 9 |
| CPddlAF; ATAATGGGGGATCATCAGTTGC | 16 | ||||
| dut | CPdutR; CTGTAGTACCAAATCCACCACG | 16 | 441 | 843,000 | 5 |
| CPdutF; TTAAGTATTTTGATAACGCAAC | 16 | ||||
| glpK | CPglpKR; CACCTTTTGCTCCAAGGTTTGC | 16 | 574 | 2,923,000 | 6 |
| CPglpKF; TGGGTTGAGCATGATCCAATGG | 16 | ||||
| gmk | CPgmkR; TACTGCATCTTCTACATTATCG | 16 | 475 | 2,025,000 | 5 |
| CPgmkF; TAAGGGAACTATTTGTAAAGCC | 16 | ||||
| recA | CPrecAR; CTCCATATGAGAACCAAGCTCC | 16 | 475 | 1,949,000 | 4 |
| CPrecAF; GCTATAGATGTTTTAGTTGTGG | 16 | ||||
| sod | CPsodR; AATAATAAGCATGTTCCCAAAC | 16 | 478 | 1,444,000 | 5 |
| CPsodF; GATGCTTTAGAGCCATCAATAG | 16 | ||||
| tpiA | CPtpiR; CATTAGCTTGGTCTGAAGTAGC | 16 | 451 | 1,542,000 | 9 |
| CPtpiF; AAATGTGAAGTTGTTGTTTGCC | 16 | ||||
| pfoS | CPpfoSR; GCAAATCTTATTTGGAACAT | This study | 560, 562, 563 | 1,190,000 | 4 |
| CPpfoSF; GTGCAGTTGCAACCACTGTT | 24 |
Gene locations were based on the published genome sequence of C. perfringens strain 13 (27).
The average number of alleles per locus in 61 isolates was 5.9.
Concatemers of all nine sequences were analyzed using BioNumerics software version 5.1 (Applied Maths, Austin, TX), and a dendrogram was created using cluster analysis with pairwise similarities and the unweighted-pair group method using arithmetic mean (UPGMA) clustering; neighbor-joining cluster analysis was also performed for comparison. Concatemers of the nine genes from three sequenced C. perfringens genomes, strain 13, ATCC 13124, and SM101 (22, 27), were generated in silico and were included as well. Additionally, sequences obtained from eight avian C. perfringens isolates previously analyzed by Jost and collaborators were also included (16). These included seven STs, 6, 7, 26, 50, 61, 68, and 80, which are listed in their publication (B. H. Jost, personal communication).
eBURST analysis of isolates was performed, using eBURSTv3, with STs assigned based on unique allele profiles (http://eburst.mlst.net) (10). Groups were defined by isolates with at least seven of the nine alleles being identical. All ST numbers were based on this analysis (data not shown).
Presence of toxin genes.
Detection of various toxin genes was carried out by PCR amplification of each C. perfringens isolate (n = 61). This included toxin typing (alpha, beta, epsilon, and iota toxin genes), which was performed as a multiplex reaction according to Yoo and collaborators (35). The beta-2 toxin gene (cpb2) and the enterotoxin gene (cpe) were detected using protocols described by Meer and Songer (21) and Herholz and collaborators (14), respectively. Additionally, a recently described NE beta-like toxin, NetB (netB) (17), was detected by the primers AKP78 and AKP79 described in the publication.
The presence of the tpeL (toxin C. perfringens large cytotoxin) gene was detected by PCR amplification of an internal fragment, using primers designed from the C. perfringens toxin type C sequence of tpeL (AB262081) supplied by Amimoto and collaborators (2). Amplifications were performed in a 25-μl total volume containing the following: 5 μl of template DNA, 1× PCR buffer with Mg2+ (New England BioLabs, Ipswich, MA), 0.2 mM deoxynucleoside triphosphate mixture, 2.5 units of Taq DNA polymerase (New England BioLabs), and 800 nM of each primer (T2875-F [ATAAGTGCCAATCAATATGA] and T3348-R [TTCTCCTGAATTAATATTAA]). After an initial denaturing step of 3 min at 94°C, the samples were subjected to 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, and final extension at 72°C for 10 min. All PCR products were visualized by horizontal agarose gel electrophoresis.
One netB and three tpeL PCR products were sequenced to confirm the specificity of the PCR. The entire tpeL gene from C. perfringens strain CP4 was amplified using primers specific to sequences 298 bp upstream of the first potential tpeL start site to 370 bp downstream of the stop codon for tpeL in C. perfringens type C (AB262081). Both strands of this entire PCR product were sequenced by primer walking, and the DNA sequence was deposited in the GenBank database (EU848493). A Southern blot to determine if tpeL is plasmid-borne was performed on plasmid preparations (plasmid midi kit; Qiagen Inc.) from four isolates (three tpeL positive and one negative), using a PCR digoxigenin-labeled probe (Roche Diagnostics, Mannheim, Germany) synthesized using the primers and PCR conditions described for tpeL detection by Amimoto and collaborators (2).
Susceptibility testing.
Susceptibility to bacitracin was determined for each isolate by using Etest strips (AB Biodisk, Solna, Sweden) following the manufacturer's instructions. Isolates were grown overnight on blood agar plates, and a 1.0 McFarland standard was suspended in Mueller-Hinton broth (Sensititre, Westlake, OH). Brucella agar (150 mm) with vitamin K and Hemin agar plates (Medox Diagnostics, Ottawa, Ontario, Canada) were swabbed with the suspension, and single strips were applied to the surface. Readings were taken after 24 h of incubation under anaerobic conditions, and end points were determined using Etest guidelines for anaerobes. C. perfringens strain ATCC 13124 was used as a control. Isolates with MICs of >16 μg/ml were considered resistant, according to Johansson and collaborators (15).
Statistical analysis.
Linkage disequilibrium analysis was performed using Arlequin version 3.11 (9) population genetics software. Polymorphic sites (n = 116) from each ST were analyzed (only one isolate per ST was used for this analysis), and linkage disequilibrium testing was estimated for all pairs of polymorphic sites. Significance was set at a P value of 0.05, with 10,000 steps in the Markov chain and 1,000 dememorization steps, resulting in a matrix of P values. A second measure of linkage disequilibrium was performed using the tool provided by MLST.net (http://linux.mlst.net/link_dis/index.htm). This method calculates an index of association (IA), which measures the degree of association between loci (28).
Nucleotide sequence accession numbers.
Each unique sequence for each locus found in this study (n = 53) has been uploaded to the GenBank database (http://www.ncbi.nlm.nih.gov) under the following accession numbers: EU605734-EU605739 for plc, the alpha toxin gene; EU605740-EU605748 for ddlA, the d-alanine-d-alanine ligase gene; EU605749-EU605753 for dut, the deoxyuridine triphosphatase gene; EU605754-EU605759 for glpK, the glycerol kinase gene; EU605760-EU605764 for gmk, the deoxyguanylate kinase gene; EU605765-EU605768 for recA, the recombinase gene; EU605769-EU605773 for sod, the superoxide dismutase gene; EU605774-EU605782 for tpiA, the triose phosphate isomerase gene; and EU605783-EU605786 for pfoS, the putative membrane protein gene.
RESULTS
C. perfringens isolates were cultured from birds from all NE outbreaks but were more difficult to recover from healthy, nonoutbreak birds. Duodenal samples were generally sufficient for culture; however, mid-small-intestinal and cecal samples from healthy birds in which concentrations of C. perfringens were found to be lower were also used (Table 1). Sixty-one isolates were selected for MLST analysis from 45 birds, which included 11 nonoutbreak birds. In a few instances (Table 1), two isolates were taken from a single sample to assess the genetic diversity between isolates within a single bird.
Amplification and sequence analysis of the 61 isolates were successful using the protocol outlined by Jost and collaborators (16). The addition of the pfoS locus did not create more STs and did not increase the discrimination of the method even though four different alleles were found (Table 2). The pfoS amplicon also showed some sequence length polymorphism of up to three base pairs, which was not observed in the other loci (Table 2). One pfoS allele (EU605786) showed a deletion within a long poly(A) sequence, introducing an internal stop codon, and resulted in a truncated PfoS protein.
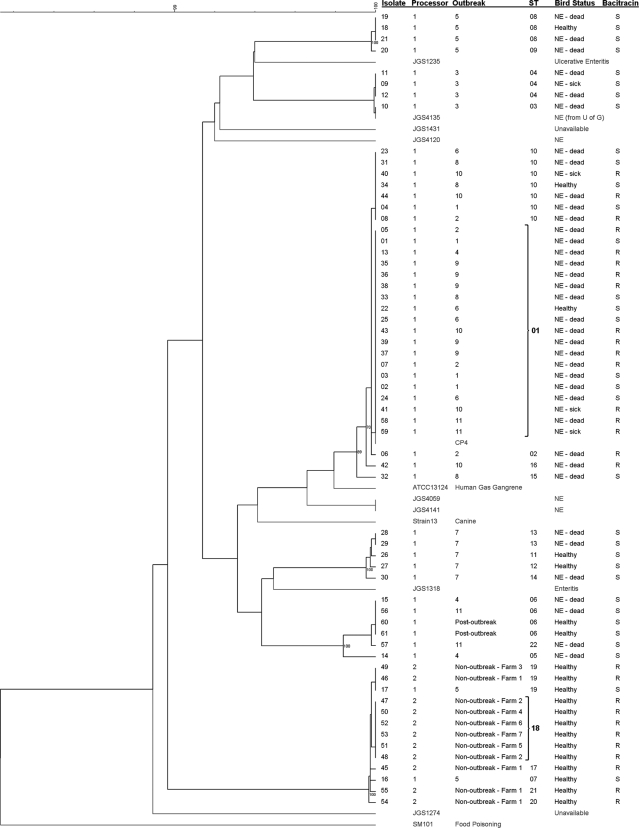
The average number of alleles per locus was 5.9, with a minimum of 4 and a maximum of 9. Twenty-two STs were identified. With a few exceptions (outbreaks 4, 5, and 11), clustering of isolates within each outbreak was observed (Fig. 1). Isolates obtained from healthy birds in the same outbreak flock (outbreaks 5, 6, 7, and 8) also clustered with isolates from dead birds affected by NE. Multiple isolates obtained from a single bird were either identical (n = 8) or very similar (n = 2) (Fig. 1). The isolates from healthy birds originating from the seven farms of processor 2 all clustered tightly together. Although a major cluster contained isolates from eight different outbreaks from processor 1 (outbreaks 1, 2, 4, 6, 8, 9, 10, and 11), the isolates from the three remaining outbreaks were part of unrelated clusters. Neighbor-joining cluster analysis produced identical major clusters of isolates compared to the UPGMA clustering.
FIG. 1.
Dendrogram of concatenated MLST sequences of 61 avian isolates from Southern Ontario generated with pairwise similarities and UPGMA clustering. It includes eight previously published avian C. perfringens MLST sequences (16). Isolates from the study by Jost and collaborators are labeled JGS and shown in gray. CP4 is a virulent NE-associated isolate from another previous study in Ontario (30). The three C. perfringens strains with published genome sequences (strains 13, ATCC 13124, and SM101) were also included, and their MLST types were generated in silico (22, 27). Selected bootstrap values are indicated at nodes. Susceptibility to bacitracin was determined using a 16-μg/ml breakpoint. STs were determined through eBURSTv3 analysis (10), and ST01 and 18 are labeled in boldface type (founders).
The eBURST analysis produced 22 STs and was in general agreement with the other clustering analysis. Six complexes (linked by single allelic differences) and two singletons were generated, with ST01 and ST18 considered founders (10) of the two major complexes (data not shown). ST01 included 19 isolates from the eight outbreaks mentioned above, all from processor 1. ST18 contained six isolates, all from processor 2, and included five nonoutbreak farms (Fig. 1).
The isolate JGS4135 from Jost and collaborators (16), which was the only one studied by these authors that clustered tightly with our own isolates, was traced back to a poultry sample from Ontario originally collected in Guelph (Fig. 1). Strain CP4, which was isolated about 10 years ago from a chicken with NE in Ontario, is also part of the major ST01 group (Fig. 1).
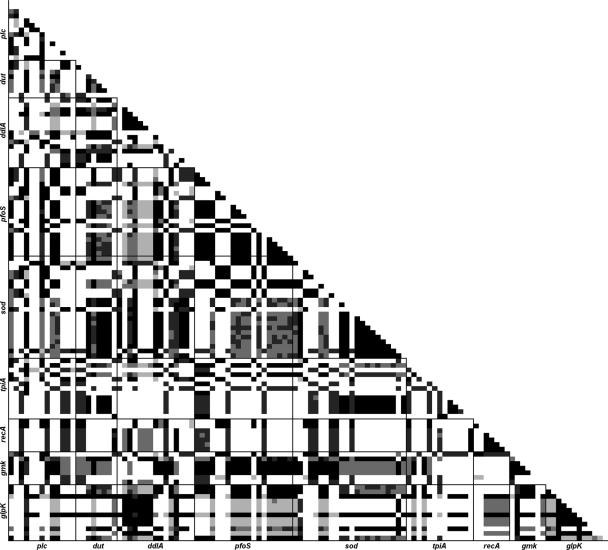
Results from the Arlequin linkage disequilibrium analysis (Fig. 2) showed a random distribution of significant values. The extent of significant disequilibrium (P ≤ 0.05) between polymorphic sites ranged between 7.2% (tpiA and ddlA) and 43.9% (glpK and ddlA), with an overall average of 23.6%. Linkage disequilibrium within each locus was higher (38.0% of polymorphic sites), as was expected. The IA value calculated using the MLST.net tool was 3.140, which implies significant linkage disequilibrium as well (28).
FIG. 2.
Linkage disequilibrium matrix of polymorphic sites in 22 STs, generated by Arlequin version 3.11 (9). Darker cells refer to more-significant P values (black cells represent values of ≤0.05 and the lightest-gray cells up to 0.20). Locus names are shown on each axis, in sequential order on the chromosome.
All isolates from our birds were found to be toxin type A and negative for the enterotoxin gene. The netB gene was found only in isolates from processor 1 (92.0%). Conversely, the cpb2 toxin gene was found only in isolates from processor 2 (72.7%).
DNA sequencing of the gene amplified from CP4 showed that the tpeL gene consisted of 4,955 bp (EU848493). The translated amino acid sequence of TpeL in strain CP4 has 97% identity with the sequence of TpeL from a C. perfringens toxin type B strain (NZ_ABDV01000024), and both are 128 amino acids longer than that in the toxin type C strain MC18 (AB262081). The complete tpeL genes from two NE isolates, 23 and 28, were sequenced and found to have 100% identity with tpeL from strain CP4. Among the 61 isolates investigated here, tpeL was detected in 11 outbreak-associated isolates (Table 1) which included eight STs and four out of eleven outbreaks. All isolates from processor 2 were negative for tpeL (n = 13). Southern blot results showed that the tpeL gene was present on a large plasmid of identical size (∼100 kb) in all three tpeL-positive isolates (13, 30, and CP4) examined (data not shown).
Etest values for susceptibility to bacitracin ranged between 1.5 and 256 μg/ml (Table 1). A very distinct bimodal distribution was present with MICs being either between 1.5 and 3 μg/ml (susceptible) or >256 μg/ml (resistant). Seventeen resistant and 33 susceptible isolates were obtained from processor 1, whereas only bacitracin-resistant isolates (n = 11) were detected in birds from processor 2. All the bacitracin-resistant isolates were part of the two major clusters containing ST01 and ST18, but not all the isolates in these two clusters were resistant (Fig. 1).
DISCUSSION
In this study, C. perfringens was typically isolated from the duodenal samples of NE-affected birds, whereas isolates from healthy birds could be recovered frequently from cecal samples only. This has been previously observed in other studies, in which concentrations of C. perfringens are higher in the intestinal tract of NE-affected birds, where the bacteria have proliferated upward into the duodenum and are often associated with coccidial coinfection (1, 3, 26).
The MLST method developed by Jost and collaborators was originally developed for a broad array of C. perfringens isolates of very diverse origins (16) and may not have been sufficient for this study, where only chicken isolates were used. Therefore, we initially attempted to increase the discriminatory power of this typing scheme by investigating potential additional loci. The loci for gyrA and rplL (24) and rpoD (primers derived from the published C. perfringens SM101 sequence [22]) showed little or no polymorphism at all in a subset of our isolates (data not shown). However, the pfoS locus did show some diversity and was added to the established protocol. Ultimately, this locus did not contribute any additional discrimination despite the presence of four different alleles at this locus in our isolates. It may also not be a suitable MLST locus because of its sequence length variability; one allele of the pfoS locus appeared to have resulted from the deletion of an “ATT” tandem repeat in the gene. Another pfoS allele encodes a truncated and probably nonfunctional protein. Therefore, it appears that the protocol described by Jost and collaborators (16) already provides a good estimate of C. perfringens diversity and was not improved with the addition of pfoS. When isolates from the three sequenced genomes of C. perfringens were included (Fig. 1), as well as the eight avian isolates from the Jost et al. study (16), these isolates generally belonged to different sequence types than to those from this study. Interestingly though, the JGS4135 isolate typed by Jost and collaborators was identical to isolate 10 in our study, and it was later revealed that the former isolate had originated from the University of Guelph about 10 years earlier. This is a convincing indication that this MLST system is epidemiologically viable and also showed that the same C. perfringens clone may have persisted for many years in Southern Ontario.
In previous studies (7, 12, 23), researchers have proposed that there are distinct clones within C. perfringens populations which cause NE outbreaks and that birds within an outbreak are usually affected by only one of these clones. Our results, which showed strong genetic clustering in isolates from NE outbreaks, both within a single bird and between birds of the same outbreak, are in agreement with this hypothesis. However, we did not observe any major differences between isolates from healthy birds and those from NE cases within any outbreak. This stresses once more the fact that NE is a multifactorial disease and that the mere presence of a specific C. perfringens strain in a bird is not a sufficient cause for the development of the disease.
The very strong clustering of isolates originating from birds of seven different farms from geographically separate areas affiliated with processor 2 is striking and was unexpected. Similarly, despite the higher diversity observed among isolates from farms with NE outbreaks and those associated with processor 1, the majority of these isolates belonged to a single major cluster (cluster containing ST01, -02, -10, -15, and -16) (Fig. 1). In the case of this processor, the clustering may be related to a higher propensity of a specific clone for causing NE. These clusters also suggest a limited degree of genetic diversity of C. perfringens throughout each processor's poultry operations. This could be the result of the dissemination of clones throughout integrated producer networks, starting at hatcheries and chick distribution systems. Selection of a limited number of resistant clones through antimicrobial agent use, either for prevention (cluster containing ST18) or therapy (cluster that contains ST01 and is associated with NE outbreaks), may also be part of the reason for the observed clustering. This latter hypothesis is supported by the fact that bacitracin resistance was clearly associated with these two major clusters and lineages.
Locus linkage was found to be high in the isolates characterized in this study, with significant linkage disequilibrium found between all loci (IA = 3.140); panmictic populations have an IA value approaching zero. This also shows that recombination rates for the core genome of C. perfringens are low or possibly absent (28). Additionally, only 22 STs were found in the sample population. When considering the number of possible allele combinations (over 5 million), the 22 STs found in 61 isolates may be considered low. These data suggest that the C. perfringens population under investigation had a highly clonal structure (28).
Since MLST examines core genes encoding house-keeping functions not usually involved in pathogenesis, there is not necessarily a correlation between virulence potential and the overall genomic relationships evidenced by MLST (20, 31). It is therefore possible that more virulent clones do exist within these outbreaks, which carry accessory genes and virulence factors not detected using MLST.
The complete absence of cpb2 in the NE outbreak isolates was surprising, since this toxin had originally been associated with virulence (11, 13, 34). However, there is a growing body of evidence that suggests that the beta-2 toxin is not a critical factor of C. perfringens infection in this particular case of poultry (4, 6, 30). The recently discovered netB toxin gene, however, was found exclusively in isolates which were all associated with NE outbreaks and was never detected in healthy control samples from processor 2. Although netB was detected in every outbreak, it was still found in healthy matched birds from outbreak flocks as well. Additionally, netB was not always detected in isolates from birds clearly affected with NE (Table 1, isolates 03 and 33). Overall, this study provided a good opportunity to observe the prevalence of the netB gene in a field situation where NE- and non-NE-associated isolates were used. The data suggest that this toxin is an important factor in C. perfringens infection, facilitating the development of NE. However, our results also show that it is neither a necessary nor a sufficient cause for the disease to occur. As the authors of the netB discovery stated (17), there may be many other unidentified factors which are also important for virulence.
The C. perfringens toxin TpeL (2) was identified in the culture supernatant of C. perfringens strain CP4, when the secreted proteins of CP4 were electrophoretically separated and reacted with the serum from broiler chickens immune to NE (18, 19), and was further identified by mass spectrometry (unpublished data). This is the first report describing the presence of tpeL genes in C. perfringens type A strains and the first report of the presence of this gene in isolates from chickens with NE. The tpeL gene identified in these toxin type A isolates is very similar to the tpeL gene in the toxin type B strain (NZ_ABDV01000024). The toxin type C strain (2), however, has a cytosine insert at position 4941, which creates a premature stop codon and thus truncates the protein. The variable presence of this toxin gene within clones and even within clonally related isolates from a single farm suggested that it is located on a mobile genetic element. Our Southern blot results confirmed that it is indeed located on large plasmids. The identification of TpeL among the secreted proteins of CP4 may be related to the greater virulence of this strain in an experimental model of NE in comparison to some other strains in which NE originated (30). Since tpeL was detected in isolates and STs associated with NE outbreaks only, it is reasonable to speculate that it contributes to the pathogenesis of NE, but further work is required to assess its contribution to the virulence of NE isolates.
In conclusion, the MLST protocol described by Jost and collaborators (16) was successfully applied in this study for typing avian C. perfringens isolates. The resulting data suggest that clustering of MLST types occurs in association with the source of the isolates and with bacitracin resistance. A major clonal lineage containing isolates from eight different outbreaks was identified, which may be either more prone to cause outbreaks than others or selected by the use of antimicrobial agents. Toxin typing suggests that NetB is a far more significant toxin than the beta-2 toxin, although there may be many more unexplored factors that are important for NE development, such as TpeL, which was identified here for the first time in NE isolates.
Acknowledgments
This work was funded by the Canadian Poultry Research Council and the Poultry Industry Council.
We thank the Animal Health Laboratory at the University of Guelph, the poultry processors, and the Canadian Food Inspection Agency for their assistance in this project. We also thank Teresa Crease for her help with the linkage disequilibrium analysis.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Al-Sheikhly, F., and R. B. Truscott. 1977. The interaction of Clostridium perfringens and its toxins in the production of necrotic enteritis of chickens. Avian Dis. 21256-263. [PubMed] [Google Scholar]
- 2.Amimoto, K., T. Noro, E. Oishi, and M. Shimizu. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 1531198-1206. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, E. M., G. C. Mead, D. A. Barnum, and E. G. Harry. 1972. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci. 13311-326. [DOI] [PubMed] [Google Scholar]
- 4.Chalmers, G., S. W. Martin, D. B. Hunter, J. F. Prescott, L. J. Weber, and P. Boerlin. 2008. Genetic diversity of Clostridium perfringens isolated from healthy broiler chickens at a commercial farm. Vet. Microbiol. 127116-127. [DOI] [PubMed] [Google Scholar]
- 5.Chalmers, G., S. W. Martin, J. F. Prescott, and P. Boerlin. 2008. Typing of Clostridium perfringens by multiple-locus variable number of tandem repeats analysis. Vet. Microbiol. 128126-135. [DOI] [PubMed] [Google Scholar]
- 6.Crespo, R., D. J. Fisher, H. L. Shivaprasad, M. E. Fernandez-Miyakawa, and F. A. Uzal. 2007. Toxinotypes of Clostridium perfringens isolated from sick and healthy avian species. J. Vet. Diagn. Investig. 19329-333. [DOI] [PubMed] [Google Scholar]
- 7.Engström, B. E., C. Fermer, A. Lindberg, E. Saarinen, V. Baverud, and A. Gunnarsson. 2003. Molecular typing of isolates of Clostridium perfringens from healthy and diseased poultry. Vet. Microbiol. 94225-235. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7482-487. [DOI] [PubMed] [Google Scholar]
- 9.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 147-50. [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, D. J., K. Miyamoto, B. Harrison, S. Akimoto, M. R. Sarker, and B. A. McClane. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56747-762. [DOI] [PubMed] [Google Scholar]
- 12.Gholamiandekhordi, A. R., R. Ducatelle, M. Heyndrickx, F. Haesebrouck, and F. Van Immerseel. 2006. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet. Microbiol. 113143-152. [DOI] [PubMed] [Google Scholar]
- 13.Gibert, M., C. Jolivet-Reynaud, and M. R. Popoff. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 20365-73. [DOI] [PubMed] [Google Scholar]
- 14.Herholz, C., R. Miserez, J. Nicolet, J. Frey, M. Popoff, M. Gibert, H. Gerber, and R. Straub. 1999. Prevalence of beta2-toxigenic Clostridium perfringens in horses with intestinal disorders. J. Clin. Microbiol. 37358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson, A., C. Greko, B. E. Engström, and M. Karlsson. 2004. Antimicrobial susceptibility of Swedish, Norwegian and Danish isolates of Clostridium perfringens from poultry, and distribution of tetracycline resistance genes. Vet. Microbiol. 99251-257. [DOI] [PubMed] [Google Scholar]
- 16.Jost, B. H., H. T. Trinh, and J. G. Songer. 2006. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet. Microbiol. 116158-165. [DOI] [PubMed] [Google Scholar]
- 17.Keyburn, A. L., J. D. Boyce, P. Vaz, T. L. Bannam, M. E. Ford, D. Parker, A. Di Rubbo, J. I. Rood, and R. J. Moore. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni, R. R., V. R. Parreira, S. Sharif, and J. F. Prescott. 2006. Clostridium perfringens antigens recognized by broiler chickens immune to necrotic enteritis. Clin. Vaccine Immunol. 131358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni, R. R., V. R. Parreira, S. Sharif, and J. F. Prescott. 2007. Immunization of broiler chickens against Clostridium perfringens-induced necrotic enteritis. Clin. Vaccine Immunol. 141070-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60561-588. [DOI] [PubMed] [Google Scholar]
- 21.Meer, R. R., and J. G. Songer. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am. J. Vet. Res. 58702-705. [PubMed] [Google Scholar]
- 22.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 161031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nauerby, B., K. Pedersen, and M. Madsen. 2003. Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet. Microbiol. 94257-266. [DOI] [PubMed] [Google Scholar]
- 24.Rooney, A. P., J. L. Swezey, R. Friedman, D. W. Hecht, and C. W. Maddox. 2006. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 1722081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawires, Y. S., and J. G. Songer. 2005. Multiple-locus variable-number tandem repeat analysis for strain typing of Clostridium perfringens. Anaerobe 11262-272. [DOI] [PubMed] [Google Scholar]
- 26.Shane, S. M., J. E. Gyimah, K. S. Harrington, and T. G. Snider III. 1985. Etiology and pathogenesis of necrotic enteritis. Vet. Res. Commun. 9269-287. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 904384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, D. R., V. R. Parreira, R. R. Kulkarni, and J. F. Prescott. 2006. Live attenuated vaccine-based control of necrotic enteritis of broiler chickens. Vet. Microbiol. 11325-34. [DOI] [PubMed] [Google Scholar]
- 31.Turner, K. M., and E. J. Feil. 2007. The secret life of the multilocus sequence type. Int. J. Antimicrob. Agents 29129-135. [DOI] [PubMed] [Google Scholar]
- 32.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11479-487. [DOI] [PubMed] [Google Scholar]
- 33.Van Immerseel, F., J. De Buck, F. Pasmans, G. Huyghebaert, F. Haesebrouck, and R. Ducatelle. 2004. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 33537-549. [DOI] [PubMed] [Google Scholar]
- 34.Waters, M., A. Savoie, H. S. Garmory, D. Bueschel, M. R. Popoff, J. G. Songer, R. W. Titball, B. A. McClane, and M. R. Sarker. 2003. Genotyping and phenotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J. Clin. Microbiol. 413584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Yoo, H. S., S. U. Lee, K. Y. Park, and Y. H. Park. 1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 35228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]