Abstract
Following a flooding event close to a shellfish production lagoon, 205 cases of gastroenteritis were linked to oyster consumption. Twelve stool samples from different individuals were collected. Analysis showed that eight samples were positive for multiple enteric viruses, and one stool sample had seven different enteric viruses. Analysis of shellfish implicated in the outbreak allowed detection of the same diversity of enteric viruses, with some viral genomic sequences being identical to those obtained from stool sample analysis. Shellfish were contaminated by as many as five different enteric viruses. For the first time in Europe, Aichi virus was identified in oyster samples. Shellfish samples collected over 3 weeks following the outbreak showed a progressive decline in the level of virus contamination as measured by the virus diversity detected and by quantitative reverse transcription-PCR.
Foods play an important role in the transmission of enteric viruses. For example, for noroviruses (NoVs), the predominant agents of nonbacterial gastroenteritis in humans, as many as 40% of cases of infections are estimated to be linked to contaminated food consumption (3, 16, 28). All other enteric viruses, including rotavirus (RV), astrovirus (AV), and Aichi virus (AiV), also cause gastroenteritis, with symptoms that are more or less similar to those caused by NoV, but their etiological importance in outbreaks is not really known. However, these viruses have occasionally been shown to be transmitted by foods (21). Food may be contaminated at different stages of production, such as by fecal contamination of shellfish-growing waters, the use of night soil to fertilize crops, the fecal contamination of water used to wash fruits after harvest, or poor hand hygiene by an infected food handler (16, 21). While a single strain is usually responsible for clinical cases associated with direct contamination by food handlers, multiple strains may be detected when a poorly functioning sewage treatment plant is responsible for contamination of foods such as oysters.
Viruses that cause gastroenteritis multiply in the intestines and are excreted in large quantities in human feces. Human waste is processed in sewage treatment plants, but the treatment procedures do not completely remove enteric viruses from the water effluents leaving the plant (8, 10, 12). Strains that cause severe symptomatic infection, as well as those that cause subclinical infection, are excreted into sewage. Thus, wastewaters reflect a wide range of virus strains circulating in the population (8, 13). When the sewage treatment process is overwhelmed, as can occur with some flooding events, accidental contamination of shellfish-growing beds may provide an opportunity for enteric viruses other than NoV to infect people and cause disease.
This study reports a recent event that occurred in France following oyster consumption. Stool analyses identified as many as seven different strains in one stool sample. The two major points of this study are, first, the analysis of shellfish related to the outbreak, allowing detection of the same human enteric viruses, and second, to report for the first time in Europe the presence of Aichi virus in oyster samples. Environmental investigations identified the leading cause of the lagoon oyster production area contamination, but prevention measures such as prolonged depuration time implemented to satisfy European regulation (Escherichia coli counts) for oysters were unable to avoid human contamination.
MATERIALS AND METHODS
Epidemiological data.
All data concerning clusters of gastroenteritis cases in the affected area during February 2006 were collected either from medical doctors or directly from sanitary services (DDASS and DDSV). A standardized questionnaire covering foods consumed, symptoms, and timing of illness was completed by each participant in the study. The association between food consumption and illness was estimated by calculation of the relative risk and its 95% confidence interval using Epi Info version 6 software.
Clinical sample analysis.
Twelve fecal samples collected from 12 patients from three clusters were analyzed. Group A rotaviruses were detected by enzyme immunoassay (EIA) with group-specific monoclonal antibodies as previously described (1). Astroviruses and adenovirus types 40 and 41 were detected with the IDEIA astrovirus (Dako Diagnostics Ltd.) EIA kit and the Adenoclone type 40/41 EIA (Meridian Diagnostics Inc.), respectively. Results were then confirmed by reverse transcription-PCR (RT-PCR) (1). For typing and other enteric virus detection, nucleic acids (NA) were extracted and purified using a QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. NoVs and sapoviruses were detected by several RT-PCRs that amplified regions of the RNA-dependent, RNA polymerase, and capsid genes (1, 19); hepatitis A virus (HAV) was identified by amplification of a VP1 gene fragment (1); and enteroviruses were identified by amplification of the 5′ untranslated region (1). Typing of AV and group G and P typing of RV type A (RV-A)-positive samples were performed as reported previously (1). Aichi virus was detected by amplification of a 519-bp fragment (1).
Shellfish sample analysis.
Sixty-six samples were collected. Four oyster (Crassostrea gigas) samples directly linked to human consumption (two from leftovers from refrigerators in private homes and from a restaurant and two from exactly the same batch in producers) were analyzed, as were 62 oyster samples collected from the same production area. Shellfish, kept at 4°C during shipment, were washed and shucked, and the total weight was recorded. The stomach and digestive diverticula (DT) were removed by dissection and divided into 1.5-g portions. For analysis, digestive tissues were homogenized, extracted with chloroform-butanol, and precipitated with Cat-floc (Calgon, Ellwood City, PA), followed by polyethylene glycol 6000 (Sigma, St. Quentin, France) precipitation (2). Viral NAs were extracted and purified using proteinase K, phenol-chloroform, and cetyltrimethylammonium bromide as previously described (19). The NA was suspended in 100 μl of RNase-free water with 20 units of RNase inhibitor (Invitrogen) and analyzed immediately or kept frozen (−80°C).
Real-time RT-PCR.
All shellfish NA extracts were first screened by real-time RT-PCR (rRT-PCR) using previously published primers and probe for NoV (8), HAV (7), and enterovirus (EV) (9). rRT-PCR was performed with an MX3000 (Stratagene, France) instrument or an ABI Prism 7000 SDS detector (Applied Biosystem, France), using a Platinum quantitative RT-PCR ThermoScript one-step system (Invitrogen, France). Briefly, 5 μl of undiluted or 10-fold-diluted RNA extract was added in duplicate to 20 μl of a mixture containing 1× Thermoscript reaction buffer, 200 nM of the probe and primers, 0.5 μM of Rox reference dye (50×), 0.5 μl of Thermoscript Plus/Platinum Taq enzyme mixture, and 2 U of RNase inhibitor (Applied Biosystems, France). RT was performed for 30 min at 50°C and denaturation for 5 min at 95°C, followed by 45 cycles of PCR amplification (denaturation at 95°C for 15 s, annealing and extension at 60°C for 1 min). The cycle threshold (CT) was defined as the cycle at which a significant increase in fluorescence occurred (i.e., when fluorescence became distinguishable from the background) (8). Precautions such as isolated rooms for various steps and the use of filter tips were taken to prevent false-positive results. Two negative amplification controls (water) were included in each amplification series, and no more than six samples were analyzed in an RT-PCR assay.
The number of NoV RNA copies present in positive samples was estimated using standard curves generated from RNA transcripts. In brief, the first two open reading frames of the GI.1 Norwalk virus (nucleotides [nt] 146 to 6935) and the sequence between nucleotides 4191 and 5863 of the GII.4 Houston virus (Hu/Houston/TCH186/2002/US; GenBank accession no. EU310927) were each cloned in the pCRII TOPO (Invitrogen) vector. In vitro transcription was performed with linearized plasmid samples, using a Promega riboprobe system. After DNase treatment, RNA was purified and quantified by optical density at 260 nm (10). A standard curve was generated using 5.3 to 530,000 and 7.5 to 750,000 copies of transcript for GI and GII, respectively, and the genomic copy number was determined by interpolation using the CT values generated from the shellfish extracts. To be evaluable, all wells had to yield a CT value of ≤41. The final concentration was then adjusted based on the NA volume analyzed (5 μl of NA extract) and reported per g of DT (1.5 g analyzed).
The presence of RT-PCR inhibitors was evaluated by coamplification of 2.5 μl of each NA extract with 2.5 μl containing 100 copies of GI or GII RNA internal controls in separate experiments (7). Amplification of RNA indicated that no more than partial inhibition was present; no adjustments to quantitative estimates were made for samples with partial inhibition.
Standard RT-PCR.
The viruses that were detected in samples by real-time RT-PCR were typed by sequencing after amplification with a standard, two-step RT-PCR format using 40 cycles of amplification (17, 19). For detecting NoV, six primer sets targeting the polymerase gene and three targeting the capsid gene were used (17, 19). In some cases, a seminested PCR was performed using the same amplification conditions, taking precautions to avoid cross-contamination (each sample was amplified alone and with negative controls) (19).
RV detection was performed by amplification of a portion of the VP6 gene and confirmed by hybridization (27). AV was detected by amplification of a small fragment in the 3′ noncoding region, and positive samples were typed using the same primers as those for stool samples (1). AiV was detected by using primers that amplify the polymerase gene (24, 29). Virus-specific amplicons were identified by liquid hybridization. Nested amplification was performed to generate an amplicon of 179 bp which could then be sequenced.
Sequence analysis.
Amplicons from virus-positive samples were purified and sequenced with a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Sequences were analyzed through the European Food-Borne Viruses Database (https://hypocrates.rivm.nl/bnwww; sequence no. FBVE QLK1-CT-1999-00594) for identification of the NoV genotypes. For other enteric viruses, sequence homologies obtained from all samples were evaluated using the BLAST search program in GenBank.
Environmental investigations.
Oysters implicated in the outbreaks were produced in several sites, all of which were located in one lagoon in southern France. Data from climate events (Meteo France) and the epidemiological status of the population (Sentiweb [http://www.sentiweb.org]) were obtained. At the same time, IFREMER surveillance network (REMI [http://www.ifremer.fr]) data for shellfish quality (Escherichia coli concentrations) were collected.
RESULTS
Epidemiological investigation.
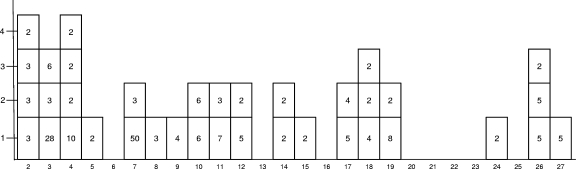
A total of 38 clusters of cases with acute gastroenteritis were traced to oyster consumption between 2 and 27 February (Fig. 1). Thirty-one clusters were reported by ill people directly to sanitary services, and seven were reported by physicians. A total of 205 cases with acute gastroenteritis were identified. Vomiting was reported in 96%, diarrhea in 92%, abdominal pain in 92%, and fever in 50% of cases. Two persons were hospitalized for 1 day. Median incubation periods were between 12 and 54 h. Twenty-two (58%) of the clusters were associated with oysters purchased at a market and consumed in private homes. Thirteen (35%) of the clusters were associated with oysters consumed in a restaurant. One event occurred after oyster consumption in a school and one at a banquet during a scientific meeting.
FIG. 1.
Clusters occurring during February. Clusters are reported for each day of February (x axis), and the number of clusters reported per day (y axis) are shown. Each cluster is represented by a square, and the number of cases per cluster is written in each square.
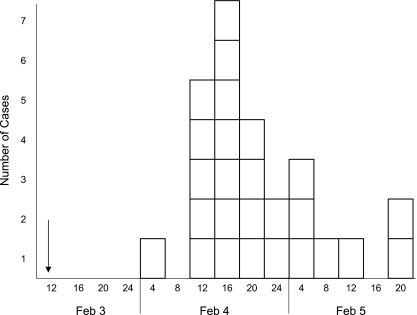
The largest cluster of cases was on 3 February at the scientific meeting with 77 attendees. Twenty-seven attendees were interviewed by phone on 10 February. Participants were adults (between 26 and 59 years old) with a sex ratio of 1:6 (male/female). Eleven attendees had symptoms of gastroenteritis with diarrhea (73%), abdominal pain (64%), vomiting (55%), nausea (82%), and fever (64%). The median incubation period was 39 h (range, 27 to 60 h) (Fig. 2). The retrospective cohort study showed that people who ate oysters had a 4.5-fold greater risk of illness than people who did not eat oysters (RR = 4.5; 95% confidence interval, 1.6 to 13.3; P = 0.003). Other foods served (fish soup, chicken, vegetables, chocolate mousse) were not associated with disease occurrence. Twenty-six of 27 Italian persons who also participated in the meeting were contacted later. Seventeen of this group ate oysters and were ill. Of the nine persons who did not consume oysters, only one reported gastroenteritis symptoms.
FIG. 2.
Scientific meeting cluster: onset of symptoms. Oysters were consumed on 3 February for lunch (arrow). Each box represents one clinical case occurring every 4 h (x axis) and the number of cases (y axis).
Stool analysis results.
A total of 12 stool samples collected from different sick patients were analyzed: two from people who ate oysters during a family dinner (cluster 1), four from people who attended the scientific meeting (cluster 2), and six from people who bought their oysters from the same producer at a market (cluster 3). All stool samples were negative for adenovirus, sapovirus, and HAV. One stool sample (E1201) had no viral pathogens detected, and the other 11 samples were positive for at least one virus (Table 1). Nine (75%) samples were positive for NoV, six for AiV (50%), six for EV (50%), three for AV (25%), and two for RV-A (17%). Eight samples were positive for multiple enteric viruses, and one stool had seven different viruses (AiV, AV type 8, EV, NoV GI.1, NoV GII.17, RV G1P[8], and RV G9P[8]). One stool sample was contaminated only with AiV and two only with NoV. Five stool samples were contaminated by two different NoV strains. One stool sample (E1203) had two RV strains, G1P[8] and G9P[8].
TABLE 1.
Results obtained from stool samples and related shellfish samples
| Cluster date | Stool sample | Shellfish sample | Virus
|
Genotype(s)a
|
||||
|---|---|---|---|---|---|---|---|---|
| AiV | AV | EV | RV | NoV GI | NoV GII | |||
| 8 February | 73 | − | − | + | − | + GI.2 | − | |
| 74 | + | − | + | − | + GI.1 | + GII.2 | ||
| 109 | + | + | − | − | + GI.4 | − | ||
| 1739 | + | + | − | + | + GI | − | ||
| 15-17 February | E1196 | − | − | + | − | − | + GII.7, GIIb | |
| E1197 | + | + | + | − | − | − | ||
| E1201 | − | − | − | − | − | − | ||
| E1202 | + | − | − | − | − | − | ||
| 93 | − | + | − | − | − | + GII.4 | ||
| 107 | + | + | + | − | + GI.1 | + GII | ||
| 110 | − | + | − | + | + GI.4 | + GII | ||
| 18 February | E1203 | + | + | + | + | + GI.1 | + GII.17 | |
| E1204 | − | − | − | − | − | + GII.4 | ||
| E1205 | − | − | − | − | + GI.1 | + GII.4 | ||
| E1206 | − | − | + | − | + GI.1 | − | ||
| E1207 | + | − | − | − | + GI.2 | + GII.7 | ||
| E1208 | + | + | − | + | − | + GIIb, | ||
| 140 | + | + | − | + | + GI | + GII.4 | ||
| 115 | + | + | − | − | − | − | ||
| 130 | − | + | − | − | + GI.2 | |||
| 131 | + | − | − | + | + GI | + GII.4 | ||
Genotypes are listed (+) if genotypes were determined; − indicates genotype was undetermined.
A large number of different virus strains were identified by sequence analysis. There were seven NoV genotypes (GI.1, GI.2, GII.2, GII.4, GII.7, GII.17, and GIIb), two AV types (4 and 8), and three AiV strains.
Shellfish analysis results.
Shellfish samples that could be linked to consumers based on consumer, producer, and environmental data were identified (Table 2). Inhibitor removal was evaluated for all samples, and only samples showing less than 50% inhibition were considered for further analysis. Two samples left over from consumers (Table 2, samples 1739 and 140) were positive for AiV, AV, NoV (one sample having both the GI and GII strains), and RV. Two other samples (138 and 139) from producers were also positive for virus but contained only AV (these samples were not included in the table as no stool specimens linked to these samples were collected). Sample 1739 had a low level of virus contamination near the limit of detection as estimated by rRT-PCR, but the same approximate value was consistently obtained with repeated testing (about 72 to 130 RNA copies/g of DT) (Table 2). Repeated analyses of samples from the same lot (except for sample 140, for which only a limited number of oysters were available, such that only one extraction could be performed) yielded quantitative estimates within fourfold (two CT) of each other, except for that obtained for sample 109 (Table 2).
TABLE 2.
NoV estimated concentrations found in shellfish related to human casesa
| Cluster | Shellfish sample | NoV GI
|
NoV GII
|
||
|---|---|---|---|---|---|
| Genotype | RNA copies/g DT | Genotype | RNA copies/g DT | ||
| 1 | 109 | + GI.4 | 150-3,700 | − | |
| 1739 | + GI | 72-130 | − | ||
| 2 | 93 | − | + GII.4 | 1,600-2,500 | |
| 107 | + GI.1 | 5,000-16,000 | + GII | DL | |
| 110 | + GI.4 | DL | + GII | DL | |
| 3 | 140 | + GI | 2,300 | + GII.4 | 1,100 |
| 130 | + GI.2 | 610-2,300 | − | ||
| 131 | + GI | 260-880 | + GII.4 | DL-79 | |
Numbers represent copy numbers observed for two separate extractions. DL, the sample was too close to the limit of detection for quantification. −, indicates no virus was detected.
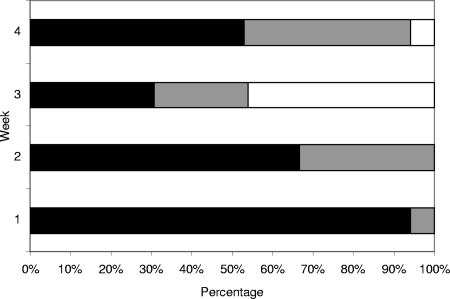
A total of 62 samples were collected from the same production area in the lagoon over a period of 30 days (6 February to 8 March 2006). Among the 62 samples analyzed within the 4-week period of time (the last 2 days were added to week 4), seven (11%) samples were negative for virus, 16 (26%) were contaminated by one enteric virus, and 39 (63%) were contaminated by at least two different enteric viruses. Sixteen of the 17 samples collected during the first week, were contaminated by two or more different enteric viruses. Five different viruses were detected in one sample and four viruses in four samples (Fig. 3). Of the 15 samples collected during the second week, one was contaminated with five different viruses and six with at least two viruses. In weeks 3 and 4, 31% of 13 samples and 40% of 17 samples, respectively, were contaminated by more than one (all with only two) virus strain. AV was detected in a large number of samples (41), primarily during the first 2 weeks (94% and 78% samples were positive for the first and second weeks, respectively) (Table 3). RV was detected in 15 samples throughout most of the study period, except during the third week (Table 3). NoV was detected in 33 samples, with 76% positive samples collected the first week, then 33, 30, and 59% for the following weeks. In the first week, 12 samples (among 17) were positive for GII NoVs, and GI NoVs were detected in 10 samples, of which 9 samples were contaminated by both genogroups (Table 3). At the end of the study, GII NoVs were detected in 3 samples and GI NoVs in 7 samples (among 17) (Table 3), and no sample was contaminated by both genogroups. In the first week, the mean concentrations were about 7 × 103 RNA copies/g for GI NoV and 1.3 ×103 RNA copies/g for GII NoV (Table 4). After 3 weeks, the concentrations decreased and reached an average of 2 × 102 RNA copies/g for each genogroup (Table 4).
FIG. 3.
Multiple contaminations observed for shellfish samples over time. Black bars indicate two or more different enteric viruses detected per sample; gray bars indicate one virus detected per sample; white bars indicate no virus detected. The x axis shows the percentage of positive samples; the y axis shows week of detection.
TABLE 3.
Viral contamination in shellfish over 4 weeks
| Week | No. of samples | No. of samples positive for:
|
Negative | |||||
|---|---|---|---|---|---|---|---|---|
| Virus
|
NoV genotype(s)
|
|||||||
| AiV | AV | RV | Both GI and GII | GI only | GII only | |||
| 1 | 17 | 2 | 16 | 6 | 9 | 1 | 3 | 0 |
| 2 | 15 | 2 | 12 | 4 | 2 | 3 | 1 | 0 |
| 3 | 13 | 1 | 6 | 0 | 1 | 2 | 1 | 6 |
| 4 | 17 | 0 | 7 | 5 | 0 | 7 | 3 | 1 |
TABLE 4.
Detection and quantification of NoV in shellfish samples
| Week | Total no. of samples | Genogroup I
|
Genogroup II
|
||
|---|---|---|---|---|---|
| Positive samples (%) | Mean concn (RNA copies/g of digestive tissue) | Positive samples (%) | Mean concn (RNA copies/g of digestive tissue) | ||
| 1 | 17 | 10 (59) | 6,900 | 12 (70) | 1,300 |
| 2 | 15 | 5 (33) | 3,100 | 3 (20) | 120 |
| 3 | 13 | 3 (23) | 120 | 2 (15) | DLa |
| 4 | 17 | 7 (41) | 220 | 3 (17) | 200 |
DL, the sample was too close to the limit of detection for quantification.
AiV was detected in five samples (two samples collected the first week, two during the second week, one during the third week, and none during the last week) (Table 3). Only one sample was found contaminated by EV, and no samples were contaminated by HAV.
Sequence comparisons.
Only one sequence (GII.4) could be obtained from sample 140, directly linked to the clinical cases, and none from sample 1739. The GII.4 sequence obtained by standard RT-PCR targeting the capsid region was identical to the sequences obtained from two stool samples (E1204 and E1205). For AiV, the same sequence was detected in two shellfish samples (Fig. 4, samples 115 and 131) and three stool samples (Fig. 4). Two other stool samples contained a virus sequence that differed at a single nt, and one shellfish sample collected during the third week contained a virus with three nucleotide changes over the 140 nt sequenced. The same type 8 AV sequence was obtained from one shellfish sample (no. 115) and two stool samples (E1203 and E1208). Two other AV sequences were identified: a type 4 strain in stool sample E1197 and a type 1 strain in shellfish sample 107. One additional shellfish sample, collected on 10 February, was also positive for AV type 1. For NoV, three genotypes with identical sequences were detected in shellfish and stool samples: two genogroup I (GI.1 and GI.2) and one genogroup II (GII.4). Other genotypes were also detected either in stool or shellfish samples (Tables 1 and 2). No sequence data could be obtained for shellfish samples positive for EV or RV.
FIG. 4.
Alignment of AiV sequences obtained from stool and shellfish samples. Sequences aligned here are those between nucleotides 6298 and 6438, based on the reference strain A846/88 (accession no. AB010145) (16) and on five sequences obtained from stool samples (numbered E1197 to E1207) and three from shellfish samples (samples 131, 115, and 152). Conserved nucleotide are represented by a dash, and nucleotide differences are shown by base changes.
Environmental data and sanitary measures.
In 2006, winter gastroenteritis outbreaks reached a peak in the population (up to 742 cases/100,000 inhabitants, with an outbreak threshold of 182 cases/100,000) in this area of France from 7 to 15 January (http://websenti.b3e.jussieu.fr/sentiweb/). One week later, heavy rain (138.2 mm) occurred in this area for 1 week (23 to 29 January), with an accumulation of 76 mm on 1 day (Meteo France data). This amount of water within 1 week or even 1 day is much more than the monthly average of 65.18 mm shown by data collected over the previous 43 years (since 1963). On 30 January, the IFREMER shellfish surveillance network (REMI) increased shellfish sampling in this area and set up an alert to inform producers. Samples collected on 30 January showed an increase of E. coli contamination in the area (concentrations between 700 and 4,800 E. coli organisms/100 g for nine samples). Based on E. coli results on 31 January, the administration advised producers about the risk and asked them to adopt depuration times to improve shellfish quality based on E. coli counts for a period of 2 weeks. On 6 February, all shellfish collected in this area met European regulation requirements (class B, i.e., less than 3,600 E. coli organisms/100 g of shellfish meat). On 7 February, the first information concerning outbreaks was received, and thus, on 10 February, producers were requested to withdraw shellfish collected between 30 January and 5 February from the market. On 13 February, E. coli controls showed no evidence of bacterial contamination. On 20 February, as human cases were still being reported, shellfish collected before 17 February were withdrawn from the market. On 1 March, 1 month after the contaminating event, the area was closed, and all marketing of shellfish collected from this lagoon was prohibited for 3 weeks. On 20 March, three shellfish samples were analyzed. One was positive for NoV GI (near the limit of detection). The area was reopened, and no additional outbreak related to shellfish consumption was reported.
DISCUSSION
Shellfish are known to be vulnerable to contamination by sewage due to their filter-feeding activities, and contamination of shellfish-growing waters has been a cause of gastroenteritis outbreaks. Food contamination by sewage has often been suspected when multiple strains are detected in food or in patient stool samples, as previously reported with shellfish (4, 5, 15, 18, 19). Only noroviruses were detected or reported in prior reports of gastroenteritis outbreaks caused by multiple strains. For example, Boxman et al. (5) described a mixed infection in four patients, with one person infected by three virus genotypes. Two of these strains were detected in implicated shellfish samples. Kageyama et al. (15) reported that the majority of mixed norovirus infections were associated with shellfish consumption. Previously, based on epidemiological investigation and analysis of the shellfish implicated in the outbreak, we detected multiple norovirus contamination (up to five different NoV strains) involved in the same area of production in southern France (19).
The outbreak reported here is remarkable for the diversity of human enteric viruses detected in both clinical and oyster samples. Up to seven different viruses were detected together in environmental or clinical samples. Few studies have reported mixed infections with several different enteric viruses. In an HAV outbreak linked to frozen clams imported from Peru, RVs and EV were also detected in shellfish samples, but no data linked to human illness were reported (4). NoVs and human EVs were detected in both oyster and stool samples in one other report (6). The EVs were suspected to be responsible for secondary symptoms such as myalgia and arthralgia. In the present study, clinical signs could not be used to discriminate among the roles of the different viruses detected in fecal samples of symptomatic persons. We are also not able to determine whether coinfection with multiple viruses contributed to the severe symptoms reported by some affected persons. However, the large number of enteric viruses involved in the outbreaks and the symptom severity may explain the unusually higher number of direct reports made to sanitary authorities.
To our knowledge, AiV was not reported previously in a shellfish-associated gastroenteritis outbreak in Europe. AiV was first recognized in 1989 as the cause of oyster-associated nonbacterial gastroenteritis in Japan (29). There is a single previous report of an AiV-associated outbreak in Europe, but no details of the outbreak, which occurred in Germany, were provided (24). AiVs have not been found in other studies of outbreak investigations in Finland (14) and The Netherlands (25). In a retrospective study performed in France, 0.9% of stool samples collected from children between 2001 and 2004 were positive for AiV (1). Our previous attempts to detect AiV in shellfish implicated in outbreaks failed (data not shown). The detection of AiV in shellfish from the contaminated harvesting area and the identification of this virus as the sole pathogen found in at least one person's stool sample suggest that AiV contributed to the illness burden seen in these outbreaks.
There is still relatively little quantitative information for the levels of NoV contamination in shellfish implicated in outbreaks. Quantification of noroviruses in shellfish is a complex procedure. It is subject to problems with inhibition of the RT-PCR by shellfish tissue components, which can cause false negatives. Several precautions were taken in this study to avoid such false negatives (persistence of RT or PCR inhibitors was evaluated) or false positives (separate room, filter tips, several negative controls). When sufficient amounts of shellfish tissues were available, two extractions were performed. All but one sample had quantitative estimates that were within fourfold (two CT values) of each other. Sample shellfish lot 109 had a larger variation, which may have been due to greater variability in levels of virus contamination among the shellfish collected for this lot. The quantitative estimates reported here are likely to be minimal values, as several factors may have led to underestimating the actual level of virus contamination. For example, nucleic extraction efficiencies using an external added virus, as proposed by Costafreda et al. (7), were not used in this study. In addition, we made no adjustments for PCR amplification efficiency, which may have been adversely affected by partial sample inhibition or by sequence variation in different norovirus genotypes, leading to mismatches with the primers used in the real-time assay. The latter concern is offset by the results of previous studies that have demonstrated the broad reactivity of the NoV-specific primers and probes used in the current study (8, 15, 25).
The level of NoV contamination in the present report is similar to levels of viral shellfish contamination described for NoV outbreaks (18, 19), for an HAV outbreak (7), and for a field production area in Japan (23). More oyster samples were found to be contaminated with GII NoVs than GI strains in the first week after the contamination event, but after 4 weeks, a greater percentage of samples contained GI NoV strains. Previous studies have shown that GI NoVs are more resistant to sewage treatment (8) and are more often implicated in food-related outbreaks than are GII NoV strains (5, 11, 15, 19). The findings in the present study support the hypothesis that GI NoVs are more stable in the environment and may explain the relatively higher frequency with which strains from this genogroup are associated with food-borne outbreaks.
Shellfish are regularly consumed in France, and it is important to avoid such outbreaks to protect consumer health. The harvesting area, classified as a B area, is known to be sensitive to rainfall events and sewage contamination (19, 22). When heavy rains were observed during the winter gastroenteritis outbreaks, conditions previously responsible for a large shellfish outbreak 4 years ago, an advisory alert was set up with increased evaluation of bacteriological controls and producer information. IFREMER and the sanitary service (DDASS) recommended closure of the production area on 30 January, but this advice was not followed by the regional authority. Instead, producers were instructed to increase depuration so that shellfish conformed to bacterial contamination regulatory requirements in accordance with European regulation. This approach was not sufficient, and the outbreaks were not prevented, as shown by data presented here. Some shellfish samples, kept for several days in depuration tanks before being sent to the laboratory for analysis, were still contaminated by different types of enteric viruses (data not shown). This is additional evidence that short-term depuration to meet recommended bacterial regulatory requirement is not efficient at removing contaminating viral pathogens. Specific binding of noroviruses to shellfish tissues may contribute to delayed depuration of these viruses (20, 26), but the importance of this mechanism for other enteric viruses is not known at this time.
In summary, this study is informative for several reasons. (i) This study presents additional evidence that coliform indicators are not reliable for viral elimination and that the depuration process as done is not efficient for enteric viruses. (ii) Closure of the harvesting area just after a flooding event as proposed by IFREMER likely would have prevented some of the outbreaks. (iii) Low levels of NoV were infectious in consumers, and the number of virus-contaminated samples decreased slowly over a 1-month period. (iv) Many different enteric viruses were found both in stool and in shellfish samples, showing the potential impact of sewage contamination by emerging strains. (v) This study presents the first documentation of Aichi virus transmission by food in Europe.
Acknowledgments
This study was supported by a European Community Food-borne Viruses in Europe grant (FBVE, QLK1-CT-1999-00594), EVENT (FP6-2002-SSP-1), and Rephepa (Virus-Safe-Seafood, QLK1-1999-00634).
We thank all contributors for their help in sampling and data collection (DSV, DDASS, CIRE, and private clinical laboratories).
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Ambert-Balay, K., M. Lorrot, F. Bon, H. Giraudon, J. Kaplon, M. Wolfer, P. Lebon, D. Gendrel, and P. Pothier. 2008. Prevalence and genetic diversity of Aichi virus in community and hospitalized patients. J. Clin. Microbiol. 461252-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 613014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35275-290. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, A., G. Sanchez, F. Le Guyader, H. Vanaclocha, L. Haugarreau, and R. M. Pinto. 2001. Human enteric viruses in coquina clams associated with a large hepatitis A outbreak. Water Sci. Technol. 4361-65. [PubMed] [Google Scholar]
- 5.Boxman, I. L. A., J. J. H. C. Tilburg, N. A. J. M. te Loeke, H. Vennema, K. Jonker, E. de Boer, and M. Koopmans. 2006. Detection of noroviruses in shellfish in the Netherlands. Int. J. Food Microbiol. 108391-396. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, B. F., D. Lees, K. Henshilwood, T. Bjergskov, and J. Green. 1998. Human enteric viruses in oysters causing a large outbreak of human food borne infection in 1996/97. J. Shellfish Res. 171633-1635. [Google Scholar]
- 7.Costafreda, I., A. Bosch, and R. M. Pinto. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 723846-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silva, A., J. C. Le Saux, S. Parnaudeau, M. Pommepuy, M. Elimelech, and F. S. Le Guyader. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 737891-7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donia, D., M. Divizia, and A. Pana. 2005. Use of armored RNA as a standard to construct a calibration curve for real-time RT-PCR. J. Virol. Methods 126157-163. [DOI] [PubMed] [Google Scholar]
- 10.El Senousy, W. M., S. Guix, I. Abid, R. M. Pinto, and A. Bosch. 2007. Removal of astrovirus from water and sewage treatment plants, evaluated by a competitive reverse transcription -PCR. Appl. Environ. Microbiol. 73164-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallimore, C., J. S. Cheesbrough, K. Lamden, C. Bingham, and J. Gray. 2005. Multiple norovirus genotypes characterised from an oyster-associated outbreak of gastroenteritis. Int. J. Food Microbiol. 103323-330. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, J. B., R. W. Litaker, and R. T. Noble. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 723960-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovi, T. 2006. Surveillance for polioviruses. Biologicals 34123-126. [DOI] [PubMed] [Google Scholar]
- 14.Jokela, P., P. Joki-Korpela, M. Maaronen, V. Glumoff, and T. Hyypia. 2005. Detection of human picornaviruses by multiplex reverse transcription-PCR and liquid hybridization. J. Clin. Microbiol. 431239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 422988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopmans, M., and E. Duizer. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 9023-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Guyader, F. S., L. Haugarreau, L. Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 663241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Guyader, F. S., F. H. Neil, E. Dubois, F. Bon, F. Loisy, E. Kohli, M. Pommepuy, and R. L. Atmar. 2003. A semi-quantitative approach to estimate Norwalk-like virus contamination of oysters implicated in an outbreak. Int. J. Food Microbiol. 87107-112. [DOI] [PubMed] [Google Scholar]
- 19.Le Guyader, F. S., F. Bon, D. DeMedici, S. Parnaudeau, A. Bertone, S. Crudeli, A. Doyle, M. Zidane, E. Suffredini, E. Kohli, F. Maddalo, M. Monini, A. Gallay, M. Pommepuy, P. Pothier, and F. M. Ruggeri. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J. Clin. Microbiol. 443878-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Guyader, F. S., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvoen-Clouet, M. Pommepuy, and J. Le Pendu. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Guyader, F. S., and R. L. Atmar. 2008. Binding and inactivation of viruses on and in food, with a focus on the roles of the matrix, p. 189-208. In M. Koopmans, A. Bosch, and D. Cleaver (ed.), Food-borne viruses: progress and challenges. ASM Press, Washington, DC.
- 22.Miossec, L., F. Le Guyader, L. Haugarreau, M. A. Comps, and M. Pommepuy. 1998. Possible relationship between a winter epidemic of acute gastroenteritis in France and viral contamination of shellfish. J. Shellfish Res. 171661-1664. [Google Scholar]
- 23.Nishida, T., H. Kimura, M. Saitoh, M. Shinohara, M. Kato, S. Fukuda, T. Munemura, T. Mikami, A. Kawamoto, M. Akiyama, Y. Kato, K. Nishi, K. Kozawa, and O. Nishio. 2003. Detection, quantitation, and phylogenetic analysis of noroviruses in Japanese oysters. Appl. Environ. Microbiol. 695782-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh, D. Y., P. A. Siva, B. Hauroeder, S. Diedrich, D. D. P. Cardoso, and E. Schreier. 2006. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 1511199-1206. [DOI] [PubMed] [Google Scholar]
- 25.Svraka, S., E. Duizer, H. Vennema, E. de Bruin, B. van der Veer, B. Dorresteijn, and M. Koopmans. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J. Clin. Microbiol. 451389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian, P., A. H. Bates, H. M. Jensen, and R. E. Mandrell. 2006. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett. Appl. Microbiol. 43645-651. [DOI] [PubMed] [Google Scholar]
- 27.Villena, C., W. M. El-Senousy, F. X. Abad, R. M. Pinto, and A. Bosch. 2003. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl. Environ. Microbiol. 693919-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widdowson, M. A., A. Sulka, S. N. Bulens, R. S. Beard, S. S. Chaves, R. Hammond, E. D. P. Salehi, E. Swanson, J. Totaro, R. Woron, P. S. Mead, J. S. Bresee, S. S. Monroe, and R. I. Glass. 2005. Norovirus and foodborne disease, United States, 1991-2000. Emerg. Infect. Dis. 1195-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita, T., M. Sugiyama, H. Tsuzuki, K. Sakae, Y. Suzuki, and Y. Miyazaki. 2000. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 382955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]