Abstract
A real-time reverse transcriptase PCR (RT-PCR) assay that targeted both the mumps virus F gene and human RNase P in clinical samples was adapted for use with the LightCycler platform. LightCycler RT-PCR is as sensitive as conventional nested RT-PCR and significantly less expensive and labor-intensive, making it ideal for mumps diagnosis during outbreaks.
Mumps is a vaccine-preventable disease caused by an RNA virus of the family Paramyxoviridae. Despite vaccination strategies, recent outbreaks in the United Kingdom, United States, and Canada highlight the need for rapid diagnosis (1, 3, 9, 12). Serological methods have limited (if any) utility for the diagnosis of mumps in partially immunized individuals (7, 10). Detection of mumps virus RNA by reverse transcriptase PCR (RT-PCR) and a subsequent second round of nested PCR (RT-n-PCR) is the most sensitive method for laboratory confirmation (4; G. Tipples et al., http://www.nml-lnm.gc.ca/guide/docs/Mumps_Lab__guide.pdf). As part of public health management of a previous mumps outbreak (12), RT-n-PCR (4) was implemented in Nova Scotia. However, a larger outbreak this year prompted investigations into faster methodologies (i.e., real-time RT-PCR).
Unlike methods targeting the highly variable small hydrophobic (SH) gene (2), a real-time PCR with a target amplifying the most conserved region of the mumps virus genome, the fusion (F) gene, was described by Uchida et al. (11). In order to decrease sample turnaround times and minimize the potential for amplicon contamination, real-time RT-PCR assays have recently evolved from a two-step process (a first round of RT-PCR followed by a round of real-time PCR) (2, 5, 11) to one-step processes (RT-PCR only) (2, 6, 8). Despite their increased sensitivity and speed, few real-time RT-PCR assays have been validated for clinical use (2, 5, 6, 8, 11). Boddicker et al. (2) developed a one-step real-time RT-PCR assay targeting both the mumps virus SH gene and the endogenous internal control Homo sapiens RNase (RNase P) that could be used on clinical specimens. However, among others, this assay was not validated for use on the LightCycler platform. Jin et al. (5) described a real-time method compatible with the LightCycler platform which targeted hemagglutinin-neuraminidase using minor-groove binder technology. However, this assay involved a two-step real-time PCR approach. This study adapted methods described by Uchida et al. (11) for one-step real-time RT-PCR detection of mumps virus RNA targeting the F gene and incorporated the internal control RNase P described by Boddicker et al. (2). This assay was evaluated and compared to conventional RT-n-PCR for the detection of mumps virus RNA in clinical specimens submitted during an outbreak in Nova Scotia.
In accordance with recent National Microbiology Laboratory guidelines (G. Tipples et al., http://www.nml-lnm.gc.ca/guide/docs/Mumps_Lab__guide.pdf), buccal swabs in universal transport media (Copan Diagnostics, Corona, CA) and urine specimens were collected from patients presenting to Nova Scotia physicians with symptoms suggestive of mumps virus infection or with epidemiological contact with a mumps virus-positive individual. Specimens were refrigerated and tested within 12 h.
Nucleic acid extraction was performed on a MagNA Pure LC (Roche Diagnostics, Branchburg, NJ) from 140 μl of specimen with a total nucleic acid isolation kit and a 60-μl elution volume. Real-time RT-PCR was performed on a LightCycler 2.0 instrument using a QuantiTect Multiplex NR RT-PCR kit in 20-μl reaction mixtures consisting of 1× master mix, 2 μl enzyme, 300 nM of primers, and 100 nM of probes. Sequences for the primers and probes have been described elsewhere (2, 11). To better complement the LightCycler platform, the RNase P probe (2) was modified (the 3′ and 5′ ends were labeled with Pulsar 650 and BHQ2, respectively). Thermocycling conditions were as follows: reverse transcription at 48°C for 45 min, initial denaturation at 95°C for 15 min, and 45 cycles of denaturation at 95°C for 15 s and combined annealing/extension steps at 59°C for 60 s. Fluorescence analyses for the F gene and RNase P were performed at 530 and 705 nm, respectively.
Ten arbitrarily chosen buccal and urine specimens were evaluated to compare primer/probe combinations targeting either the SH (2) or F (11) gene. Crossing point (Cp; cycle in which the fluorescence crosses the threshold) values for the SH target were consistently lower than those for the F gene, resulting in failure to detect mumps virus in three cases. Where highly conserved targets like the F gene would be able to detect mumps virus RNA (6, 11), sequence variation in primer or probe binding sites in highly variable targets such as the SH gene (4) could lead to false-negative results. In light of these results, all subsequent analyses were performed using the F gene.
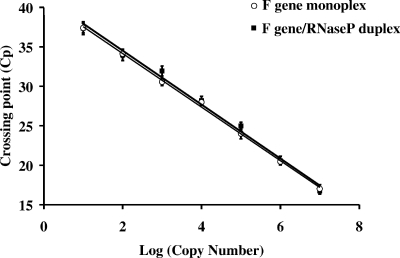
The analytical sensitivities of the monoplex (F gene only) and duplex (F gene and RNase P) assays were evaluated using RNA extracted from 10-fold serial dilutions (n = 5) of cultured mumps virus. Viral copy numbers were estimated using a standard curve generated with known concentrations (determined by spectrophotometry) of a linearized plasmid. The plasmid harbored the F gene target sequence cloned using primers FgFOR (CGCCTCGAGCTGCCAATACAATGAGGCAG) and FgREV (CGCTCTAGACCGTCCAAGGACAATGTTTG). Both monoplex (y = −3.39x + 40.93; R2 = 0.999) and duplex (y = −3.41x + 41.34; R2 = 0.992) reactions displayed an inverse linear relationship (Fig. 1). In both cases, Cp values ranged from 17 to 37 and copy numbers ranged from 107 to 101. Therefore, no significant decrease in sensitivity was observed when primers and the probe targeting RNase P (2) were included in the assay. An internal control offers the ability to monitor the presence of template RNA during RT-PCR, as well as PCR inhibitors potentially present in clinical specimens.
FIG. 1.
Standard curve comparison between monoplex and duplex real-time RT-PCR assays. The monoplex (F gene only) and duplex (F gene and RNase P) assays were compared by using equivalent amounts of RNA extracted from serially diluted viral cultures. Copy number was estimated in relation to the standard curve generated using a known concentration of a linearized plasmid harboring the F gene target sequence.
The analytical specificity of the real-time RT-PCR assay was assessed by testing nucleic acids extracted from various other viruses, including adenovirus; cytomegalovirus; enterovirus; herpes simplex virus types 1 and 2; influenza virus types A and B; norovirus; measles virus; parainfluenza virus types 1, 2, and 3; respiratory syncytial virus; severe acute respiratory syndrome coronavirus; varicella-zoster virus; and West Nile virus. No cross-reactions with other viruses were observed. It should be noted that the real-time RT-PCR assay was able to detect not only the circulating genotype G5 strain but also other mumps virus genotypes (A, C, D, G1, G2, G6, H, and J).
To compare real-time RT-PCR and conventional RT-n-PCR (Table 1), parallel testing was prospectively performed on specimens submitted during the Nova Scotia outbreak. RT-n-PCR was performed using a Qiagen One-Step RT-PCR kit by following a well-established protocol (4). Of 478 specimens, 125 (26%) samples were positive, representing 73 (of 261) buccal swabs, 51 (of 214) urine specimens, and 1 (of 3) cerebrospinal fluid specimen. A single discordant result was observed: a urine specimen was positive by real-time RT-PCR (F gene) and negative by RT-n-PCR (SH gene). Since mumps virus RNA was also detected by an alternative method (2) and the paired buccal swab in this patient was also positive, it was concluded that this specimen was likely a true positive. Analysis of the data suggests that results should be interpreted as follows: positive if Cp values are below 37, equivocal if Cp values are between 37 and 40, and negative if the Cp values for the F gene are above 40 (or undetected) and the Cp value for RNase P is below 37. A sample would have been considered indeterminate if it was negative for the F gene target and amplification of RNase P did not occur; however, RNase P was detected in all cases.
TABLE 1.
Comparison of RT-n-PCR results against real-time RT-PCR
| Sample type and RT-n-PCR result | No. of specimens with real-time RT-PCR result of:
|
Total no. of specimens | |
|---|---|---|---|
| Positive | Negative | ||
| Buccal swabs | |||
| Positive | 73 | 0 | 73 |
| Negative | 0 | 188 | 188 |
| Total | 73 | 188 | 261 |
| Urine | |||
| Positive | 50 | 0 | 50 |
| Negative | 1 | 163 | 164 |
| Total | 51 | 163 | 214 |
| Cerebrospinal fluid | |||
| Positive | 1 | 0 | 1 |
| Negative | 0 | 2 | 2 |
| Total | 1 | 2 | 3 |
Real-time RT-PCR technology offers a number of advantages compared to conventional RT-n-PCR. Following extraction, an average experiment by real-time one-step RT-PCR can be performed in approximately 2.5 h, compared to 8 h for conventional RT-n-PCR, and the average cost per specimen (without labor) for a typical run of 32 samples with controls was $15.60 for conventional RT-n-PCR compared to $9.31 for real-time RT-PCR. When labor cost, in addition to faster turnaround times, is accounted for, real-time RT-PCR undoubtedly offers additional cost savings.
In summary, the real-time RT-PCR assay described herein is sensitive, specific, and far more rapid than conventional RT-n-PCR. In addition, it is significantly less expensive and labor-intensive, making it ideal for mumps diagnosis. Since mumps outbreaks have been documented worldwide, the one-step real-time RT-PCR validated for the Roche LightCycler instrument could easily be implemented in other laboratories using this platform.
Acknowledgments
We thank Li Jin (Health Protection Agency, United Kingdom) for providing various genotypes of mumps virus used in the specificity panel. Secondly, we recognize the efforts of all staff that were instrumental in data collection and managing the Nova Scotia outbreak, including members of Public Health Agency of Canada, the NML, and the microbiology division at the QEII HSC.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Bloom, S., and M. Wharton. 2005. Mumps outbreak among young adults in UK. BMJ 331E363-E364. [DOI] [PubMed] [Google Scholar]
- 2.Boddicker, J. D., P. A. Rota, T. Kreman, A. Wangeman, L. Lowe, K. B. Hummel, R. Thompson, W. J. Bellini, M. Pentella, and L. E. DesJardin. 2007. Real-time reverse transcription-PCR assay for detection of mumps virus RNA in clinical samples. J. Clin. Microbiol. 452902-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, C., J. M. White, E. J. Savage, J. R. Glynn, Y. Chol, N. Andrews, D. Brown, and M. E. Ramsay. 2007. Vaccine effectiveness estimates 2004-2005 mumps outbreak, England. Emerg. Infect. Dis. 1312-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin, L., S. Beard, and D. W. Brown. 1999. Genetic heterogeneity of mumps virus in the United Kingdom: identification of two new genotypes. J. Infect. Dis. 180829-833. [DOI] [PubMed] [Google Scholar]
- 5.Jin, L., Y. Fengt, R. Parry, A. Cui, and Y. Lu. 2007. Real-time PCR and its application to mumps rapid diagnosis. J. Med. Virol. 791761-1767. [DOI] [PubMed] [Google Scholar]
- 6.Krause, C. H., K. Eastick, and M. M. Ogilvie. 2006. Real-time PCR for mumps detection on clinical specimens—comparison with results of conventional methods of virus detection and nested PCR. J. Clin. Virol. 37184-189. [DOI] [PubMed] [Google Scholar]
- 7.Krause, C. H., P. J. Molyneaux, D. O. Ho-Yen, P. McIntyre, W. F. Carman, and K. E. Templeton. 2007. Comparison of mumps-IgM ELISAs in acute infection. J. Clin. Virol. 38153-156. [DOI] [PubMed] [Google Scholar]
- 8.Kubar, A., M. Yapar, B. Besirbellioglu, I. Y. Avci, and C. Guney. 2004. Rapid and quantitative detection of mumps virus RNA by one-step real-time RT-PCR. Diagn. Microbiol. Infect. Dis. 4983-88. [DOI] [PubMed] [Google Scholar]
- 9.Peltola, H., P. S. Kulkarni, S. V. Kapre, M. Paunio, S. S. Jadhav, and R. M. Dhere. 2007. Mumps outbreaks in Canada and the United States: time for new thinking on mumps vaccines. Clin. Infect. Dis. 45459-466. [DOI] [PubMed] [Google Scholar]
- 10.Sanz, J. C., M. D. M. Mosquera, J. E. Echevarria, M. Fernandez, N. Herranz, G. Palacios, and F. DeOry. 2006. Sensitivity and specificity of immunoglobulin G titer for the diagnosis of mumps virus in infected patients depending on vaccination status. APMIS 114788. [DOI] [PubMed] [Google Scholar]
- 11.Uchida, K., M. Shinohara, S. Shimada, Y. Segawa, R. Doi, A. Gotoh, and R. Hondo. 2005. Rapid and sensitive detection of mumps virus RNA directly from clinical samples by real-time PCR. J. Med. Virol. 75470-474. [DOI] [PubMed] [Google Scholar]
- 12.Watson-Creed, G., A. Saunders, J. Scott, L. Lowe, J. Pettipas, and T. F. Hatchette. 2006. Two successive outbreaks of mumps in Nova Scotia among vaccinated adolescents and young adults. Can. Med. Assoc. J. 175483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]