Abstract
The efficiency of transmission of a pathogen within families compared with that between unrelated persons can affect both the strategies needed to control or eradicate infection and how the pathogen evolves. In industrialized countries, most cases of transmission of the gastric pathogen Helicobacter pylori seems to be from mother to child. An alternative model, potentially applicable among the very poor in developing countries, where infection is more common and the sanitary infrastructure is often deficient, invokes frequent transmission among unrelated persons, often via environmental sources. In the present study, we compared the genotypes of H. pylori from members of shantytown households in Peru to better understand the transmission of H. pylori in developing-country settings. H. pylori cultures and/or DNAs were obtained with informed consent by the string test (a minimally invasive alternative to endoscopy) from at least one child and one parent from each of 62 families. The random amplified polymorphic DNA fingerprints of 57 of 81 (70%) child-mother strain pairs did not match, nor did the diagnostic gene sequences (>1% DNA sequence difference), independent of the child's age (range, 1 to 39 years). Most strains from siblings or other paired family members were also unrelated. These results suggest that H. pylori infections are often community acquired in the society studied. Transmission between unrelated persons should facilitate the formation of novel recombinant genotypes by interstrain DNA transfer and selection for genotypes that are well suited for individual hosts. It also implies that the effective prevention of H. pylori infection and associated gastroduodenal disease will require anti-H. pylori measures to be applied communitywide.
How a pathogen is transmitted in host populations can affect the strategies needed to prevent or eradicate infection and disease. It can also affect the relative contributions to genome evolution of interstrain recombination versus mutation and of the selection for increased fitness versus random genetic drift. At one extreme are the epidemic strains of Vibrio cholerae, which thrive in South Asian aquatic ecosystems and which cause debilitating, potentially lethal diarrheal disease in many thousands of people annually (20). Several new strains swept through at-risk human populations following the transfer of genes that increased their infectivity in partially immune hosts or their proliferation or persistence in environmental reservoirs or that conferred resistance to commonly used antibiotics (21, 39). At another extreme are obligate endosymbionts, such as those of nematodes and insects (35, 36), which evolve solely by mutation and selection for phenotypes that may benefit their hosts or that are at least benign. Other microbes, including Helicobacter pylori (7, 8, 19), which was examined in this study have lifestyles intermediate between the extremes of an epidemic, highly virulent pathogen and a benign endosymbiont.
H. pylori chronically infects the gastric mucosa of billions of people worldwide, typically for decades, despite inflammation and other host defenses. Such chronic H. pylori infections constitute major risk factors for duodenal and gastric ulcers and for gastric cancer, one of the most frequently lethal malignancies (3, 13, 33). Especially among the poor in developing countries, H. pylori infection also increases susceptibility to other food- and waterborne pathogens and contributes to iron deficiency anemia and malnutrition (10, 37, 46). This said, most infections seem to be benign; and some, it has been argued, can even be beneficial (7), although the latter view is controversial (25).
Most people in developing countries become chronically infected by H. pylori in infancy (8, 19) and may be exposed to it repeatedly thereafter. For example, in the shantytown in Peru evaluated for the present study, we had estimated that adults are exposed to infectious doses (doses sufficient to reestablish chronic infection after therapeutic eradication of prior infection) at rates of about 20% per person per year. This reinfection rate is far higher than that seen in equivalent clinical trials in industrialized nations (<1% per person per year) (44). Although it is not feasible to monitor transmission directly, it is often ascribed to fecal or vomitus contamination of food or water or perhaps utensils, fingers, etc. (8, 9, 19, 38). The fact that the prevalence of H. pylori infection has declined markedly in recent decades in industrialized countries but not among the poor in developing countries can be ascribed to better sanitation and hygiene in industrialized countries and a negligible risk of reinfection among individuals in such countries once a prior infection has been cured (1, 8, 19, 38).
A consensus “all-in-the-family” model (12, 27), involving mostly mother-to-child transmission, was proposed to explain new infections in industrialized countries. This was based on serologic and urea breath test survey data showing a statistical association between infection in children and in their parents (22, 24, 48) and DNA fingerprint data that revealed identical or nearly identical strains in different members of individual families (15, 28, 32, 41). The DNA analyses are particularly compelling because H. pylori is genetically diverse, and isolates from unrelated persons are usually readily distinguished by random amplified polymorphic DNA (RAPD) fingerprinting or the sequencing of one or a few genes (typical divergence in DNA sequence, ≥3%, on average). As a consequence, any finding of closely matched strains indicates transmission within families or from a common source.
Although H. pylori is typically spoken of as human specific, it was not clear a priori if most established infections in developing countries would also stem from direct transmission from mothers to their infants rather than from other sources. An early finding that the risk of infection in shantytowns in Peru is inversely related to household water quality (30) is consistent with either explanation. However, a model involving at least transient environmental niches for H. pylori that allows its spread between unrelated persons gained plausibility with laboratory findings of H. pylori survival for a few days in unchlorinated mineral water or seawater (31, 40), biofilm formation on inanimate surfaces (11, 50), and proliferation when it was cultured with amoebae (51).
We studied the genotypes of H. pylori strains from members of households in a shantytown in Peru. We found that only ∼30% of strains from children were closely matched to those from their mothers and that the frequency of matched strains did not depend on the child's age. This outcome suggests that H. pylori transmission among unrelated persons is particularly important in developing countries, an outcome with important evolutionary and disease prevention implications, as will be discussed below.
MATERIALS AND METHODS
Setting.
Our study focused on H. pylori strains from residents of Las Pampas de San Juan de Miraflores (PSJM), a periurban shantytown of 40,000 persons of low income (≤$1,500 per family per year, on average) in Lima, Peru, that is representative of many developing-country societies. The health of the residents of PJSM has been under surveillance for more than 20 years by physicians and health care and social workers of the Peruvian nongovernmental organization Proyectos en Informt́ica, Salud, Medicina y Agricultura (PRISMA; http://www.prisma.org.pe/nwWeb/Portal/) (23, 29, 30, 37). Most residents of PJSM become chronically infected with H. pylori in early childhood (30), as is typical in developing countries worldwide (8, 19). All strains and DNAs analyzed here were from persons specifically recruited for this study with informed consent or assent, as appropriate. None were samples of convenience that might have initially been obtained for other purposes.
One hundred eighty-one families in PJSM were chosen at random for possible inclusion from among families found in a recent census to contain at least one parent and one child in the same household. Ninety-eight families met the criteria of agreement to participate and the availability of at least one offspring and one parent. The participants were scheduled for minimally invasive string tests (42, 49) to obtain culturable H. pylori or DNA for PCR, when culture was unsuccessful. Families in which none of the children or either parent tested seemed to be H. pylori positive were excluded (36 families). Sixty-two families met our final inclusion criteria.
Participant characteristics.
For each of the 62 participating families, each family member included in the study lived in the same house. The age range of the participating children was 1 to 39 years, with a median of 9.5 years and a median interquartile range (between the first and the fourth quartiles) of 12 years. The age range of the participating parents was 19 to 59 years, with a median of 37 years and an interquartile range of 12.75 years. Among the parents, 34% had ≤6 years of education. Seventy-nine percent of the families had domestic animals in and about their houses; 74% of the houses were connected to the Lima municipal water system and had a sewage connection, and all but one family lived in a houses with electricity. This distribution of features is typical for residents of this shantytown (Table 1 and Table 2).
TABLE 1.
Sociodemographic characteristics of participating shantytown families
| Sociodemographic characteristic | Parents | Children |
|---|---|---|
| Median age (yr) | 37 (12.75)a | 9.5 (12)a |
| No. (%) with the following level of education: | ||
| Primary school or less | 23 (34) | 49 (58) |
| Secondary school | 32 (47) | 25 (28) |
| Technical school | 8 (12) | 7 (8.2) |
| University | 5 (7.4) | 5 (6.7) |
| No. (%) in the following age range (yr): | ||
| 0-5 | 30 | |
| 6-10 | 19 | |
| 11-15 | 16 | |
| 16-20 | 1 | 10 |
| 21-30 | 15 | 14 |
| 31-40 | 27 | 2 |
| 41-50 | 18 | |
| 51-60 | 7 |
The median interquartile range (between the first and the fourth quartiles) is given in parentheses in this row.
TABLE 2.
Housing characteristics of participating shantytown families
| Housing characteristic | No. (%) of households |
|---|---|
| Running water | 46 (74) |
| Water closet | 50 (80.6) |
| Dirt floor | 21 (33.9) |
| Electricity | 61 (98.4) |
| Food consumption in comedor populara | 20 (32.2) |
| Domestic animalsb | 49 (79) |
| Animal feces inside the house | 33 (53.2) |
| Houseflies | 57 (91.9) |
Comedor popular is a community facility where families buy cooked food.
A domestic animal is any animal in the house noted by the participant during the interview. The animals include cats, dogs, chickens, pigs, goats, etc.
Ethical considerations.
Our study protocol, including the use of Spanish-language informed consent and assent forms, was approved by three human studies or ethics committees: those of the Universidad Peruana Cayetano Heredia and the Asociacion Benefica PRISMA (both of Lima, Peru) and The Johns Hopkins Bloomberg School of Hygiene and Public Health (Baltimore, MD).
Sociodemographic assessment.
Standard demographic information, including sex, age, marital status, employment, and educational level, was obtained in initial interviews (Table 1). Housing characteristics were also evaluated (Table 2).
String test procedure.
All consenting subjects were screened for H. pylori by using the minimally invasive string test procedure (42, 49), which has been approved for use with adults and children who are at least 1 year old by the three institutional ethics committees listed above. String test participants swallow a gelatin capsule containing an absorbent cotton string with a protruding end, which is taped to the subject's cheek. The string is gently withdrawn 90 min later, and the distal 20 to 30 cm, which had been in the stomach, is put in 500 μl of transport medium at 4°C and brought to the microbiology laboratory within 4 h of collection. Our earlier studies had confirmed that the H. pylori strains cultured from strings match those from traditional biopsy specimens (49).
H. pylori culture.
Tubes containing string samples in liquid were vortexed for 3 min, and 150-μl aliquots were spread on brain heart infusion agar containing 5% defibrinated sheep blood and Skirrow supplement (Oxoid) and also on Columbia-colistin-nalidixic acid agar with Dent supplement (Oxoid). The plates were incubated under microaerobic conditions (10% CO2, 5% O2) at 37°C for 4 to 10 days. H. pylori colonies were identified by characteristic morphology, a positive urease test, and Gram staining (49).
ureB PCR.
PCR with primers specific for a 463-bp ureB (urease) gene segment was used to detect H. pylori (44) in string samples and was performed in parallel with H. pylori culturing. To prepare DNA from the strings for PCR tests, ∼300 μl of phosphate-buffered saline was added to the remaining liquid in the sample tube, which was then centrifuged at 13,000 × g for 10 min. DNA was extracted from the pellets with a QIAamp DNA minikit (Qiagen). PCR was carried out under standard conditions with the primers shown in Table 3; product formation was assayed by electrophoresis in agarose gels.
TABLE 3.
Primers used for PCR-based sequencing
| Gene | Primer namea | Sequence | Product length (bp) |
|---|---|---|---|
| glr | 5733 | 5′-CACATCGCCGCTCGCATGA-3′ | 800 |
| 6544 | 5′-AAGCTTTGTGTATTCTAAAATGCAAC-3′ | ||
| glr-peru-F | 5′-TTTTGCTGCAAATATTCCAC-3′ | 762 | |
| glr-peru2-R | 5′-ATGCAACAAACTAAGGACAA-3′ | ||
| cysS | cysS-F | 5′-CTACGGTGTATGATGACGCTCA-3′ | 1,287 |
| cysS-R | 5′-CCTTGTGGGGTGTCCATCAAAG-3′ | ||
| cys-peru-F | 5′-GATTTGTTAGGCGCACGCTT-3′ | 880 | |
| cys-peru2-R | 5′-TTTTGAGGGCGTCTTTAATG-3′ | ||
| atpA | atpA1 | 5′-GCTTAAATGGTGTGATGTCG-3′ | 1,200 |
| atpA6 | 5′-CTTATTCGCCCTTGCCCATT-3′ | ||
| atp-peru-F | 5′-GGGATAAGGCGTTGCGGCTA-3′ | 1,072 | |
| atp-peru2-R | 5′-AAACCTGCTTGGTCGCTTTG-3′ | ||
| glmM | glmM F | 5′-TTTGGGACTGATGGCGTGAGGG-3′ | 1,303 |
| glmM R | 5′-TCTTTTAATTCTTGCATTTTGGATTCTA-3′ | ||
| glm-peru2-F | 5′-GTTGTGAGCGTTGGGCATTG-3′ | 881 | |
| glm-peru-R | 5′-AAGCCGCGCCATTAGCCGTA-3′ | ||
| ppa | ppa8 | 5′-CCCCTAGAAAATCCTATTTTGATAATC-3′ | 902 |
| ppa9 | 5′-AGTGGTGAGCTTTAGCGACGCTC-3′ | ||
| ppa-peru-F | 5′-TATTATTCACAGGCGATTGC-3′ | 719 | |
| ppa-peru2-R | 5′-TAAAGCCATTTTACACCAAC-3′ | ||
| recA | recA1 F | 5′-GCGTTGGTACGCCTTGGGGATAAGCAA-3′ | 833 |
| recA4 R | 5′-GCCTTGCCCTAGCTTTTTATCCTGGT-3′ | ||
| rec-peru2-F | 5′-GATCGGGGGCGTTCCAAAAG-3′ | 728 | |
| rec-peru2-R | 5′-GCCATGCCCCACTCTTATCC-3′ | ||
| ureB | UreB-F | 5′-CGTCCGGCAATAGCTGCCATAGT-3′ | 463 |
| UreB-R | 5′-GTAGGTCCTGCTACTGAAGCCTTA-3′ |
Paired forward (F) and reverse (R) primers were used in a first PCR and then, if needed, a second (nested) PCR. The names of the primers used for sequencing are indicated in boldface.
H. pylori strain typing by fingerprinting.
RAPD fingerprinting of H. pylori genomic DNAs was used to assess the relatedness of strains from household members whenever pure cultures were available by using at least two of the following primers in separate reactions: primer 1254 (5′-CCGCAGCCAA), primer 1281 (5′-AACGCGCAAC), primer 1283 (5′-GCGATCCCCA), and primer 1290 (5′-GTGGATGCGA) (14, 49). RAPD fingerprinting resulted in informative profiles, such as those illustrated in Fig. 1. RAPD fingerprinting was not attempted when only DNAs from string tests were available because the abundance of other DNAs (the DNAs of the human host and other microbes) results in RAPD profiles that do not reliably indicate which H. pylori strains are related and which are not related to one another.
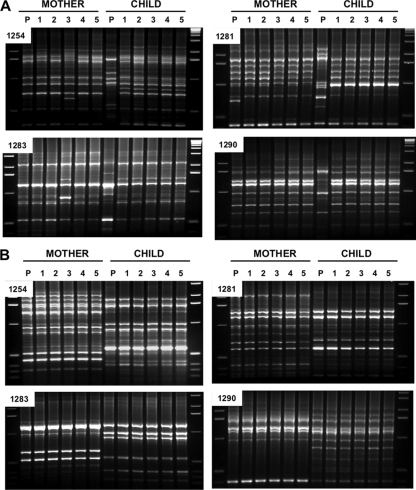
FIG. 1.
Representative RAPD profiles from pools of H. pylori colonies (lanes P) and from individual single colonies (lanes 1 though 5) from child-mother pairs from individual households. The sequences of the four RAPD primers used (primers 1254, 1281, 1283, and 1290) are given in Materials and Methods. (A) Profiles in which predominant strains from the child and mother are closely matched (as was seen in about 30% of the cases) and also the profile of a rare instance in which the child seemed to carry more than one strain in abundance (compare the pooled and the multiple single colonies). The glr gene fragments from the isolates from child 1 and mother 1 (767 bp) were 100% identical. (B) Profiles showing that predominant strains from the child and the mother are unrelated (as was seen in about 70% of the cases) and also that just one strain is predominant in each person. The glr gene fragments (768 bp) from the isolates child 1 and mother 1 were 3.4% divergent (26 base substitution differences), as is typical of unrelated strains (see Fig. 2).
Gene sequencing.
Determination of the sequences from at least one H. pylori housekeeping gene (glr [glutamate racemase] and/or cysS [cysteinyl tRNA synthetase]) from each participating household member was used as an independent test of strain relatedness either as a quantitative complement to RAPD profiling or, when pure H. pylori cultures were not available, as the sole means of determining relatedness. Segments containing these genes were amplified by PCR from DNAs extracted from cultures grown from single colonies or from string test samples (14). In cases in which PCR products were made from string test sample DNA, to obtain unambiguous H. pylori gene sequences it was sometimes necessary to reamplify the segments with new nested gene-specific primers whose 3′ ends were just interior to those of the primers used in the first amplifications. The PCR products were purified and sequenced at the University of Washington (Seattle) Sequencing Center or the Washington University (St. Louis, MO) PNACL core facility. Percent sequence identities were scored by using ClustalW software.
Determining strain relatedness.
When culture isolates were available, the strains were considered closely related genetically if the RAPD profiles were similar (no more than 1 or 2 of the ∼30 bands did not match, as illustrated in Fig. 1) and if the glr DNA sequences were ≤0.5% divergent (>99.5% identical). This cutoff was based on the frequency distribution of divergent sequences from paired strains shown in Fig. 2, which reveals well-separated peaks centered on <0.5% and on 3% to 4.5% divergence. The glr sequences of most strain pairs with closely matched RAPD patterns also matched, and vice versa. When H. pylori culture was not successful, the cysS gene was also sequenced to test the validity of inferences from tests of just one gene. Additional housekeeping genes (atpA, glmM, ppa, recA, and/or cysS) (14) were also sequenced when the RAPD analysis and glr data were discordant (i.e., when the data matched by one criterion but not another) to better understand the relatedness of strains from pairs of family members.
FIG. 2.
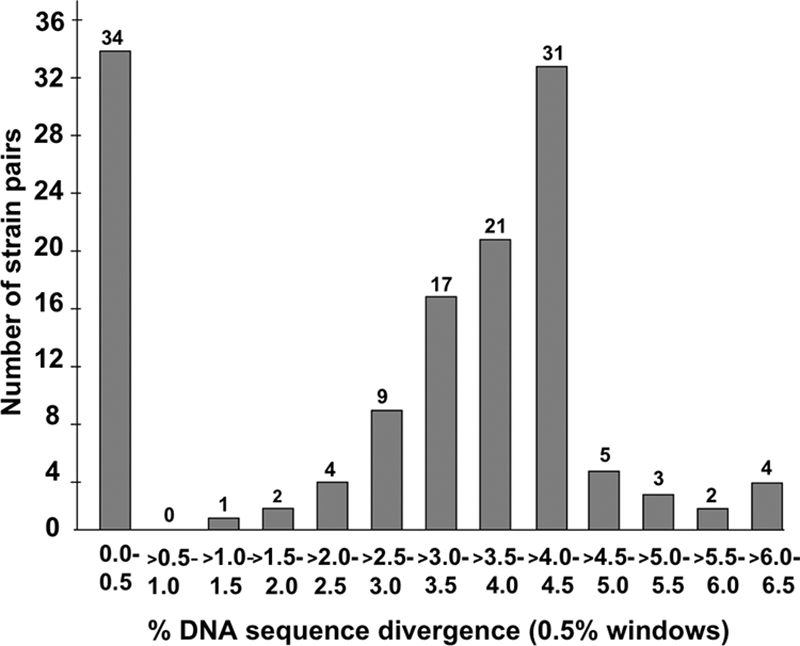
Distribution of percent DNA sequence divergence in glr genes in pairs of H. pylori strains from a child and a parent. The numbers on top of each vertical bar indicate the number of strain pairs from family members that exhibited a particular level of divergence.
To determine the frequencies of alleles that were ≥99.5% identical in the community at large, a list of every possible pair of strains that came from members of different families was assembled. Pairwise alignments of the sequences of glr genes were scored at each position as matched (score, +1) or mismatched (score, 0) for both transitions and transversions by using a pairwise alignment program implemented in the Perl program (http://www.perl.org/). The statistical significance of differences between the frequencies of matched alleles within households and the community at large was calculated by using the Stata program (www.stata.com).
Epidemiologic data analysis.
H. pylori strains from mother-offspring pairs and also from father-offspring, sibling-sibling, and spouse-spouse pairs were classified as concordant or discordant, as defined above. To seek the factors associated with intrafamilial transmission, we analyzed contingency tables and modeled the probability that the pairs of each of the four categories shared a concordant strain by multiple logistic regressions. The covariates (possible causal determinants) of interest were relationship classes and range of age in each pair compared. We adjusted for potential confounders such as sociodemographic variables, housing characteristics, place of food consumption by the families, and the presence or the absence of domestic animals and animal feces inside houses and tested for interactions and second-order effects.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were deposited in the NCBI GenBank database under accession numbers EU930438 to EU930645.
RESULTS
Experimental goals and design.
We tested if the H. pylori strains predominating in children in a Peruvian shantytown were often related genetically to those of their parents, in particular, their mothers, as is the case in industrialized societies. Samples were obtained from participants as young as 1 year of age by using the minimally invasive string test (42, 49), with informed consent or assent, as appropriate, in protocols approved by Peruvian and U.S. human studies committees. H. pylori was cultured from 135 of 166 (81%) individuals that were found to be H. pylori infected by ureB gene PCR with string test sample DNA. There was less success in culturing H. pylori from children under age 5 years (∼10%) than from older individuals (∼48%) (P < 0.01); the reasons for this difference are not known.
Occasional findings of two unrelated H. pylori strains in the same person are interesting in epidemiologic and evolutionary contexts (26, 41) and could be important to the present study. To assess if mixed infections are common in this shantytown population, we fingerprinted by RAPD analysis five single colonies and a pool of five or more additional colonies from the first dozen mother-child pairs from whom H. pylori was cultured. All but 2 of these 24 people seemed to carry just one strain per person. The only exceptions are shown in Fig. 1A: an indication of a second strain was seen by comparing the profiles obtained from the DNAs derived from a pool of colonies and from the five separate single colonies of H. pylori culture from the child; in addition, profiles obtained with two primers (primers 1254 and 1283) but not the other two primers indicated that isolate 3 from this child's mother may have diverged from the other isolates. In each of the other 22 cases analyzed in this way, however, each participant seemed to carry just one predominant strain, as illustrated by the profiles shown in Fig. 1B, which were obtained with isolates from a mother-child pair from a different household. The generally high uniformity inferred from these RAPD data were supported by the homogeneity (the absence of mixed peaks) of the sequence tracings from H. pylori gene PCR products from total string test sample DNA (data not shown). Also in agreement, we had previously found uniform H. pylori populations (no mixed infections) in each of another 20 shantytown residents in similar RAPD tests of single colonies and pools (49). Accordingly, for efficiency in identifying the predominant or only strain that a person carried in the great majority of cases, we focused further analyses on DNA from one colony per person or, when culture was not successful, on the sequences of the PCR products generated from DNA extracted from strings.
Relatedness of H. pylori strains within and between families.
H. pylori strains from 133 pairs of infected family members were analyzed: 103 pairs by RAPD fingerprinting and PCR-based sequencing of the glr gene and 30 pairs by PCR-based sequencing of the glr and cysS genes. For the latter group, the genes were from DNAs extracted from strings for cases in which samples for culture were not obtained from one or both family members. In the glr sequence comparisons, 99 of 133 pairs (74%) formed one peak of >1% divergence (average, 3% to 4.5%), and the other 34 (26%) pairs formed a second distinct peak of <0.5% divergence (Fig. 2). A threshold of <0.5% DNA sequence divergence was therefore used to infer whether the alleles of a given gene from different isolates were closely related or not. Similarly, divergent RAPD patterns (≥3 bands different among the ∼30 bands scored, on average) were found for 77 of 103 (75%) pairs, and identical or nearly identical RAPD patterns (≤2 different bands) were found for 26 of 103 (25%) pairs.
Only 12 of 133 pairs were discordant (different indications from the glr sequence and the RAPD profile or from the glr and cysS sequences). Sequencing of four or five additional loci from discordant pairs revealed sequence identity at these other loci. Strain pairs that were discordant at only one of the six loci sequenced were considered to be closely related derivatives of a recent common ancestor, with one isolate potentially diverged from the other by interstrain gene transfer. These pairs were counted as nearly matched in the tally in Fig. 3.
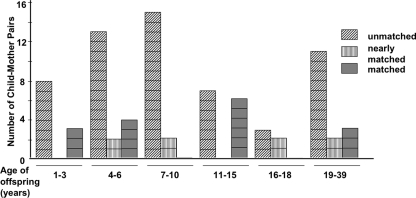
FIG. 3.
Distribution of relatedness among child-mother H. pylori strain pairs according to the age of the offspring. Taken across all age groups, 24 of 81 (30%) pairs of strains from the child and the mother were perfectly matched or nearly matched (closely related). Similarly, 2 of 11 (18%), 1 of 4 (25%), and 12 of 37 (32%) pairs of strains from the child and father, the mother and the father, and one sibling and another sibling, respectively, were perfectly matched or nearly matched (closely related). All other pairs of strains were not related in their RAPD patterns and gene sequences.
In terms of distributions within families, on the basis of the RAPD profiles and the glr or other housekeeping gene sequences, 24 of 81 strain pairs (30%) from children and mothers were closely related (matched or nearly matched) and the other 70% were not (Fig. 3). Similarly, 2 of 11 (18%) strain pairs from children and fathers, 12 of 37 (32.4%) strain pairs from siblings, and 1 of 4 (25%) strain pairs from spouses were closely related (see the Fig. 3 legend). These results further emphasize that there was no obvious relationship between the strains from pairs of family members in most families.
There were 13,512 possible pairs of glr sequences from members of different households in the broader community in our study. Only 63 (0.4%) of them had glr alleles whose sequences were ≤0.5% divergent. Thus, our results indicate both that the sharing of closely related strains among family members is markedly less common in this shantytown than in industrialized society settings and, equally, that strains from members of the same family are far more likely to be related than random strains from the community are.
Association analyses.
No significant association was seen between related strains and any of several likely covariates in any family category (between the mother or father and the child, between siblings, or between spouses). Mother-child and sibling pairs were pooled to increase the sample size (118 pairs) and statistical power; and the data were adjusted for sociologic, demographic, and housing characteristic variables and assessed for interactions and second-order effects. Again, no covariate tested was significantly associated with the probability of the sharing of a related strain.
DISCUSSION
It is only in developing countries that most people are still chronically infected by H. pylori, that frequent exposure makes eradication therapy ineffective, and that infection-associated gastric cancer remains one of the most frequently lethal of malignancies (8, 13, 33, 44). In contrast, in industrialized countries, less than half of all people are currently infected with H. pylori, and an essentially H. pylori-free future is anticipated (25). Thus, as with many pathogens, the epidemiology of H. pylori infection and associated diseases varies markedly among human populations. In the study described here, we examined the relatedness of H. pylori strains from members of families in a shantytown in Peru to test the applicability of two types of transmission models in a representative developing country setting: (i) the all-in-the-family model (12, 27), which was developed with data from industrialized countries, and (ii) a community-at-large/environmental reservoir model, which is encouraged by recent laboratory findings of the survival of H. pylori for at least a few days in several nonchlorinated waters (5, 31, 40), H. pylori biofilm formation (11, 50), and H. pylori proliferation during coculture with amoebae (51). We found that ∼70% of strains from children in our study were not closely related to those from their mothers, the parent implicated in most transmission in industrialized societies, or to those from other family members. This suggested that many infections were community acquired. However, since strains from children were still more frequently closely related to those of another family member (∼30%) than they were to those from the community at large (<1%), transmission within the family or from common sources must also be significant in this society.
A theoretical model of frequent transmission from mother to infant, with occasional displacement of the strain in the child by superinfecting strains in subsequent years, would have predicted that closely related mother-child strain pairs would be far more common in cases of very young children than of older children. Remarkably, however, this was not observed: matched pairs were about as common with older children as with younger children. This suggests that when transmission is intrafamilial, each household member may be equally prone to serve as the source of infection for other family members, independent of age, and that even cases of closely related mother-child strain pairs need not stem from early parent-to-child transmission and then the lifelong persistence of the initial strain.
We do not imagine, however, that most children had never been exposed to their mothers' strains. Prior studies have indicated that infections are often transient in infants (29, 47) and also in an experimentally infected monkey model (17), that individual hosts can differ in their susceptibilities to particular H. pylori strains, and that one strain can sometimes gradually displace another (16). These features may reflect differences among hosts and changes over time in traits that can be important to individual strains, e.g., the types of host glycans used as receptors for H. pylori adherence, the nature and intensity of inflammatory responses, and gastric pH (4, 6, 18). Such host features encourage the thinking that resident H. pylori strains must continuously adapt to their individual hosts during long-term chronic infection, such that a strain that has adapted to one person for many years may often be less well suited for colonization of the next person who ingests it (16, 45). Since exposure to infectious doses of H. pylori is relatively frequent in this community (the rate is estimated to be ∼20% per person per year for adults and higher for infants [29, 44]), this framework predicts that many of the first strains to infect infants (presumably often those of their mothers), if they get established at all, will often be displaced by superinfecting unrelated strains that happen to be better suited for the infant's physiology. Such a dynamic might reflect the differences between the juvenile stomach, which may have little gastritis and normal acidity, and the stomach of an adult, which may have severe gastritis, often with hypochlorhydria, due to the years of chronic H. pylori infection.
Just one H. pylori strain per person was found to predominate in the great majority of shantytown residents (Fig. 1) (49). This high degree of uniformity suggests either that most superinfecting strains fail to establish themselves because already established strains are difficult to displace or that displacement by a more fit superinfecting strain tends to be rather rapid and complete. Such displacement of one strain by another has been seen in experimentally infected monkeys (16) and mice (M. Zhang and D. E. Berg, unpublished data). We suggest that only rarely in this environment would two strains be so well balanced in fitness or tissue tropism that they would coexist for long periods, each at a high abundance, as was seen in mice with one exceptional strain pair (2).
How easily one strain is completely replaced by another in developing-country settings and how effectively a resident strain persists despite repeated invasion by competitors and further adapts by mutation or interstrain recombination with superinfecting strains is not known. The rapid emergence of new high-capacity, low-cost genome sequencing, the discovery of single-nucleotide polymorphisms, and transcript profiling technologies (34, 43) and the sets of strains found here should be particularly useful for the examination of the mechanisms of adaptive evolution in high-risk human populations and the loci at which selection for change operates. Most immediately, however, this study indicates that H. pylori is often transmitted from outside the family in a developing-country setting. In this setting, only community-based eradication measures will succeed in markedly reducing the prevalence of H. pylori infection and the burden of gastric cancer, peptic ulcer, and other associated diseases.
Acknowledgments
We thank the members of the community of PSJM for their collaboration and Paula Maguiña for her able assistance with the administrative aspects of this study.
This research was supported by grants from the U.S. National Institutes of Health (grant RO1 DK63041 to D.E.B. and grants T35 1007646 and D43TW006581 to R.H.G.). Other studies in D.E.B.'s laboratory, distinct from those reported here, are funded by a contract from Ondek Pty.; Washington University may receive income based on a license of technology to Ondek.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Abu-Mahfouz, M. Z., V. M. Prasad, P. Santogade, and A. F. Cutler. 1997. Helicobacter pylori recurrence after successful eradication: 5-year follow-up in the United States. Am. J. Gastroenterol. 922025-2028. [PubMed] [Google Scholar]
- 2.Akada, J. K., K. Ogura, D. Dailidiene, G. Dailide, J. M. Cheverud, and D. E. Berg. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 1491901-1909. [DOI] [PubMed] [Google Scholar]
- 3.Amieva, M. R., and E. M. El-Omar. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134306-323. [DOI] [PubMed] [Google Scholar]
- 4.Aspholm-Hurtig, M., G. Dailide, M. Lahmann, A. Kalia, D. Ilver, N. Roche, S. Vikström, R. Sjöström, S. Lindén, A. Bäckström, C. Lundberg, A. Arnqvist, J. Mahdavi, U. J. Nilsson, B. Velapatiõ, R. H. Gilman, M. Gerhard, T. Alarcon, M. López-Brea, T. Nakazawa, J. G. Fox, P. Correa, M. G. Dominguez-Bello, G. I. Perez-Perez, M. J. Blaser, S. Normark, I. Carlstedt, S. Oscarson, S. Teneberg, D. E. Berg, and T. Borén. 2004. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305:519-522. [DOI] [PubMed] [Google Scholar]
- 5.Azevedo, N. F., N. Guimarães, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. A new model for the transmission of Helicobacter pylori: role of environmental reservoirs as gene pools to increase strain diversity. Crit. Rev. Microbiol. 33157-169. [DOI] [PubMed] [Google Scholar]
- 6.Bergman, M., G. Del Prete, Y. van Kooyk, and B. Appelmelk. 2006. Helicobacter pylori phase variation, immune modulation and gastric autoimmunity. Nat. Rev. Microbiol. 4:151-159. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J. 2006. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 7956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L. M. 2000. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol. Rev. 22283-297. [DOI] [PubMed] [Google Scholar]
- 9.Bunn, J. E., W. G. MacKay, J. E. Thomas, D. C. Reid, and L. T. Weaver. 2002. Detection of Helicobacter pylori DNA in drinking water biofilms: implications for transmission in early life. Lett. Appl. Microbiol. 34450-454. [DOI] [PubMed] [Google Scholar]
- 10.Cardenas, V., Z. Mulia, M. Ortiz, and D. Graham. 2006. Iron deficiency and Helicobacter pylori infection in the United States. Am. J. Epidemiol. 163127-134. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 1863124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, B. J. 1996. Helicobacter pylori: is it all in the family? Gut 39768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa, P., and J. Houghton. 2007. Carcinogenesis of Helicobacter pylori. Gastroenterology 133659-672. [DOI] [PubMed] [Google Scholar]
- 14.Dailidiene, D., G. Dailide, K. Ogura, M. Zhang, A. K. Mukhopadhyay, K. A. Eaton, G. Cattoli, J. G. Kusters, and D. E. Berg. 2004. Helicobacter acinonychis: genetic and rodent infection studies of a Helicobacter pylori-like gastric pathogen of cheetahs and other big cats. J. Bacteriol. 186356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delport, W., M. Cunningham, B. Olivier, O. Preisig, and S. W. van der Merwe. 2006. A population genetics pedigree perspective on the transmission of Helicobacter pylori. Genetics 1742107-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois, A., D. E. Berg, E. T. Incecik, N. Fiala, L. M. Heman-Ackah, J. Del Valle, M. Yang, H. P. Wirth, G. I. Perez-Perez, and M. J. Blaser. 1999. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology 11690-96. [DOI] [PubMed] [Google Scholar]
- 17.Dubois, A., D. E. Berg, E. T. Incecik, N. Fiala, L. M. Heman-Ackah, G. I. Perez-Perez, and M. J. Blaser. 1996. Transient and persistent experimental infection of nonhuman primates with Helicobacter pylori: implications for human disease. Infect. Immun. 642885-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Omar, E. M., M. Ng, and G. L. Hold. 2008. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene 27244-252. [DOI] [PubMed] [Google Scholar]
- 19.Everhart, J. E. 2000. Recent developments in the epidemiology of Helicobacter pylori. Gastroenterol. Clin. N. Am. 29559-578. [DOI] [PubMed] [Google Scholar]
- 20.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11505-510. [DOI] [PubMed] [Google Scholar]
- 21.Faruque, S. M., and G. B. Nair. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 4659-66. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto, Y., N. Furusyo, K. Toyoda, H. Takeoka, Y. Sawayama, and J. Hayashi. 2007. Intrafamilial transmission of Helicobacter pylori among the population of endemic areas in Japan. Helicobacter 12:170-176. [DOI] [PubMed] [Google Scholar]
- 23.Gilman, R. H., G. S. Marquis, E. Miranda, M. Vestegui, and H. Martinez. 1988. Rapid reinfection by Giardia lamblia after treatment in a hyperendemic Third World community. Lancet i343-345. [DOI] [PubMed] [Google Scholar]
- 24.Goodman, K., and P. Correa. 2000. Transmission of Helicobacter pylori among siblings. Lancet 355358-362. [DOI] [PubMed] [Google Scholar]
- 25.Graham, D. Y., Y. Yamaoka, and M. H. Malaty. 2007. Contemplating the future without Helicobacter pylori and the dire consequences hypothesis. Helicobacter 12(Suppl. 2)64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 3131-43. [DOI] [PubMed] [Google Scholar]
- 27.Kivi, M., and Y. Tindberg. 2006. Helicobacter pylori occurrence and transmission: a family affair? Scand. J. Infect. Dis. 38407-417. [DOI] [PubMed] [Google Scholar]
- 28.Kivi, M., Y. Tindberg, M. Sörberg, T. H. Casswall, R. Befrits, P. M. Hellström, C. Bengtsson, L. Engstrand, and M. Granström. 2003. Concordance of Helicobacter pylori strains within families. J. Clin. Microbiol. 415604-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein, P. D., R. H. Gilman, R. Leon-Barua, F. Diaz, E. O. Smith, and D. Y. Graham. 1994. The epidemiology of Helicobacter pylori in Peruvian children between 6 and 30 months of age. Am. J. Gastroenterol. 89:2196-2200. [PubMed] [Google Scholar]
- 30.Klein, P. D., D. Y. Graham, A. Gaillour, A. R. Opekun, E. O. Smith, and The Gastrointestinal Physiology Working Group. 1991. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Lancet 337:1503-1506. [DOI] [PubMed] [Google Scholar]
- 31.Konishi, K., N. Saito, E. Shoji, H. Takeda, M. Kato, M. Asaka, and H. K. Ooi. 2007. Helicobacter pylori: longer survival in deep ground water and sea water than in a nutrient-rich environment. APMIS 1151285-1291. [DOI] [PubMed] [Google Scholar]
- 32.Konno, M., N. Fujii, S. Yokota, K. Sato, M. Takahashi, K. Sato, E. Mino, and T. Sugiyama. 2005. Five-year follow-up study of mother-to-child transmission of Helicobacter pylori infection detected by a random amplified polymorphic DNA fingerprinting method. J. Clin. Microbiol. 432246-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lochhead, P., and E. M. El-Omar. 2007. Helicobacter pylori infection and gastric cancer. Best Pract. Res. Clin. Gastroenterol. 21281-297. [DOI] [PubMed] [Google Scholar]
- 34.Mardis, E. R. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24133-141. [DOI] [PubMed] [Google Scholar]
- 35.Moran, N. A., and G. R. Plague. 2004. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14627-633. [DOI] [PubMed] [Google Scholar]
- 36.Nutman, T. B. 2001. Lymphatic filariasis: new insights and prospects for control. Curr. Opin. Infect. Dis. 14539-546. [DOI] [PubMed] [Google Scholar]
- 37.Passaro, D. J., D. N. Taylor, R. Meza, L. Cabrera, R. H. Gilman, and J. Parsonnet. 2001. Acute Helicobacter pylori infection is followed by an increase in diarrheal disease among Peruvian children. Pediatrics 108E87. [DOI] [PubMed] [Google Scholar]
- 38.Perry, S., M. de la Luz Sanchez, S. Yang, T. D. Haggerty, P. Hurst, G. I. Perez-Perez, and J. Parsonnet. 2006. Gastroenteritis and transmission of Helicobacter pylori infection in households. Emerg. Infect. Dis. 121701-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruzzo, C., L. Vezzulli, and R. R. Colwell. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 101400-1410. [DOI] [PubMed] [Google Scholar]
- 40.Queralt, N., and R. Araujo. 2007. Analysis of the survival of H. pylori within a laboratory-based aquatic model system using molecular and classical techniques. Microb. Ecol. 54771-777. [DOI] [PubMed] [Google Scholar]
- 41.Raymond, J., J. M. Thiberg, C. Chevalier, N. Kalach, M. Bergeret, A. Labigne, and C. Dauga. 2004. Genetic and transmission analysis of Helicobacter pylori strains within a family. Emerg. Infect. Dis. 101816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuels, A. L., H. M. Windsor, G. Y. Ho, L. D. Goodwin, and B. J. Marshall. 2000. Culture of Helicobacter pylori from a gastric string may be an alternative to endoscopic biopsy. J. Clin. Microbiol. 382438-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shendure, J. 2008. The beginning of the end for microarrays? Nat. Methods 5:585-587. [DOI] [PubMed] [Google Scholar]
- 44.Soto, G., C. T. Bautista, D. E. Roth, R. H. Gilman, B. Velapatiõ, M. Ogura, G. Dailide, M. Razuri, R. Meza, U. Katz, T. P. Monath, D. E. Berg, D. N. Taylor, and The Gastrointestinal Physiology Working Group in Peru. 2003. Helicobacter pylori reinfection is common in Peruvian adults after antibiotic eradication therapy. J. Infect. Dis. 188:1263-1275. [DOI] [PubMed] [Google Scholar]
- 45.Suerbaum, S., and C. Josenhans. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5441-452. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, J. E., A. Dale, J. E. Bunn, M. Harding, W. A. Coward, T. J. Cole, and L. T. Weaver. 2004. Early Helicobacter pylori colonisation: the association with growth faltering in The Gambia. Arch. Dis. Child. 891149-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, J. E., A. Dale, M. Harding, W. A. Coward, T. J. Cole, and L. T. Weaver. 1999. Helicobacter pylori colonization in early life. Pediatr. Res. 45218-223. [DOI] [PubMed] [Google Scholar]
- 48.Tindberg, Y., C. Bengtsson, F. Granath, M. Blennow, O. Nyrén, and M. Granström. 2001. Helicobacter pylori infection in Swedish school children: lack of evidence of child-to-child transmission outside the family. Gastroenterology 121:310-316. [DOI] [PubMed] [Google Scholar]
- 49.Velapatiõ, B., J. Balqui, R. H. Gilman, A. Bussalleu, W. Quino, S. A. Finger, L. Santivaẽz, P. Herrera, A. Piscoya, J. Valdivia, J. Cok, and D. E. Berg. 2006. Validation of string test for diagnosis of Helicobacter pylori infections. J. Clin. Microbiol. 44976-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, J. C., K. A. McInnis, and T. L. Testerman. 2008. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl. Environ. Microbiol. 741255-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winiecka-Krusnell, J., K. Wreiber, A. von Euler, L. Engstrand, and E. Linder. 2002. Free-living amoebae promote growth and survival of Helicobacter pylori. Scand. J. Infect. Dis. 34253-256. [DOI] [PubMed] [Google Scholar]